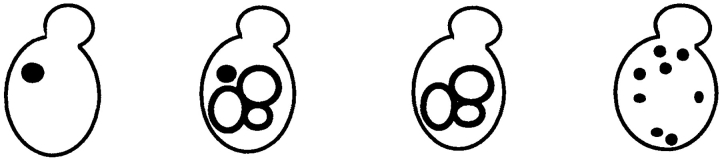Retrieval of Resident Late-Golgi Membrane Proteins from the Prevacuolar Compartment of Saccharomyces cerevisiae Is Dependent on the Function of Grd19p (original) (raw)
Abstract
The dynamic vesicle transport processes at the late-Golgi compartment of Saccharomyces cerevisiae (TGN) require dedicated mechanisms for correct localization of resident membrane proteins. In this study, we report the identification of a new gene, GRD19, involved in the localization of the model late-Golgi membrane protein A-ALP (consisting of the cytosolic domain of dipeptidyl aminopeptidase A [DPAP A] fused to the transmembrane and lumenal domains of the alkaline phosphatase [ALP]), which localizes to the yeast TGN. A grd19 null mutation causes rapid mislocalization of the late-Golgi membrane proteins A-ALP and Kex2p to the vacuole. In contrast to previously identified genes involved in late-Golgi membrane protein localization, grd19 mutations cause only minor effects on vacuolar protein sorting. The recycling of the carboxypeptidase Y sorting receptor, Vps10p, between the TGN and the prevacuolar compartment is largely unaffected in grd19Δ cells. Kinetic assays of A-ALP trafficking indicate that GRD19 is involved in the process of retrieval of A-ALP from the prevacuolar compartment. GRD19 encodes a small hydrophilic protein with a predominantly cytosolic distribution. In a yeast mutant that accumulates an exaggerated form of the prevacuolar compartment (vps27), Grd19p was observed to localize to this compartment. Using an in vitro binding assay, Grd19p was found to interact physically with the cytosolic domain of DPAP A. We conclude that Grd19p is a component of the retrieval machinery that functions by direct interaction with the cytosolic tails of certain TGN membrane proteins during the sorting/budding process at the prevacuolar compartment.
The internal organization of eukaryotic cells is characterized by a variety of distinct membranous subcompartments. Despite the continuous flow of membrane components through the secretory pathway, the localization of resident proteins of the different organelles must be maintained (for review see Rothman and Wieland, 1996). Two mechanisms are proposed to confer proper localization to membrane proteins in the secretory pathway: (a) they are separated from exiting proteins during the vesicle budding process and therefore never leave the compartment (retention), or (b) they leave the organelle together with other proteins but are recognized in a subsequent compartment and selectively transported back (retrieval; Pelham and Munro, 1993; Pelham, 1995, 1996). The TGN is the site of multiple vesicular sorting processes separating proteins destined for the plasma membrane or endosomes/lysosomes (Griffiths and Simons, 1989). The targeting and localization of resident membrane proteins to the TGN is therefore a complex process and not well understood. Most membrane proteins studied so far seem to use a combination of retrieval and retention mechanisms to be localized efficiently (Luzio and Banting, 1993; Machamer, 1993). In mammalian cells, where the destination for membrane proteins is the plasma membrane (Pfeffer and Rothman, 1987), it could be demonstrated that two membrane proteins of the TGN, furin and TGN38, are predominantly localized by a retrieval process from the plasma membrane (for review see Wilsbach and Payne, 1993_a_ ) using endocytotic internalization signals in the cytosolic tail domains (Bos et al., 1993; Humphrey et al., 1993; Wong and Hong, 1993; Voorhees et al., 1995).
The late-Golgi compartment in Saccharomyces cerevisiae is the functional equivalent of the mammalian TGN (Graham et al., 1994). It is defined by the presence of three enzymes, dipeptidyl aminopeptidase A (DPAP A;1 encoded by STE13 gene), Kex2p, and Kex1p (Franzusoff et al., 1991; Redding et al., 1991; Cooper and Bussey, 1992). All three proteins are integral membrane proteins involved in the proteolytic processing of the secreted mating pheromone α-factor (Fuller et al., 1988). The carboxypeptidase Y (CPY) sorting receptor, Vps10p, also predominantly localizes to the yeast TGN (Marcusson et al., 1994; Cereghino et al., 1995; Cooper and Stevens, 1996). In contrast to mammalian cells, yeast membrane proteins, which carry no further targeting information, by default are transported to the vacuole (Roberts et al., 1992), which is the equivalent of the mammalian lysosome (for review see Nothwehr and Stevens, 1994). The signals for localization of DPAP A, Kex2p, and Vps10p to the TGN were found to reside in their cytosolic domains (Wilcox et al., 1992; Nothwehr et al., 1993; Cereghino et al., 1995; Cooper and Stevens, 1996). Aromatic amino acids, forming a sequence motif resembling the coated pit localization signals of mammalian plasma membrane receptors, have been identified as being critical components of localization motifs within these domains specifying their retrieval from a prevacuolar compartment (Wilcox et al., 1992; Nothwehr and Stevens, 1994; Cereghino et al., 1995; Cooper and Stevens, 1996). In addition to the aromatic amino acid– containing motif, the cytosolic domain of DPAP A also contains a static retention signal, preventing it from leaving the TGN as rapidly as proteins en route to the vacuole (Bryant and Stevens, 1997). In contrast to the processing enzymes that function exclusively in the TGN, the function of the cargo receptors of lysosomal or vacuolar hydrolases, like the mammalian mannose-6-phosphate receptor (Kornfeld and Mellman, 1989; Ludwig et al., 1995) and the yeast CPY receptor Vps10p (Cereghino et al., 1995; Cooper and Stevens, 1996), requires them to cycle continuously between the TGN and a post-Golgi compartment for an efficient sorting process.
Little is known about the machinery that acts in the localization of late-Golgi membrane proteins. CHC1, the gene encoding the clathrin heavy chain protein, and VPS1, which encodes a yeast homologue of the mammalian protein dynamin implicated in budding of transport vesicles from the late-Golgi membrane, have been shown to be required for the correct localization of late-Golgi membrane proteins (Seeger and Payne, 1992_a_ ,b; Wilsbach and Payne, 1993_b_ ; Nothwehr et al., 1995; Redding et al., 1996_b_ ). Several genetic approaches have been undertaken in yeast to identify additional proteins involved in TGN membrane protein localization. Using the properties of a mutation in the retention signal of Kex2p, three allele-specific suppressor mutants, _soi1_-soi3, which increased the efficiency of retention of the mutated Kex2p, were discovered (Redding et al., 1996_a_ ). A genetic screen based on the mislocalization of A-ALP (a model late-Golgi membrane protein consisting of the cytosolic domain of DPAP A fused to the transmembrane and lumenal domains of ALP) to the vacuole identified 18 complementation groups, named grd for Golgi retention deficient (Nothwehr et al., 1996). Most of these mutants also exhibited vacuolar protein sorting defects, and significant overlap was found between the GRD and VPS (vacuolar protein sorting) genes (Banta et al., 1988; Raymond et al., 1992_a_ ,b). Interestingly, a subset of grd mutants exhibited normal vacuolar protein sorting and thus seemed to be defective specifically in the retention of DPAP A and/or Kex2p.
By performing a second round of the grd screen we have identified a new gene, GRD19, which is specifically involved in the localization of DPAP A and Kex2p. Sequence analysis showed that Grd19p contains a PX domain that is conserved among a family of proteins that includes sorting nexin-1 (SNX1), Mvp1p, and Vps5p (Ponting, 1996). GRD19 is not required for vacuolar protein sorting or the recycling of the CPY cargo receptor Vps10p. Assays of the exit and retrieval rates of a TGN membrane reporter protein revealed that GRD19 is required for the retrieval step. In vitro binding assays revealed that Grd19p binds to the cytosolic domain of DPAP A but not Vps10p.
Materials and Methods
Materials
Enzymes used in DNA manipulations were from New England Biolabs (Beverly, MA), Boehringer Mannheim Biochemicals (Indianapolis, IN), Bethesda Research Laboratories (Gaitherburg, MD), or U.S. Biochemicals (Cleveland, OH). FITC-conjugated streptavidin, Texas red–conjugated goat anti–rabbit, and biotin-conjugated goat anti–rabbit were purchased from Jackson Immunoresearch Inc. (West Grove, PA). The anti-CPY and ALP polyclonal antibodies have been reported previously (Raymond et al., 1990; Bryant and Stevens, 1997). The anti-ALP monoclonal antibody 1D3 (Molecular Probes, Eugene, OR) was used as reported previously (Bryant and Stevens, 1997). Fixed Staphylococcus aureus cells (IgG Sorb) were obtained from The Enzyme Center (Malden, MA). 35S-Express label was from New England Nuclear (Boston, MA). Oxalyticase was from Enzogenetics (Corvallis, OR). All other chemicals were of high purity commercial grade.
Plasmids
A DNA fragment encoding the cytosolic tail of DPAP A comprising the amino-terminal 117 amino acids was synthesized by PCR. The templates used were pSN55, encoding the fusion protein A-ALP with the full-length tail (for construction of pWV11), and pSN37, which contains the DPAP A cytosolic tail with a deletion of the FXFXD retrieval signal between amino acids 85 and 92 (for construction of pWV12). The fragments were digested with BamHI/BglII (both sites were introduced with the primer sequences) and were cloned into pQE70 (Qiagene, Chatsworth, CA) so that a 6x histidine tag was added in frame at the carboxy terminus of the DPAP A tail fragment. pWV16 contains the full-length open reading frame (ORF) of GRD19 including 500 bp of genomic sequences in both the 5′ and 3′ direction. The DNA fragment was obtained by PCR amplification from genomic DNA purified from strain SNY36. The produced fragment was cut with BamHI and SalI (sites introduced by PCR primers) and ligated in pRS313 (Sikorski and Hieter, 1989). A PCR fragment generated from genomic DNA of strain WVY12, which included the ORF with the HA epitope tag and 500 bp each of 5′- and 3′-flanking DNA, was cloned in YEp352 to generate the plasmid pWV32 for overexpression of Grd19p-HA. pWV36 was constructed by removing the insert of pNB81 by digestion with SacI/EcoRV and ligation into pRS315 (Sikorski and Hieter, 1989), which was cut by SacI/SmaI.
Strains
For the construction of the grd19Δ deletion strain WVY4, a PCR fragment was generated using oligonucleotides complementary to 45 bp directly up- and downstream of the GRD19 ORF, followed by a sequence complementary to sequences of the plasmid pRS304 flanking the marker gene TRP1. The fragment was transformed into strain SNY36, and Trp+ colonies were analyzed by the Grd plate assay (Nothwehr et al., 1996). Deletion of the GRD19 ORF was confirmed by PCR-Southern blot. Strain WVY5 was obtained by deletion of the PEP4 gene in strain WVY4 by using a PCR-generated deletion fragment amplified from genomic DNA of strain JHRY20. To obtain strain WVY12 the triple-hemagglutinin (HA) epitope was integrated into the ORF of GRD19 directly in front of the stop codon using a PCR-based method (Schneider et al., 1995). Strains WVY10, WVY20, and WVY21 were made by transforming strains WVY4, WVY12, and SNY36, respectively, with the vps27Δ disruption cassette (BamHI/EcoRI fragment from plasmid pCKR203) as described (Piper et al., 1995). WVY19 was obtained by disrupting the complete ORF of GRD19 with a PCR fragment using the initial disruption oligonucleotides but amplifying the Escherichia coli kanamycin-resistance gene kanr from plasmid pFA6-kanMX2 (Wach et al., 1994),\Q which was transformed into strain NBY67. Strains were constructed using standard genetic techniques and grown in rich media (1% yeast extract, 1% peptone, 2% dextrose; YPD) or standard minimal medium (S) with appropriate supplements. A summary of yeast strains used in this study is given in Table II.
Table II.
Plasmids Used in This Study
| Plasmid | Description | Reference |
|---|---|---|
| pSN37 | pBluescript SK containing Ste13p cytoplasmic tail with deletion of aa 85-92 | T.H. Stevens |
| pSN55 | CEN plasmid encoding A-ALP | (Nothwehr et al., 1993) |
| pSN97 | CEN plasmid encoding RS-ALP | (Nothwehr et al., 1993) |
| pSN100 | CEN plasmid encoding (F/A)A-ALP | (Nothwehr et al., 1993) |
| pNB81 | CEN plasmid encoding A-ALP with residues 2–11 deleted (Δ10-A-ALP) | (Bryant and Stevens, 1997) |
| pRSQ304 | derivative of pRS304 containing part of LEU2 | T.H. Stevens |
| pHY5 | CEN plasmid harboring VPS27 under GAL1 control | T.H. Stevens |
| pFvM8 | pGEX2 expressing the carboxy-terminal tail of Vps10p fused to GST | T.H. Stevens |
| pWV11 | pQE70 expressing the DPAP A 117-aa amino-terminal tail as 6xHis fusion protein | This study |
| pWV12 | pQE70 expressing the DPAP A tail with aa 85–92 deleted as 6xHis fusion protein | This study |
| pWV16 | CEN plasmid harboring GRD19 under own promoter | This study |
| pWV32 | 2μ plasmid harboring GRD19::HA under own promoter | This study |
| pWV36 | CEN plasmid encoding Δ10-A-ALP containing LEU2 marker | This study |
grd Mutant Screen and ALP Activity Assays
Strain SNY36 carrying plasmid pSN55 was mutagenized by a recombinative integration of a yeast genomic library with random integrations of a Tn3-LacZ transposon construct (Seifert et al., 1986; Burns et al., 1994). The plasmid library was cut with NotI before transformation. The activity of ALP was measured on the plate as described (Chapman and Munro, 1994; Nothwehr et al., 1996), and mutant colonies were identified. After a rescreen procedure, mutants were analyzed for CPY secretion with a colony overlay assay (Roberts et al., 1991) and subsequently subjected to complementation analysis with the vps and grd mutant collections (Rothman and Stevens, 1986; Robinson et al., 1988; Raymond et al., 1992_a_ ; Nothwehr et al., 1996). Diploid phenotypes were analyzed either for CPY secretion or processing of A-ALP, in cases where no Vps− phenotype could be detected. Mutants chosen for further analysis were backcrossed against the parental wild-type strain NBY17, sporulated, and the resulting tetrads checked for linkage of the transposon integration and the grd phenotype. Mutants were transformed with plasmid pRSQ304 cut by BamHI/ SacI to introduce a bacterial origin of replication at the position of the transposon integration. Yeast genomic DNA was recovered, cut with EcoRI, religated, and transformed in E. coli. A plasmid was recovered that contained part of the transposon sequence and adjacent genomic sequence from the insertion site. This DNA sequence was determined by automated sequencing (ABIPRISM; Pharmacia Fine Chemicals, Piscataway, NJ) using the “−40” sequencing primer (U.S. Biochemicals), and the corresponding reading frame was found by data base search (BLAST at the Saccharomyces genome database).
Pulse–Chase Labeling and Immunoprecipitation
Labeling and immunoprecipitation experiments for determining the stability of A-ALP constructs, Kex2p, Vps10p, and CPY were performed as described previously (Piper et al., 1994; Cooper and Stevens, 1996; Nothwehr et al., 1996). Labeled proteins were detected and quantified using storage phosphor technology (PhosphorImager; Molecular Dynamics). Half-times of processing or degradation were determined by linear regression analysis, plotting percentage of total and processed protein as a function of time.
Immunofluorescence
Indirect immunofluorescence microscopy for the localization of ALP, Vph1p, and Vps10p was performed as described previously (Roberts et al., 1991). Cells were grown in YPD at 30°C before fixation. For cells harboring plasmids, cells were grown to 1 OD/ml in minimal media and then resuspended in YPD to 0.25 OD/ml and allowed to grow for 2 h before fixation. For experiments using induction from the GAL1 promoter, cells were grown overnight at 30°C in synthetic media containing 2% raffinose. For induction of expression, 2% galactose was added, and samples were removed at the indicated time points for direct fixation. Cells were fixed in 3% formaldehyde for 30 min, followed by incubation in 2% paraformaldehyde/50 mM KPO4, pH 7.0 for 18 h. Cells were spheroplasted and permeabilized with 5% SDS for 5 min. After washing in 1.2 M sorbitol, cells were allowed to adhere to poly-l-lysine–coated slides. Incubation of cells with the primary antibody was performed at 4°C overnight followed by 1-h incubations of secondary and tertiary antibodies at 22°C. For experiments requiring labeling of A-ALP and Vph1p in the same cells, the anti-ALP 1D3 monoclonal antibody was visualized using biotinylated secondary antibody in combination with FITC-labeled streptavidin, and the rabbit anti-Vph1p was visualized using Texas red–labeled antibody. For double labeling of A-ALP and Vps10p, the anti-ALP monoclonal antibody was visualized with FITC-labeled goat anti–mouse antibody. Vps10p was detected using an affinity-purified rabbit antibody preparation, which was adsorbed against fixed vps10Δ cells and visualized using the biotin-enhancement protocol as described above. Images were captured using the Kodak DCS20 digital camera system with a 100× oil immersion lens (Carl Zeiss, Oberkochen, Germany) on a fluorescence microscope (Axioplan; Carl Zeiss). Images were adjusted with standard settings using Adobe Photoshop™.
Cell Fractionation
Intracellular localization studies were performed by a differential sedimentation procedure essentially as described (Piper et al., 1994). The following subcellular fractions were obtained: low speed pellet (P13), by centrifugation at 13,000 g for 10 min); membrane pellet (P100), by centrifugation at 100,000 g for 30 min, and the remaining supernatant (S100). Equal portions of each fraction were separated by SDS-PAGE and analyzed by Western blot.
Expression of the Cytosolic Domains and Binding Assay
200 ml of E. coli cells (XL1 blue; Stratagene, La Jolla, CA) containing either plasmid pQE70, pWV11, pWV12, pGEX2X, or pFvM8 were grown in LB-Medium with 100 μg/ml Ampicillin to 0.7 to 0.8 OD/ml at 37°C, and protein expression was induced by adding 1 mM IPTG. After 5 h incubation, cells were harvested and resuspended in 25 ml LP60 (30 mM Tris-HCL, pH 7.5, 300 mM NaCl, 60 mM imidazole, and 1 mM PMSF). Cells were lysed by a French Press, and cell debris and insoluble proteins were removed by 15 min centrifugation with 15,000 g at 4°C. Cell extracts were then incubated with 2 ml Ni-NTA sepharose (Qiagen) for 2 h at 4°C with gentle agitation. Unbound proteins were removed by washing the sepharose matrix with 20 ml LP60.
For binding assays with yeast proteins, the sepharose matrix with the bound expressed proteins was equilibrated to binding buffer (0.2 M sorbitol, 50 mM Tris-HCl, pH 7.5, 150 mM KCl) containing 5 mM magnesiumacetate, 1 mM DTT, and 0.1% Triton X-100. 30 μl wet volume of the matrix was incubated with 50 μl (250 μg total protein/sample) yeast cytosol extract (S100) in a total volume of 200 μl binding buffer. Extracts were prepared from SNY36-9a containing plasmid pWV32 or pAH37 as described above, except that spheroplasts were lysed in binding buffer. After 1.5 h incubation at 4°C the sepharose matrix was washed 4 times with 300 μl binding buffer, and all bound proteins were eluted by incubating in 300 μl LP250 (same as LP60 but containing 250 mM imidazole). Proteins were separated by SDS-PAGE and analyzed by Western blot. Mvp1p was detected directly by antibodies, whereas Grd19p and Vps5p were detected by the introduction of 3xHA tags and decoration with antibody directed against the tag.
Results
Identification and Cloning of GRD19
Yeast mutants defective in Golgi membrane protein retention (grd) were identified using A-ALP, a fusion protein that contains the cytosolic tail of DPAP A (encoded by the STE13 gene) and the transmembrane and lumenal domains of ALP (encoded by the PHO8 gene) as a reporter molecule. In wild-type cells A-ALP is localized to the late-Golgi compartment but becomes mislocalized to the vacuole in localization-defective strains (Nothwehr et al., 1996). Mislocalization of A-ALP to the vacuole results in processing by vacuolar proteases (PEP4 dependent) at the carboxy-terminal end of A-ALP and gain of alkaline phosphatase activity. Mutations were introduced into strain SNY36 (pho8Δ) by transformation of a genomic plasmid library containing random transposon integrations into yeast DNA fragments (Seifert et al., 1986). Colonies with alkaline phosphatase activity were detected by a colorimetric assay (Nothwehr et al., 1996). Selected mutants were backcrossed with wild-type strains to confirm linkage of mutant phenotype with the transposon insertion (Leu+). The obtained mutant alleles were tested by complementation analysis for genetic overlap with previously identified grd and vps mutants.
One mutant, subsequently called grd19, complemented each representative of the grd and vps mutant collections and therefore represents a new grd complementation group. Genomic sequences neighboring the transposon insertion site were recovered and sequenced. Comparison with the S. cerevisiae genome database revealed that the transposon insertion took place in the previously unidentified ORF YOR357c. To obtain a well-defined null mutant, the entire ORF YOR357c was replaced by the TRP1 gene, yielding a grd19Δ strain which was used for further analysis. The deletion of GRD19 did not result in any growth defects (data not shown).
GRD19 Is a Member of a Protein Family Involved in Protein Sorting and Vesicular Traffic
GRD19 encodes a small hydrophilic protein of 162 amino acids (Fig. 1 A) with a predicted molecular mass of 18.7 kD. The amino acid sequence shows no obvious modification motifs or targeting signals. Protein sequence comparisons (BLAST at NCBI) revealed significant homology to several other yeast and mammalian proteins (Fig. 1 B). Interestingly, the most similar protein found in the database is Mvp1p, which is also implicated in vesicle transport processes between the Golgi compartment and the vacuole (Ekena and Stevens, 1995). The homology between Grd19p and Mvp1p is mainly restricted to a sequence, ∼100 amino acids long, near the carboxy terminus of Grd19p. A similar homologous region was found in Vps5p (Horazdovsky et al., 1997), which was also identified as Grd2p (Nothwehr and Hindes, 1997). Grd2p/Vps5p is also involved in vacuolar sorting processes and is required for the localization of A-ALP, Kex2p, and Vps10p. An extensive set of experiments was performed to determine whether there are any genetic interactions between the GRD19, VPS5, and MVP1 genes. The Golgi localization defects of grd19Δ cells could not be suppressed by overexpression of VPS5 or MVP1. Additionally, analysis of the double deletion mutants grd19Δ vps5Δ and grd19Δ mvp1Δ did not reveal any synthetic phenotypes concerning growth, CPY secretion, or A-ALP localization (data not shown).
Figure 1.
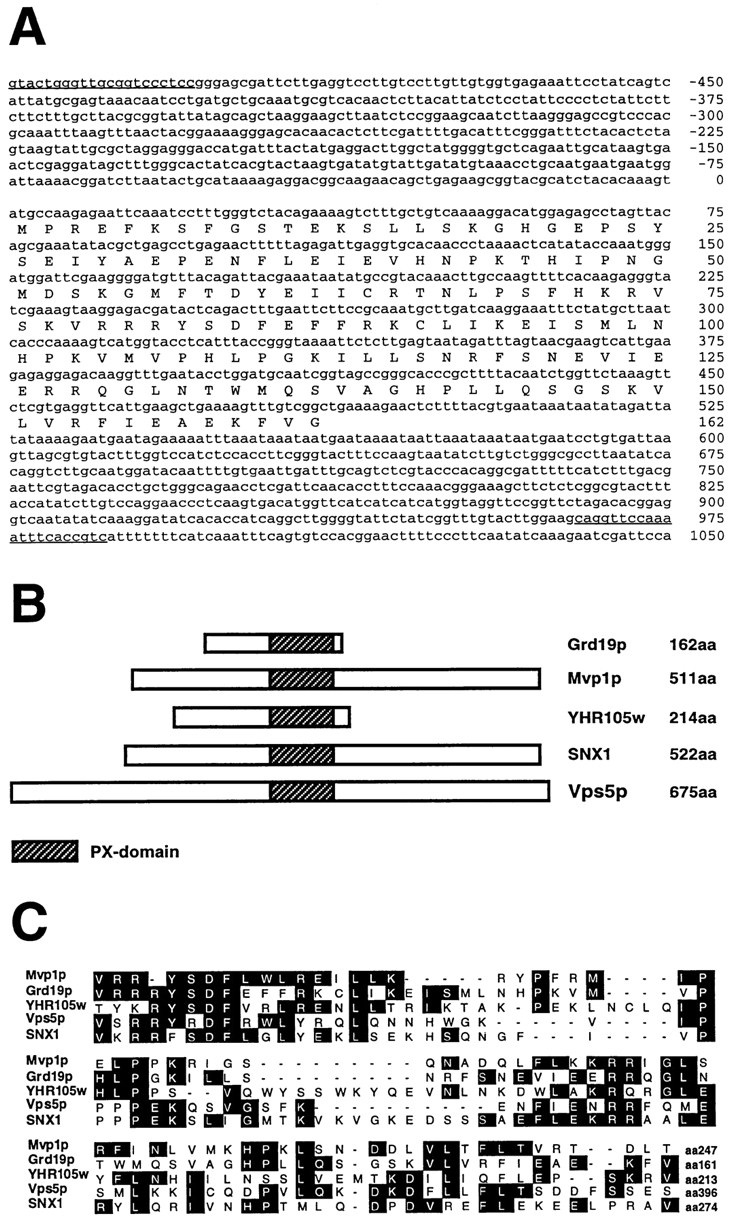
Nucleotide and amino acid sequence of GRD19 gene region. (A) Underlined nucleotide sequence represents the sequence of the oligonucleotides used for cloning the full-length GRD19 gene. The region was identified in the yeast genome database as ORF YOR357c. (B) Schematic drawing showing proteins related to Grd19p and the relative position of the conserved PX domain. (C) Sequence comparison between the PX domains of several proteins that were aligned by the ClustalW program. The sequence data are available from EMBL/DDBJ/Genbank under accession numbers: Mvp1p (U16137), Vps5p (U84735), SNX1 (U53225), and Grd19p (AF016101).
A mammalian protein, sorting nexin-1 (SNX1), shares homology with Grd19p, Mvp1p, and Vps5p. SNX1 was shown to interact with the cytosolic domain of the EGF receptor and influences its rate of degradation (Kurten et al., 1996). All the proteins show on average an amino acid identity of 30 to 40% (50–60% similarity) within this domain (Fig. 1 C) but differ substantially in other regions. YHR105w, another yeast ORF with unknown function, exhibited the greatest overall similarity to Grd19p, with respect to size and sequence homology (26% identity, 48% similarity). A null mutant of this gene was constructed, but cells deleted for ORF YHR105w showed no defects in morphology, vacuolar protein sorting, or Golgi membrane protein retention (data not shown). In addition, deleting ORF YHR105w in a grd19Δ strain did not amplify or suppress any of the phenotypes due to the grd19Δ mutation (data not shown). The region of homology shared between Grd19p, Mvp1p, Vps5p, and SNX1p, called PX domain, classifies them as members of a large group of proteins (Ponting, 1996).
Resident Late-Golgi Membrane Proteins Are Degraded in the Vacuole in grd19Δ Cells
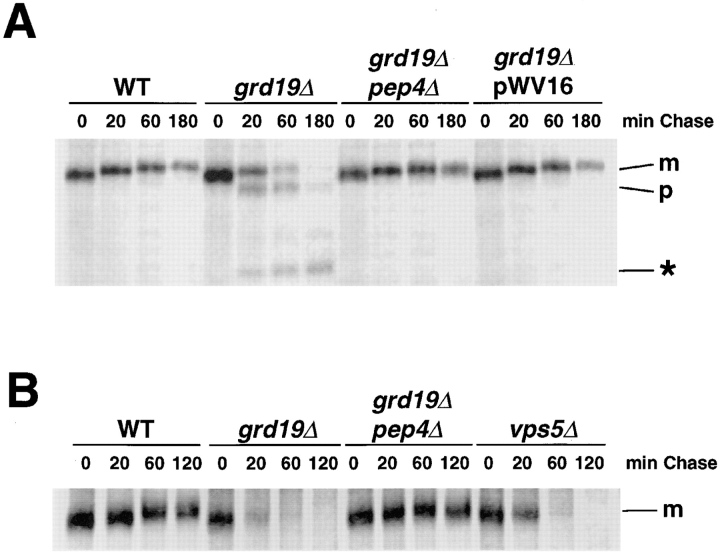
Kex2p and A-ALP reside in the yeast TGN, and a failure in retention leads to the proteolytic processing of A-ALP (Nothwehr et al., 1993) and degradation of Kex2p by vacuolar proteases (Wilcox et al., 1992). These processes can be quantitatively assessed by pulse-chase labeling and immunoprecipitation experiments. We determined the stability of A-ALP and Kex2p in grd19Δ cells. As expected, A-ALP was a stable protein in wild-type cells with a half-time of processing of >180 min. In grd19Δ cells the stability of A-ALP was greatly reduced with its half-time of processing decreased to 55 to 60 min (Fig. 2 A). Kex2p also showed a significantly increased rate of degradation in grd19Δ cells with a half-time of 20 to 25 min as compared to wild-type cells in which the half-time of Kex2p degradation was 120 min (Fig. 2 B). To determine whether this processing or degradation reaction was dependent on the action of vacuolar proteases, identical pulse–chase experiments were performed using grd19Δ pep4Δ double mutants, which lack vacuolar protease activity. Those cells showed virtually no degradation of A-ALP and Kex2p, confirming the mislocalization of the two proteins to the vacuole in grd19Δ cells. We conclude that the function of Grd19p is necessary for stabilization of the resident late-Golgi membrane proteins A-ALP and Kex2p.
Figure 2.
grd19Δ cells mislocalize late-Golgi membrane proteins. (A) Wild-type (SNY36), grd19Δ (WVY4), and grd19Δ pep4Δ (WVY5) cells carrying plasmid pSN55 (encoding the reporter fusion protein A-ALP) were analyzed by pulse–chase labeling and immunoprecipitation with antibodies against ALP. pWV16 carries the full-length GRD19 gene under its own promoter on a CEN-based plasmid (pRS313). Indicated are the mature protein (m), the vacuolar processing product (p) and an additional typical degradation product (*). (B) Labeling and immunoprecipitation were performed as above, except that antibodies against Kex2p were used for immunoprecipitation. As a positive control for rapid Kex2p degradation the strain vps5Δ (AHY41) was included.
grd19Δ Cells Sort the Vacuolar Hydrolase CPY and the CPY Sorting Receptor Vps10p Normally
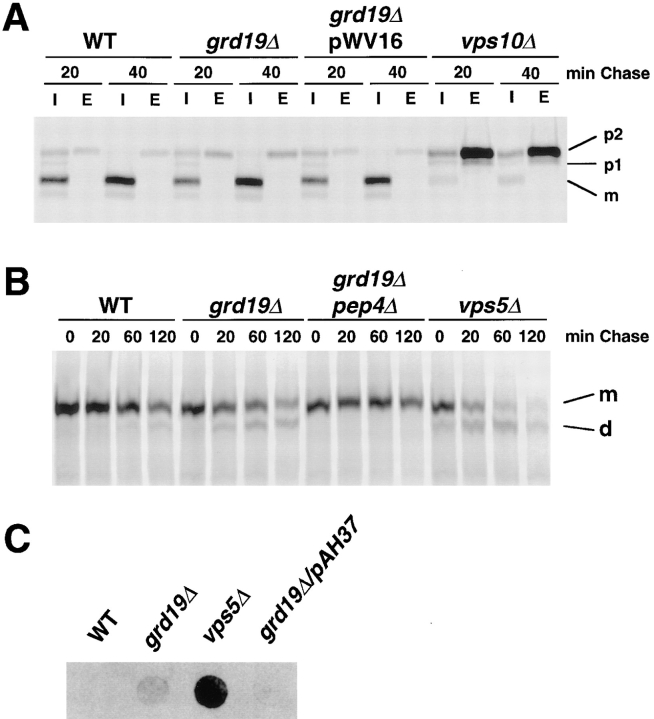
Since many of the recently identified grd mutants also exhibit Vps− phenotypes (Nothwehr et al., 1996), we tested whether a deletion of GRD19 also affects CPY sorting using pulse–chase labeling and immunoprecipitation experiments. Newly synthesized CPY was immunoprecipitated from internal and external fractions of grd19Δ and wild-type cells, as well as cells carrying a wild-type copy of GRD19 on a centromere-based plasmid. As a control for a strain with severe defects in CPY sorting, the experiment included a vps10Δ strain. As shown in Fig. 3 A, grd19Δ cells exhibited only a minor defect in the sorting of CPY. grd19Δ cells missorted ∼16% of the Golgi-modified (p2) form of CPY to the cell surface, compared to 6% in wild-type cells. By constrast, vps10 cells secreted 86% of CPY, and virtually none of the CPY reached the vacuole.
Figure 3.
Sorting of CPY and the cycling of Vps10p are normal in grd19Δ cells. (A) Wild-type (SNY36), grd19Δ (WVY4), and vps10Δ (AACY30) cells were labeled for the indicated times and analyzed for sorting of CPY by immunoprecipitation from intracellular (I) and extracellular (E) fractions as described. Indicated are the mature protein (m), the ER/early-Golgi precursor form (p1) and the fully glycosylated form (p2). (B) The same strains as in Fig. 2 B were analyzed for the stability of Vps10p by pulse– chase labeling and immunoprecipitation using antibodies against Vps10p. Indicated are the mature form (m) and the vacuolar degradation product (d). (C) Colony overlay assay with wild-type cells carrying pAH37 (2μ-VPS5) show relative rates of CPY secretion.
Since the sorting of CPY is closely coupled to the recycling of its cargo receptor Vps10p between late-Golgi and prevacuolar compartments, we also analyzed the stability of Vps10p directly by pulse–chase labeling and immunoprecipitation (Fig. 3 B). Compared to wild-type cells, grd19Δ cells show only a slight decrease in Vps10p stability due to mislocalization to the vacuole. The half-life of Vps10p in wild-type cells was typically much longer than 180 min (Cooper and Stevens, 1996). In grd19Δ cells the half-life was slightly decreased to 150 to 160 min. By contrast, in vps5Δ cells, which have been shown to have severe defects in both A-ALP localization and CPY sorting (Nothwehr and Hindes, 1997), the stability of Vps10p was decreased significantly, to a half-time of ∼40 min. Interestingly, while overexpression of Vps5p did not suppress the mislocalization of A-ALP (data not shown), elevated levels of Vps5p did suppress the low level of CPY secretion in grd19Δ cells (Fig. 3 C). We conclude that the slight effect of mutations in GRD19 on the sorting of CPY is likely to be an indirect effect, since the overexpression of a protein more directly involved in CPY sorting (Vps5p) reverses the slight CPY sorting defect without suppressing the A-ALP cycling defect. Therefore, in comparison to the previously identified grd genes (Nothwehr et al., 1996), GRD19 is involved in late-Golgi membrane protein localization in a very specific way, affecting the resident proteins A-ALP and Kex2p, but not the recycling cargo receptor Vps10p.
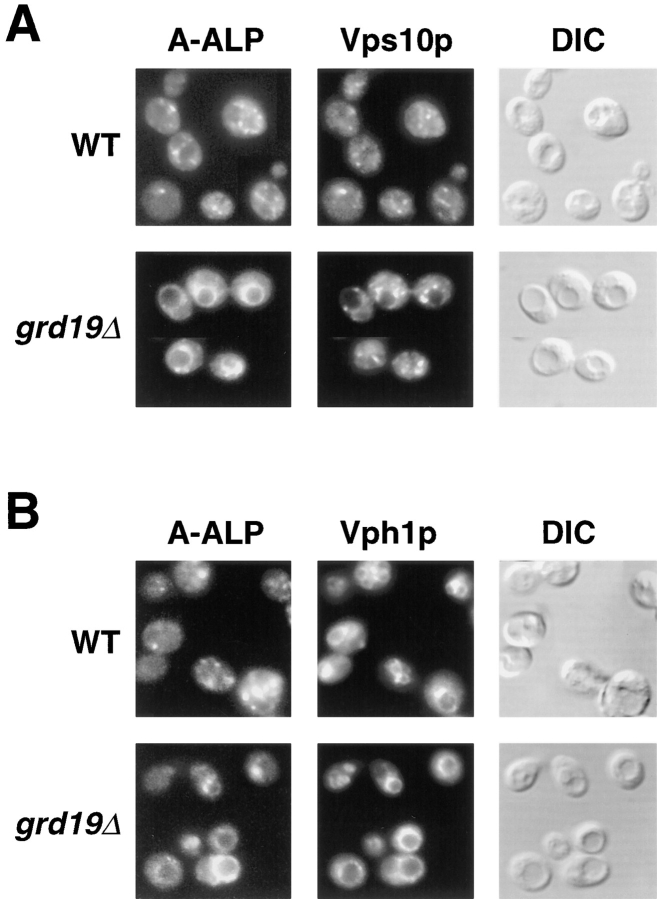
Since the degradation of the late-Golgi membrane proteins is an indirect assay for mislocalization to the vacuole, we performed immunofluorescence experiments to determine the actual intracellular localization of several membrane proteins. Double staining of wild-type cells with antibodies against A-ALP and Vps10p revealed a colocalization of both proteins to disperse, punctuate structures in the yeast cytosol indicative of the yeast TGN (Fig. 4 A; Nothwehr et al., 1993; Cooper and Stevens, 1996). However, in grd19Δ cells A-ALP was localized to ring-shaped structures indicative of vacuolar membranes. The vacuolar localization was confirmed by double staining experiments using antibodies against the 100-kD subunit of the vacuolar ATPase, encoded by the VPH1 gene (Kane et al., 1992; Manolson et al., 1992; Fig. 4 B). In contrast to the vacuolar localization of A-ALP in grd19Δ cells, Vps10p was still localized to Golgi structures in these same cells as shown by the punctuate staining pattern (Fig. 4 A). The essentially wild-type–like staining of grd19Δ cells with antibodies to the vacuolar membrane protein Vph1p (Fig. 4 B) also demonstrates that mutations in GRD19 do not generally perturb vacuolar morphology or vacuolar biogenesis. These data reveal that Grd19p is not required for Golgi localization of Vps10p or vacuolar biogenesis in general, but instead suggest that Grd19p function is more specifically involved in the localization mechanism of A-ALP and Kex2p to the TGN.
Figure 4.
A-ALP, but not Vps10p is mislocalized to the vacuole in grd19Δ cells. Wild-type (SNY63) and grd19Δ pep4Δ (WVY5) cells containing pSN55 were fixed, spheroplasted, and analyzed by indirect immunofluorescence. (A) Cells were double stained with antibodies against ALP and Vps10p. After treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by differential interference contrast optics (DIC) and by epifluorescence through filters specific for fluorescein and Texas red. (B) Cells were double-stained and analyzed as described in (A) using antibodies against ALP and the 100-kD subunit of the vacuolar ATPase (encoded by VPH1) which is an integral membrane protein of the vacuole.
Grd19p Is Required for the Retrieval from the Prevacuolar Compartment
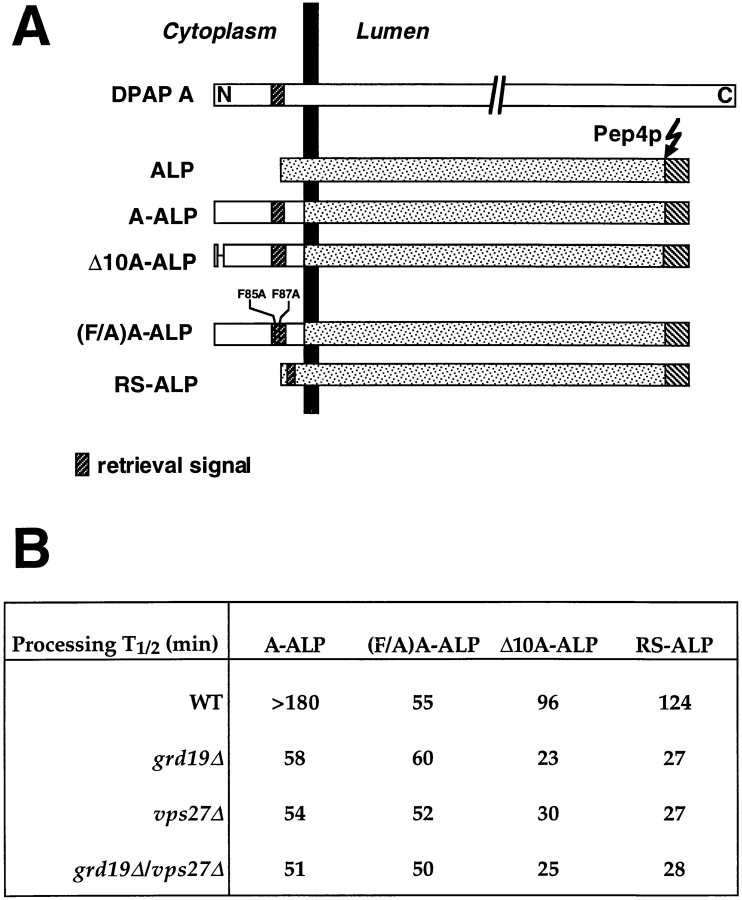
VPS27 is 1 of 13 genes required for membrane traffic out of the prevacuolar compartment (Raymond et al., 1992_a_ ; Piper et al., 1995), and vps27Δ cells accumulate an exaggerated prevacuolar compartment (Raymond et al., 1992_a_ ; Piper et al., 1995). The localization of A-ALP to the yeast TGN has been shown to involve both static retention and retrieval from the prevacuolar compartment (Bryant and Stevens, 1997). To assess whether Grd19p functions in static retention or retrieval, we employed assays that allow us to distinguish between these functions. The prevacuolar compartment that accumulates in vps27Δ mutant cells contains active forms of vacuolar proteases, and thus the processing half-time of A-ALP and related proteins in these cells has been shown to reflect the rate at which they exit Golgi membranes (Bryant and Stevens, 1997). Different variants of A-ALP with mutations in the cytosolic domain were analyzed for their processing kinetics in vps27Δ and grd19Δ strains. A schematic drawing of the analyzed fusion proteins is shown in Fig. 5 A. In wild-type cells, the full-length A-ALP is localized to the late-Golgi compartment by a combination of retention and retrieval mechanisms and has a processing half-time of >180 min (Bryant and Stevens, 1997). In vps27Δ cells, membrane traffic from the prevacuolar compartment back to the Golgi is blocked and the processing half-time of A-ALP is reduced to 60 min, reflecting its rate of exit from the late-Golgi compartment (Bryant and Stevens, 1997). Similar reductions in stability could be observed in wild-type cells with the construct (F/A)A-ALP, in which the retrieval signal has been rendered nonfunctional by mutating the phenylalanine residues in the retrieval signal (FXFXD) to alanines (Nothwehr et al., 1993; Bryant and Stevens, 1997). A different construct, which has a 10 amino acid deletion at the amino-terminal end (Δ10A-ALP), showed slightly reduced stability in wild-type cells but a very fast processing half-time of ∼20 min in vps27Δ cells (Bryant and Stevens, 1997). This deletion in the A-ALP cytosolic domain results in a loss of static retention and hence a fast exit rate of A-ALP from the late-Golgi compartment without affecting the efficiency of retrieval (Bryant and Stevens, 1997).
Figure 5.
Processing kinetics of mutant A-ALP fusion constructs in grd19Δ cells. (A) Schematic drawing of the analyzed fusion proteins between the cytosolic domain of DPAP A and the transmembrane and lumenal domains of ALP. The dark-shaded box represents the sequence in the DPAP A domain, which was previously identified as necessary and sufficient for localization to the late-Golgi compartment. (B) Table represents the halftimes of _PEP4_-dependent processing of the respective fusion proteins in wild-type (SNY36), grd19Δ (WVY4), vps27Δ (WVY21), and grd19Δ vps27Δ strains. Values were determined by pulse–chase labeling and immunoprecipitation experiments as described in the legend of Fig. 2, and represent the mean value of at least three independent experiments.
To gain insight into which of the localization mechanisms is affected in grd19Δ cells, the processing kinetics of the different A-ALP reporter proteins were now analyzed in _grd19Δ_-mutant and _grd19Δ vps27Δ_-double mutant cells and compared to cells carrying the vps27Δ mutation alone. As shown in Fig. 5 B, the processing kinetics of all three fusion protein constructs in grd19Δ cells were indistinguishable from those in vps27Δ cells. In addition, neither A-ALP nor (F/A)A-ALP were processed more rapidly in _grd19Δ vps27Δ_-double mutant cells compared to either single mutant alone, suggesting that Grd19p is not involved in static retention of A-ALP. Moreover, the processing kinetics of A-ALP in grd19Δ were identical to those of (F/A)A-ALP in wild-type cells, which strongly suggests that Grd19p is involved in the retrieval pathway from the prevacuolar compartment back to the TGN.
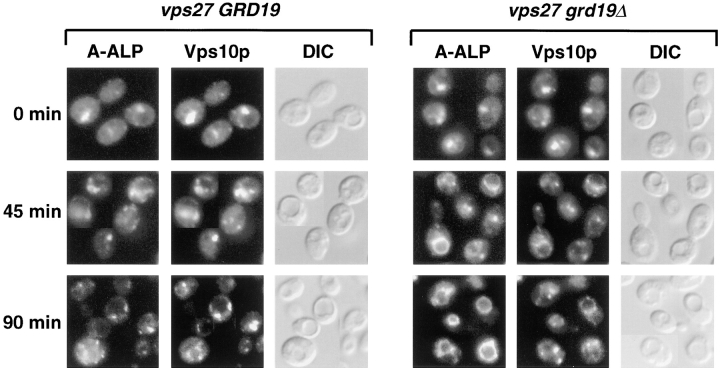
Since the processing experiments allow only an indirect assessment about the involvement of grd19Δ in the retrieval process, we used a second assay that directly studies the redistribution of membrane proteins from the accumulated prevacuolar compartment in _vps27_-mutant cells (Bryant and Stevens, 1997). We used a _vps27_-mutant strain where we could accumulate the late-Golgi membrane proteins Δ10A-ALP and Vps10p in the prevacuolar compartment. Exit from the prevacuolar compartment was then induced by expressing wild-type Vps27p under the control of the GAL1 promoter. Δ10A-ALP was used since it localizes almost exclusively to the prevacuolar compartment in _vps27_-mutant cells, yet Δ10A-ALP shows essentially the same behavior as A-ALP in the redistribution from the prevacuolar compartment (Bryant and Stevens, 1997). The induction of Vps27p was performed in both _vps27_-mutant and _vps27 grd19Δ_-double mutant cells to directly assess the involvement of Grd19p on the retrieval reaction. Samples were taken 0, 45, and 90 min after induction of Vps27p synthesis; cells were fixed; and the distribution of both Δ10A-ALP and Vps10p or Δ10A-ALP and Vph1p, respectively, was analyzed in the same cells by indirect immunofluorescence (Fig. 6). The amount of cells displaying a particular staining pattern indicative of the morphological features of Golgi membranes, vacuolar membranes, and the exaggerated prevacuolar compartment (class E compartment; Raymond et al., 1992_a_ ) was quantified (Table III).
Figure 6.
Retrieval of A-ALP but not Vps10p from the prevacuolar compartment is blocked in grd19Δ cells. Cells of the strains NBY67 (GRD19 vps27) and WVY19 (grd19Δ vps27), each containing the plasmids pH Y5 (GAL1::VPS27) and pWV36 (expressing Δ10A-ALP), were grown on synthetic medium with raffinose as carbon source. Expression of wild-type Vps27p was induced by the addition of 2% galactose. After the indicated incubation times, aliquots were taken and prepared for indirect immunofluorescence as described. Cells were stained with antibodies against ALP and Vps10p and viewed by epifluorescence and differential interference contrast optics (DIC).
Table III.
Quantification of Retrieval Assay
Before induction >80% of both cell types clearly showed the accumulation of Δ10A-ALP, Vps10p, and Vph1p in the class E prevacuolar compartment (Table III). 45 min after induction of Vps27p activity, a redistribution of the staining pattern was already observed for both proteins, and the redistribution was essentially complete 90 min after induction of Vps27p synthesis. In >80% of the GRD19 cells, both Δ10A-ALP and Vps10p localized to the same smaller multiple dots, which are typically seen for late-Golgi membrane proteins, whereas Vph1p traveled to the vacuole as expected. However, in grd19Δ cells, Δ10A-ALP exited the prevacuolar compartment and traveled to the vacuole together with the vacuolar membrane protein Vph1p instead of redistributing back to the TGN. Almost 80% (65 + 13% and 58 + 20%) of the grd19Δ cells exhibited predominantly the ring-shaped vacuolar staining pattern for both Δ10A-ALP and Vph1p 90 min after induction of Vps27p expression. On the other hand, the majority of Vps10p redistributed to Golgi structures in grd19Δ cells. More than 80% of the cells eventually showed a Golgi staining pattern for Vps10p. A small portion of Vps10p also appeared to be mislocalized to vacuolar structures in grd19Δ cells, consistent with the observed slight decrease in its stability compared to wild-type cells, as shown in Fig. 3 B. We conclude that both retrograde and anterograde traffic pathways from the prevacuolar compartment are essentially intact in grd19Δ cells, but Grd19p function is indeed required specifically for the retrieval of A-ALP from the prevacuolar compartment back to the TGN.
Grd19p Is a Cytosolic Protein Found on the Surface of the Prevacuolar Compartment in vps27Δ Cells
To assess the intracellular localization of Grd19p, we introduced an influenza hemeagglutinin epitope (HA) at the carboxy terminus of the protein. The GRD19::HA fusion was integrated at the genomic locus of GRD19, and by all criteria the GRD19::HA gene fully complemented the grd19Δ mutation (data not shown). Using antibodies against the HA epitope we detected Grd19p–HA as a 28-kD protein by Western blot analysis (Fig. 7 A). The HA-tagged Grd19p was found predominantly together with the cytosolic marker protein PGK in subcellular fractionation experiments, but a minor amount was found associated with membrane fractions in the low speed pellet (Fig. 7 B).
Figure 7.
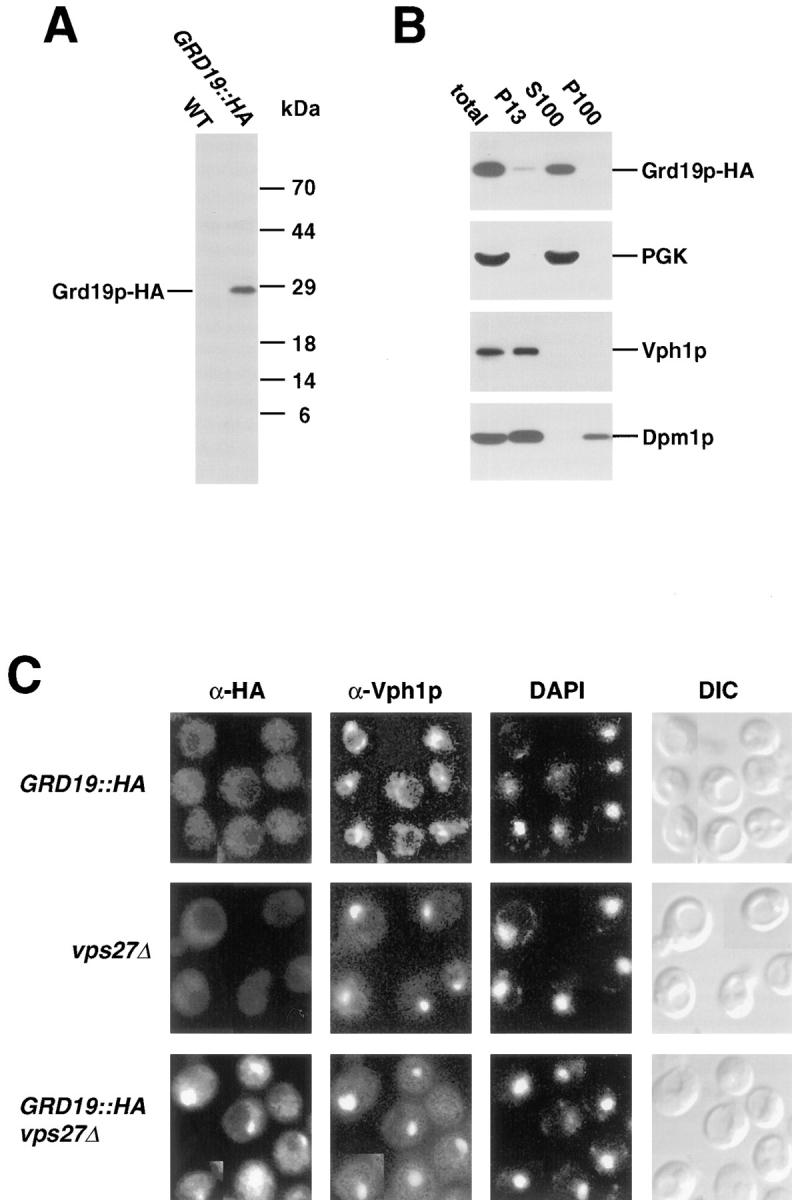
Grd19p is a soluble cytosolic protein. (A) Total cell extracts of wild-type (SNY36) and cells expressing Grd19p with an amino-terminal HA-epitope tag (WVY12) were prepared. Proteins were separated by SDS-PAGE and analyzed by Western blot. The epitope-tagged Grd19p–HA was detected by immunodecoration with antibodies against the HA epitope. (B) Subcellular fractions were prepared from GRD19::HA (WVY12) cells as described in Materials and Methods, separated by SDS-PAGE, and analyzed by Western blot. The fractions were decorated with antibodies against the HA epitope, phosphoglycerate kinase (PGK) as a cytosolic marker protein, Dpm1p (ER membranes), and Vph1p (vacuolar membrane). (C) Grd19p associates with the prevacuolar compartment in vps27Δ cells. Cells from the strains WVY12 (GRD19::HA), WVY20 (GRD19::HA vps27Δ) and WVY20 (vps27Δ) were analyzed by indirect immunofluorescence as described in the legend of Fig. 4. The intracellular distribution of Grd19p and vacuolar membranes was visualized by double-staining with antibodies against the HA epitope of Grd19p-HA and against Vph1p (100-kD subunit of the vacuolar ATPase). Nuclear DNA was viewed by staining with DAPI. Yeast cells were viewed by differential interference contrast optics (DIC).
Consistent with a predominantly cytosolic localization for Grd19p, immunofluorescence experiments using antibodies against the HA epitope revealed only a low level and diffuse staining in GRD19::HA cells (Fig. 7 C). Since the correct localization of late-Golgi membrane proteins has been found to involve recycling between the late-Golgi membrane and a post-Golgi prevacuolar compartment (Nothwehr and Stevens, 1994), we also analyzed the localization of Grd19p in a yeast mutant (vps27Δ) that accumulates an exaggerated prevacuolar compartment. Interestingly, a distinct staining pattern of Grd19p-HA was observed in vps27Δ cells, and this staining pattern overlapped with that of the 100-kD subunit of the V-ATPase, the protein marker for the accumulated prevacuolar compartment (Fig. 7 C). To exclude that the Grd19p–HA staining pattern is not the result of an artificial accumulation of HA-crossreactive structures due to the vps27Δ mutation, we included a vps27Δ mutant that expresses no HA epitope as a control. This control showed no staining, and as can be seen from the DAPI stained panels, the staining pattern of Grd19p–HA was also clearly distinct from nuclear staining. The subcellular localization experiments indicate that Grd19p is localized predominantly in the cytosol, but becomes associated with the prevacuolar compartment in vps27Δ cells, suggesting that Grd19p may function at the periphery of this organelle.
Grd19p Interacts Physically with the Cytosolic Domain of DPAP A
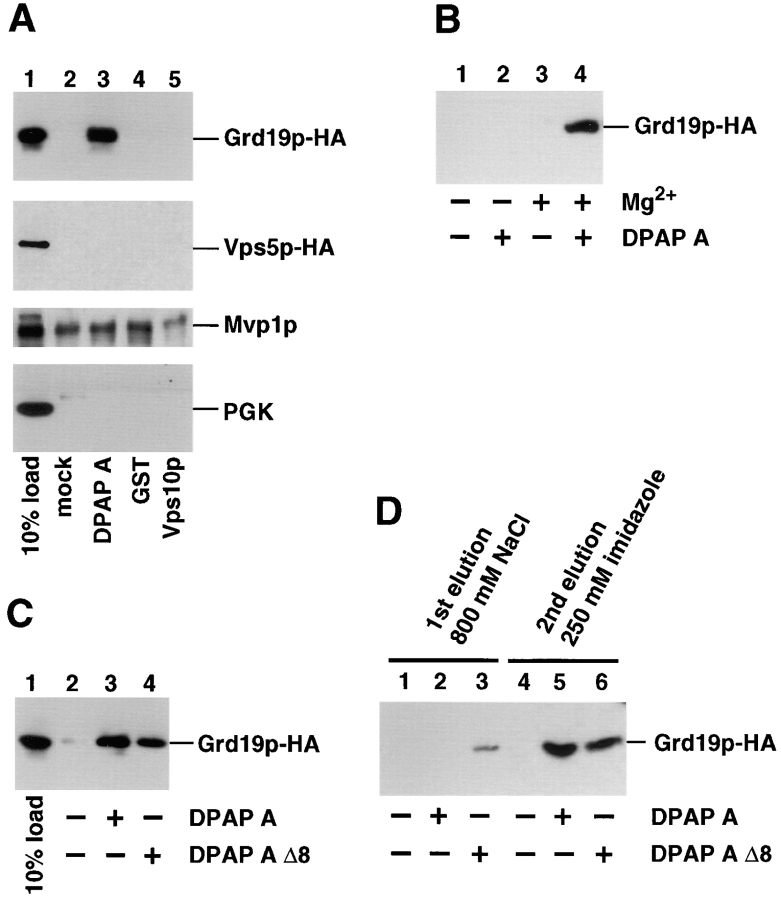
The previous experiments established that Grd19p is likely to function in sorting and/or transport of certain TGN membrane proteins from the prevacuolar compartment back to the TGN. Additionally, Grd19p seems to confer a certain specificity to the sorting reaction of membrane proteins since it is not necessary for the cycling of the late-Golgi cargo receptor Vps10p but rather is required for the retrieval of the proteases DPAP A and Kex2p. To determine whether Grd19p functions by physically interacting with the cytosolic domain of DPAP A, we tested whether it can bind to the protein in vitro. We used a fusion protein consisting of the first 117 amino acids of DPAP A (the entire cytosolic domain) and a 6xHis tag, which was introduced in place of the transmembrane domain to enhance the formation of the native conformation of the cytosolic domain. The fusion protein was expressed in E. coli and purified using a Ni-Agarose resin under native conditions (see Materials and Methods). The immobilized DPAP A tail was incubated with yeast cytosolic extracts obtained from a strain expressing Grd19p-HA. In addition, a Vps10p cytosolic domain expression construct was used in the same binding assay. In this case, the carboxy-terminal tail domain was fused to glutathione-S-transferase (GST). To determine if Grd19p might bind directly to the FXFXD retrieval signal motif within the DPAP A cytosolic domain, we also analyzed a DPAP A–tail expression construct that had an eight amino acid residue deletion that removes the FXFXD motif. After extensive washing of the matrix material, all bound proteins were eluted, and the presence of Grd19p was detected by SDS-PAGE followed by Western blot analysis using anti-HA antibodies.
As shown in Fig. 8 A, Grd19p bound to the expressed cytosolic domain of DPAP A (lane 3). In contrast, similar amounts of expressed Vps10p tail domain did not show any binding of Grd19p in the in vitro binding assay (Fig. 8 A, lane 5). As control reactions, a mock sample that contained empty Ni-NTA resin and a sample expressing the GST moiety alone were included. No significant binding could be detected in the control samples (Fig. 8 A, lanes 2 and 4), confirming the specificity of the binding reaction. The related proteins Vps5p and Mvp1p, which show partial sequence homology to Grd19p, the PX domain (Ponting, 1996), were also tested for binding to the cytosolic domains of DPAP A or Vps10p under conditions where the interaction with Grd19p could be detected. Vps5p could not be detected in the eluate fractions, and Mvp1p showed only a low-level, nonspecific binding to the columns whether protein was attached or not (Fig. 8 A). To exclude the possibility that cytosolic proteins were nonspecifically retained during the incubations and washing steps, we probed all samples with antibodies against the abundant cytosolic protein phosphoglycerate kinase (PGK) but could not detect it in the elution fractions. The binding of Grd19p to the DPAP A cytosolic domain was dependent on the presence of Mg2+ ions in the medium (Fig. 8 B). Under the conditions used, ∼10% of the added Grd19p in the yeast extract bound to the DPAP A tail. Under the chosen conditions, Grd19p also bound to the DPAP A tail construct missing the FXFXD retrieval sequence (DPAP A–tail Δ8; Nothwehr et al., 1993) as well as the full-length tail construct (Fig. 8 C). However, the complex with Grd19p and the DPAP A–tail Δ8 construct seemed to be less stable since incubations in buffer with elevated salt concentrations led to the release of significant amounts of the bound Grd19p from the column, whereas the binding of Grd19p to the full-length DPAP A tail was not affected by 800 mM NaCl (Fig. 8 D).
Figure 8.
Grd19p physically interacts with the cytosolic domain of DPAP A. (A) Cytosolic extracts of yeast cells carrying plasmid pWV32 were prepared and incubated with empty Ni-NTA resin (mock), E. coli expressed DPAP A cytosolic tail (DPAP A), GST or Vps10p cytosolic tail fused to GST (Vps10p) as described in Materials and Methods. Bound proteins were either eluted with 250 mM imidazole or 5 mM Glutathione, TCA-precipitated and separated by SDS-PAGE. The presence of Grd19p-HA, Vps5p-HA, Mvp1p, and PGK in the eluate was detected by Western blot. To compare the efficiency of binding 10% of the starting material was included in the analysis (10% load). (B) Binding assays were performed as described, except that the binding and washing reactions were done in the presence of 5 mM EDTA (−Mg2+) or in the presence of 5 mM magnesium acetate (+Mg2+). (C) Binding assays were performed as described above in the presence of Mg2+. In addition, E. coli expressed DPAP A tail with a deletion of the retrieval motif (DPAP A Δ8) was included. (D) Proteins were bound to cytosolic domains of DPAP A as described above. The column material containing the bound proteins was first washed by 15 min incubation with 800 mM NaCl (1st elution). Bound proteins were then eluted by the addition of 250 mM imidazole (2nd elution).
We conclude that Grd19p interacts specifically with the cytosolic domain of DPAP A in the in vitro binding assay. The related proteins Vps5p and Mvp1p are not part of the complex between Grd19p and the cytosolic domain of DPAP A. The presence of the FXFXD retrieval motif contributes to the stability of that interaction. The specificity of Grd19p involvement in the retrieval of resident late-Golgi membrane proteins is correlated with its ability to physically interact with the cytosolic domain of DPAP A but not with the cytosolic domain of the CPY sorting receptor Vps10p.
Discussion
In this study we report the identification of a novel yeast gene, GRD19, and provide data to support a model for its function in the retrieval process of membrane proteins of the TGN from the prevacuolar compartment. GRD19 is a nonessential gene that encodes a small hydrophilic protein with a calculated molecular mass of 18.7 kD. In grd19Δ cells the resident late-Golgi membrane proteins A-ALP and Kex2p are mislocalized to the vacuole. Mutations in GRD19 did not affect vacuolar biogenesis, vacuolar protein sorting, or vacuolar morphology (Raymond et al., 1992_a_ ). The vacuolar hydrolase CPY was sorted with almost wild-type efficiency. The observed slight increase in CPY secretion in grd19Δ cells correlates well with a minor decrease in the stability of the CPY cargo receptor Vps10p (Cereghino et al., 1995; Cooper and Stevens, 1996). Since overexpression of Vps5p suppresses the slight CPY sorting defect in grd19Δ cells, this defect is likely to result indirectly from the loss of Grd19p function. The steady-state localization of Vps10p in the late-Golgi compartment and its ability to redistribute back to late-Golgi structures from an accumulated prevacuolar compartment suggest that in grd19Δ cells Vps10p cycles normally between the late-Golgi and prevacuolar compartments and is able to sort and transport most of the newly synthesized CPY to the vacuole. Therefore, mutations in GRD19 do not affect the general vesicle traffic between the yeast Golgi and vacuole, but rather specific sorting processes involving the resident late-Golgi membrane proteins DPAP A and Kex2p. These properties make GRD19 unique among the components required for TGN membrane protein localization, since all other genes identified so far are also required for vacuolar protein sorting (Seeger and Payne, 1992_a_ ; Nothwehr et al., 1996; Redding et al., 1996_a_ ,b; Nothwehr and Hindes, 1997; Seaman et al., 1997).
Since the correct localization of A-ALP in the yeast TGN has been shown to involve both static retention and retrieval from the prevacuolar compartment, we investigated whether GRD19 might be involved specifically in one of these modes of A-ALP localization. An analysis of the exit rates of A-ALP from the late-Golgi compartment (Bryant and Stevens, 1997) showed that A-ALP exits the TGN with normal kinetics in grd19Δ cells. The processing kinetics of A-ALP in grd19Δ cells were very similar to those measured in a vps27Δ mutant, a mutant already shown to be defective for membrane traffic from the prevacuolar compartment (Piper et al., 1995). The processing rate of A-ALP was not further accelerated in _grd19Δ vps27Δ_-double mutant cells. In addition, the half-time of processing of A-ALP in grd19Δ cells is similar to that of the retrieval defective (F/A)A-ALP construct in wild-type cells (Nothwehr and Stevens, 1994). Taken together, these data indicate a role for GRD19 in the retrieval of specific TGN membrane proteins from the prevacuolar compartment.
The function of GRD19 in the retrieval process could be directly demonstrated by an assay based on the redistribution of membrane proteins accumulated in the prevacuolar compartment of _vps27_-mutant cells (Bryant and Stevens, 1997). Cells carrying the grd19Δ mutations were not able to retrieve A-ALP from the prevacuolar compartment upon restoration of Vps27p function. Instead it redistributed to the vacuole like the vacuolar membrane protein Vph1p. The defect in A-ALP retrieval is not caused by a complete block of membrane traffic from the prevacuolar compartment to the Golgi in grd19Δ cells, since the retrieval of Vps10p, and the vacuolar delivery of Vph1p were unaffected. A role for Grd19p in TGN membrane protein retrieval from the prevacuolar compartment was further supported by experiments analyzing its intracellular distribution. Grd19p is a hydrophilic protein that localizes predominantly to the yeast cytosol. However, in vps27Δ cells, in which membrane traffic from the prevacuolar compartment is blocked, Grd19p immunolocalized to the accumulated prevacuolar compartment. The block of membrane traffic in vps27 cells traps cargo molecules such as DPAP A and Vps10p, which presumably then leads to the accumulation of associated cytosolic components such as Grd19p. The small amount of Grd19p that was observed in fractions containing membrane structures in subcellular fractionation experiments is consistent with at least a transient association of Grd19p with the prevacuolar compartment even in wild-type cells.
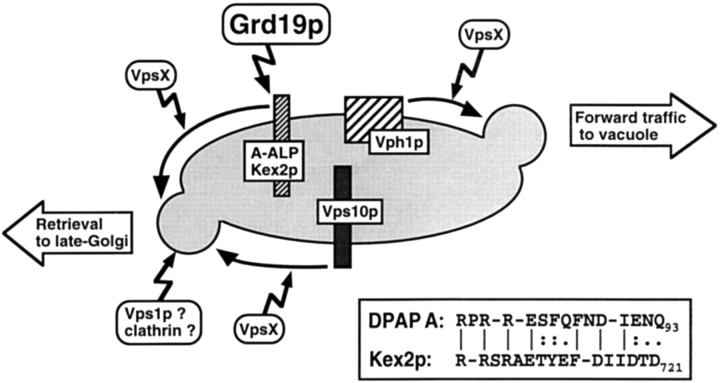
The involvement of a common subset of VPS and GRD genes in the localization of Vps10p, A-ALP, and Kex2p indicates that all three proteins use a similar pathway for cycling between the late-Golgi and the prevacuolar compartment. The apparent specificity of Grd19p for the retrieval of the resident proteins DPAP A and Kex2p may reflect the fact that the retrieval signals in the cytosolic domains of these two proteins exhibit significant similarity (as shown in the inset of Fig. 9), that is not found in the cytosolic domain of Vps10p. The DPAP A and Kex2p retrieval signals consist of two aromatic residues, preceded by a cluster of basic amino acids and followed by some acidic residues (Wilcox et al., 1992; Nothwehr et al., 1993). Together, these regions of ∼15 to 20 amino acids could constitute a conserved Grd19p-binding site.
Figure 9.
Sorting at the prevacuolar compartment of S. cerevisiae. Schematic drawing of the function of Grd19p at the surface of the prevacuolar compartment showing its requirement for the retrieval of DPAP A and Kex2p but not Vps10p. (Inset) Alignment of the putative retrieval signal regions of DPAP A and Kex2p.
A shared and saturable protein machinery for the localization of DPAP A and Kex2p was initially indicated by the observation that overexpression of Kex2p leads to an increased vacuolar degradation of A-ALP (Nothwehr et al., 1993). Because of its differential behavior influencing the localization of DPAP A/Kex2p but not Vps10p, Grd19p might be a candidate for a protein interacting with the retrieval signal motif of the resident membrane proteins. As our experiments show, Grd19p indeed physically interacts with the expressed tail domain of DPAP A in vitro. The binding is specific, dependent on the presence of magnesium ions and also stable in the presence of high salt. Grd19p also bound to a DPAP A tail construct in which the FXFXD motif had been deleted, although the complex displayed a slightly lower resistance to elevated salt conditions. Grd19p binding to the DPAP A–Δ8 tail could reflect the fact that the Grd19p-binding site extends well outside the FXFXD sequence, so that removal of the FXFXD portion of the signal only slightly reduces the binding affinity of Grd19p to the DPAP A–Δ8 tail domain. Whereas this decrease might not be apparent in our nonequilibrium binding assay, this difference in binding could translate into a significant degree of mislocalization in vivo. Alternatively, Grd19p might be part of a multiprotein complex interacting with the cytosolic domain of DPAP A and another unknown factor confers the specific recognition of the aromatic amino acid–based retrieval signal.
Analysis of the protein sequence of Grd19p revealed it to be a member of a diverse protein family sharing a common sequence motif, the PX domain (Ponting, 1996). Several members of this protein family have been implicated in vesicular protein traffic. In mammalian cells a member of the PX protein family, the SNX1 protein, was found to interact with the cytosolic domain of the EGF receptor and regulate its degradation (Kurten et al., 1996). It is thus tempting to speculate that the Grd19p PX domain functions to either directly bind to the DPAP A/Kex2p cytosolic domains, or alternatively the PX domain might allow interaction with other proteins required to recognize the DPAP A/Kex2p retrieval signals as a multiprotein complex. However, the PX domain proteins Vps5p and Mvp1p, which share a similar localization and effects on TGN protein localization with Grd19p (Ekena and Stevens, 1995; Nothwehr and Hindes, 1997) could not be detected bound to the cytosolic domain of DPAP A in contrast to Grd19p itself. This renders the second possibility unlikely, suggesting that Vps5p and/or Mvp1p function in the same pathway as Grd19p but at a different step of the localization process.
Based on our results we propose the following model for Grd19p function in late-Golgi membrane protein localization. DPAP A and Kex2p get sorted into vesicles destined for traffic to the vacuole together with the cargo receptor Vps10p. The budding process requires Vps1p function (Rothman et al., 1990; Vater et al., 1992; Wilsbach and Payne, 1993_a_ ,b; Nothwehr et al., 1995) as well as the formation of a clathrin coat (Seeger and Payne, 1992_a_ ,b; Redding et al., 1996). Numerous VPS gene products are involved in the recycling of Vps10p from the prevacuolar compartment to the yeast TGN (Stack et al., 1995; Cooper and Stevens, 1996; Redding et al., 1996; Seaman et al., 1997). The same set of VPS gene products is involved in the retrieval of the mislocalized TGN membrane proteins DPAP A and Kex2p (Nothwehr et al., 1996; Voos, W., and T.H. Stevens, unpublished observations), except the retrieval of these proteins additionally requires the function of Grd19p. This is likely accomplished by a specific recognition and/or sorting event that involves the direct interaction of Grd19p with the exposed cytosolic tail domains of these proteins. The reason for the use of an additional recognition factor might be based on the functional difference between Vps10p and DPAP A/Kex2p. After becoming mislocalized to the prevacuolar compartment, DPAP A and Kex2p must use the general mechanism evolved for the cycling of the cargo receptors (Cereghino et al., 1995; Cooper and Stevens, 1996; Seaman et al., 1997), and it is reasonable that at least one additional component may be required to make a functional connection to the general retrieval machinery. Grd19p presumably performs a very specialized and important function in this process since overexpression of the PX-domain proteins Vps5p or Mvp1p did not compensate for the loss of Grd19p. Future experiments will be directed at determining whether Grd19p is part of a protein complex that recruits certain TGN membrane proteins into vesicles leaving the prevacuolar compartment for transport back to the TGN.
The importance of Grd19p function is further supported by the identification of a similar protein in the human EST (expressed sequence tag) database (Conibear, E., personal communication). The identification of a specific retrieval component interacting with the cytosolic tail domain of DPAP A should provide the basis for a more detailed, molecular understanding of the retrograde sorting and transport processes between the late-Golgi and the prevacuolar compartment.
Table I.
Yeast Strains Used in This Study
| Strain | Genotype | Source |
|---|---|---|
| SNY36 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 | (Nothwehr et al, 1993) |
| SNY63 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ::LEU2 pep4-3 | (Nothwehr and Hindes, 1997) |
| NBY17 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::TRP1 | T.H. Stevens |
| NBY67 | MATα vps27-59 pep4-3 leu2-3,112 ura3-52 his4-519 gal2 pho8ΔX | (Bryant et al., 1997) |
| AHY41 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::LEU2 vps5Δ::HIS3 | (Nothwehr and Hindes, 1997) |
| AACY30 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 suc2-Δ9 vps10Δ::URA3 | (Cooper et al., 1996) |
| JHRY20 | MATa leu2-3,112 ura3-52 his3-Δ200 can1 pep4Δ::LEU2 | T.H. Stevens |
| WVY4 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 grd19Δ::TRP1 | This study |
| WVY5 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 grd19Δ::TRP1 pep4Δ::LEU2 | This study |
| WVY10 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 grd19Δ::TRP1 vps27Δ::LEU2 | This study |
| WVY12 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 GRD19::HA | This study |
| WVY19 | MATα vps27-59 pep4-3 leu2-3,112 ura3-52 his4-519 gal2 pho8ΔX grd19Δ::kanr | This study |
| WVY20 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 GRD19::HA vps27Δ::LEU2 | This study |
| WVY21 | MATa leu2-3,ura3-52 his3-Δ200 trp1-901 ade2-101 suc2-Δ9 pho8Δ::ADE2 vps27Δ::LEU2 | This study |
Acknowledgments
We thank N.J. Bryant for the introduction to the grd screen. We acknowledge M. Snyder for providing the yeast transposon library, J. Horecka for plasmid pRSQ304, N.J. Bryant for the antisera against ALP and Vps10p, S.F. Nothwehr for supplying strain AHY41, plasmid pAH37 and antibodies against Kex2p, and N. Zufall for expert technical assistance. G. Fischer von Mollard provided plasmid pFvM8. E. Conibear, N.J. Bryant, G. Fischer von Mollard, and S. Wizigmann-Voos are thanked for stimulating discussions and critically reading the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (W. Voos) and from the National Institutes of Health (T.H. Stevens; GM38006).
Abbreviations used in this paper
ALP
alkaline phosphatase
CPY
carboxypeptidase Y
DPAP A
dipeptidyl aminopeptidase A
HA
hemagglutinin
ORF
open reading frame
Footnotes
Address all correspondence to Tom Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403-1229. Tel.: (541) 346-5884. Fax: (541) 346-4854. E-mail: stevens@molbio.uoregon.edu
Wolfgang Voos's present address is Institut für Biochemie und Molekularbiologie, Universität Freiburg, Hermann-Herder-Strasse 7, D-79104 Freiburg, Germany.
References
- Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos K, Wraight C, Stanley KK. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late-Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi E-Y, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. . Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO (Eur Mol Biol Organ) J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K, Stevens TH. The Saccharomyces cerevisiaeMVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol Cell Biol. 1995;15:1671–1678. doi: 10.1128/mcb.15.3.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Graham TR, Seeger M, Payne GS, MacKay VL, Emr SD. Clathrin-dependent localization of α 1,3-mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. . J Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Simons K. The trans-Golgi network: sorting at the exit site of the Golgi complex. Science. 1989;234:438–443. doi: 10.1126/science.2945253. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MN, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1993;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane PM, Kuehn MC, Howald-Stevenson I, Stevens TH. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:447–454. [PubMed] [Google Scholar]
- Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- Ludwig T, LeBorgne R, Hoflack B. Roles for mannose-6-phosphate receptors in lysosomal enzyme sorting, IGF-II binding and clathrin-coat assembly. Trends Cell Biol. 1995;5:202–206. doi: 10.1016/s0962-8924(00)89000-5. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Banting G. Eukaryotic membrane traffic: retrieval and retention mechanisms to achieve organelle residence. Trends Biochem Sci. 1993;18:395–398. doi: 10.1016/0968-0004(93)90097-7. [DOI] [PubMed] [Google Scholar]
- Machamer CE. Targeting and retention of Golgi membrane proteins. Curr Opin Cell Biol. 1993;5:606–612. doi: 10.1016/0955-0674(93)90129-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Warren G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269:10185–10188. [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2gene encodes a sorting nexin-1–like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRDgenes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr Opin Cell Biol. 1995;7:530–535. doi: 10.1016/0955-0674(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Pelham HR. The dynamic organisation of the secretory pathway. Cell Struct Funct. 1996;21:413–419. doi: 10.1247/csf.21.413. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR, Rothman JE. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a precvacuolar compartment in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? . Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, O'Hara PJ, Eichinger G, Rothman JH, Stevens TH. Molecular analysis of the yeast VPS3gene and the role of its product in vacuolar protein sorting and vacuolar segregation during the cell cycle. J Cell Biol. 1990;111:877–892. doi: 10.1083/jcb.111.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vpsmutants. Mol Biol Cell. 1992a;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C.K., C.J. Roberts, K.E. Moore, I. Howald, and T.H. Stevens. 1992_b._ Biogenesis of the vacuole in Saccharomyces cerevisae. Int. Rev. Cytol. 139: 59–120. [DOI] [PubMed]
- Rayner JC, Palham HR. Transmembrane domain–dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO (Eur Mol Biol Organ) J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Brickner JH, Marschall LG, Nichols W, Fuller RS. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol Cell Biol. 1996a;16:6208–6217. doi: 10.1128/mcb.16.11.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996b;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for the study of the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Raymond CK, Gilbert T, O'Hara PJ, Stevens TH. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- Schäfer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern H F, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO (Eur Mol Biol Organ) J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. . Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar proteins sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO (Eur Mol Biol Organ) J. 1992a;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. . J Cell Biol. 1992b;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS, Chen EY, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaper JH, Shaper NL. Enzymes associated with glycosylation. Curr Opin Struct Biol. 1992;2:701–709. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and a GTP-binding protein. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees, P., E. Deignan, D.E. van, J. Humphrey, M.S. Marks, P.J. Peters, and J.S. Bonifacino. 1995. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO (Eur. Mol. Biol. Organ.) J. 14: 4961–4975. [DOI] [PMC free article] [PubMed]
- Wach A, Brachat A, Pohlman R, Philipsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Dynamic retention of TGN membrane proteins in Saccharomyces cerevisiae. . Trends Cell Biol. 1993a;3:426–432. doi: 10.1016/0962-8924(93)90031-u. [DOI] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1993b;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Hong W. The SXYQRL sequence in the cytoplasmic domain of TGN38 plays a major role in trans-Golgi network localization. J Biol Chem. 1993;268:22853–22862. [PubMed] [Google Scholar]