Ezrin/Radixin/Moesin (ERM) Proteins Bind to a Positively Charged Amino Acid Cluster in the Juxta-Membrane Cytoplasmic Domain of CD44, CD43, and ICAM-2 (original) (raw)
Abstract
Abstract. CD44 has been identified as a membrane-binding partner for ezrin/radixin/moesin (ERM) proteins, plasma membrane/actin filament cross-linkers. ERM proteins, however, are not necessarily colocalized with CD44 in tissues, but with CD43 and ICAM-2 in some types of cells. We found that glutathione-S-transferase fusion proteins with the cytoplasmic domain of CD43 and ICAM-2, as well as CD44, bound to moesin in vitro. The regions responsible for the in vitro binding of CD43 and CD44 to moesin were narrowed down to their juxta-membrane 20–30–amino acid sequences in the cytoplasmic domain. These sequences and the cytoplasmic domain of ICAM-2 (28 amino acids) were all characterized by the positively charged amino acid clusters. When E-cadherin chimeric molecules bearing these positively charged amino acid clusters of CD44, CD43, or ICAM-2 were expressed in mouse L fibroblasts, they were co-concentrated with ERM proteins at microvilli, whereas those lacking these clusters were diffusely distributed on the cell surface. The specific binding of ERM proteins to the juxta-membrane positively charged amino acid clusters of CD44, CD43, and ICAM-2 was confirmed by immunoprecipitation and site-directed mutagenesis. From these findings, we conclude that ERM proteins bind to integral membrane proteins bearing a positively charged amino acid cluster in their juxta-membrane cytoplasmic domain.
Ezrin/radixin/moesin (ERM)1 proteins are thought to function as general cross-linkers between plasma membranes and actin filaments (Bretscher, 1983; Pakkanen et al., 1987; Lankes et al., 1988; Tsukita et al., 1989; Algrain et al., 1993; Arpin et al., 1994; Tsukita et al., 1997_a_ ,b). In cultured cells, ERM proteins are mostly coexpressed and concentrated just beneath specialized domains of plasma membranes such as microvilli and cell– cell or cell–substrate adhesion sites, where actin filaments are densely associated. In differentiated tissues, however, their expression levels are specifically regulated (Bretscher, 1983; Pakkanen et al., 1987; Lankes et al., 1988; Tsukita et al., 1989, 1992; Sato et al., 1991, 1992; Berryman et al., 1993; Franck et al., 1993; Amieva et al., 1994; Takeuchi et al., 1994_b_ ; Henry et al., 1995). The suppression of ERM protein expression with antisense oligonucleotides in cultured cells destroys cell surface structures such as microvilli and cell adhesion sites (Takeuchi et al., 1994_b_ ).
Sequencing of cDNAs has revealed that the amino acid sequence identity among ERM proteins is 70–80% (Gould et al., 1989; Turunen et al., 1989; Funayama et al., 1991; Lankes and Furthmayr, 1991; Sato et al., 1992). The sequences of their amino-terminal halves are highly conserved (∼85% identity) and homologous to the amino-terminal ends of some membrane-associated proteins, such as band 4.1 protein, talin, merlin–schwannomin (a tumor suppressor molecule for neurofibromatosis type II), indicating that the ERM family is included in the band 4.1 superfamily (Conboy et al., 1986; Rees et al., 1990; Rouleau et al., 1993; Trofatter et al., 1993; Takeuchi et al., 1994_a_ ; Arpin et al., 1994; Tsukita et al., 1997_a_ ,b). Because the amino-terminal domain in band 4.1 protein is responsible for its direct association with the integral membrane protein, glycophorin C (Bennet, 1989), ERM proteins were thought to associate with integral membrane proteins through their amino-terminal halves (Algrain et al., 1993). Subsequently, immunoprecipitation studies using cultured cells revealed that CD44, a widely distributed integral membrane protein, is associated with ERM proteins (Tsukita et al., 1994). Recently, in vitro binding analyses revealed that the cytoplasmic domain of CD44 directly binds to the amino-terminal half of ERM proteins (Hirao et al., 1996).
An actin filament–binding domain is located at the carboxyl-terminal half of each ERM protein, especially the carboxyl-terminal 34–amino acid residues (Algrain et al., 1993; Edwards et al., 1994; Turunen et al., 1994; Henry et al., 1995; Martin et al., 1995; Pestonjamasp et al., 1995). In full-length native ERM proteins, however, the actin-binding site as well as the CD44-binding site were reported to be masked through intramolecular and/or intermolecular head-to-tail association (Gary and Bretscher, 1993; Algrain et al., 1993; Andréoli et al., 1994; Gary and Bretscher, 1995; Berryman et al., 1995; Bretscher et al., 1995; Martin et al., 1995; Tsukita et al., 1997_a_ ). Within cells, some signal must release this masking mechanism for ERM proteins to function as cross-linkers just beneath the plasma membrane. Phosphorylation and phosphoinositide turnover are supposed to be involved in this activation (Bretscher, 1989; Urushidani et al., 1989; Gary and Bretscher, 1995; Berryman et al., 1995; Chen et al., 1995; Nakamura et al., 1995; Hirao et al., 1996). In addition, several lines of evidence indicate that Rho regulates this activation in vivo (Hirao et al., 1996; Mackay et al., 1997; Tsukita et al., 1997_a_ ,b).
Although CD44 is precisely colocalized with ERM proteins in cultured fibroblasts (Tsukita et al., 1994), the expression of CD44 varies among tissues and its distribution is not necessarily identical to those of ERM proteins in tissues or in cultured epithelial cells (Berryman et al., 1995; von Andrian et al., 1995; Nakamura and Ozawa, 1996). Furthermore, targeted disruption of CD44 in MDAY-D2 lymphosarcoma cells had no effect on their growth or metastatic capacity, suggesting that the actin filament/plasma membrane linkage was normal in these CD44-deficient cells (Driessens et al., 1995). In some types of cells, integral membrane proteins such as the H+/K+ ATPase, CD43, and ICAM-2 are precisely colocalized with ERM proteins (Hanzel et al., 1991; Yonemura et al., 1993; Helander et al., 1996). In this study, we showed that moesin binds in vitro to the cytoplasmic domains of CD43 and ICAM-2 as well as CD44. The regions responsible for moesin binding in their cytoplasmic domains were narrowed down to the juxta-membrane regions that were characterized by positively charged amino acid clusters. The involvement of these juxta-membrane regions in their ERM binding was also confirmed by transfection and immunoprecipitation. Based on these findings, we concluded that ERM proteins bind to integral membrane proteins bearing a positively charged amino acid cluster just beneath the plasma membrane.
Materials and Methods
Cells and Antibodies
Sf9 and High Five cells (Invitrogen, Carlsbad, CA) were cultured in TC-100 medium (GIBCO BRL, Gaithersburg, MD) supplemented with tryptose phosphate broth (GIBCO BRL) and 10% FCS at 27°C. Mouse fibroblastic L cells (Earle, 1943) were cultured in DME with 10% FCS.
We detected ERM proteins, using mouse anti-ERM mAb, CR22 (Sato et al., 1991), which has higher affinity for moesin than for ezrin and radixin, rat antiezrin mAb (M11), rat antiradixin mAb (R2-1), rat antimoesin mAb (M22), and rabbit anti-ERM pAb (TK89) (Takeuchi et al., 1994_b_ ). TK89 recognizes ezrin, radixin, and moesin both by immunoblotting and immunofluorescence microscopy. Rat anti–E-cadherin mAb (ECCD-2; Shirayoshi et al., 1986) was provided by M. Takeichi (Kyoto University, Kyoto, Japan).
Production and Purification of Glutathione-S-Transferase (GST) Fusion Proteins with Cytoplasmic Domains of Integral Membrane Proteins
Various cDNA fragments for mouse CD44 and mouse ICAM-2 were obtained by reverse transcriptase-polymerase chain reaction, using mouse lung total RNA as a template, and those for rat CD43 were generated by PCR, using LSP-1 (Yonemura et al., 1993) as a template. They were then subcloned into pBluescript SK− (Stratagene, La Jolla, CA) and confirmed by sequencing with an ABI PRISM cycle sequencing kit (Perkin-Elmer Corp., Foster City, CA).
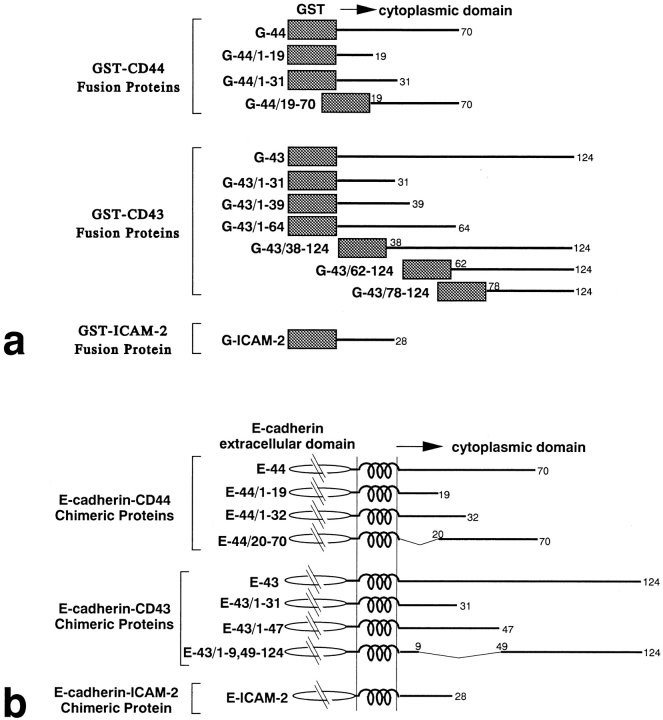
The PCR products subcloned into pBluescript SK− were excised and subcloned into pGEX2T vector (Pharmacia Diagnostics AB, Uppsala, Sweden) to produce GST fusion proteins with full-length and various truncated cytoplasmic domains of CD44 and CD43. The cytoplasmic domain of mouse CD44 contains 70 amino acids (He et al., 1992), and the following GST–CD44 cytoplasmic domain fusion proteins were produced (Fig. 1 a): G-44 containing the whole cytoplasmic domain of amino acids (a.a.)1–70, G-44/1–19 containing a.a.1–19, G-44/1–31 containing a.a.1–31 plus RN at the carboxyl terminus as a result of construction, and G-44/19–70 containing a.a.19–70. The cytoplasmic domain of rat CD 43 contains 124 amino acids (Killeen et al., 1987), and the following GST–CD43 cytoplasmic domain fusion proteins were produced (see Fig. 1 a): G-43 containing the entire cytoplasmic domain of a.a.1–124, G-43/1–31 containing a.a.1–31 plus INSS at the carboxyl terminus, G-43/1–39 containing a.a.1–39 plus EFIVTD, G-43/1–64 containing a.a.1–64 plus EFIVTD, G-43/38–124 containing a.a.38–124, G-43/62–124 containing a.a.62–124, and G-43/78– 124 containing a.a.78–124. The cytoplasmic domain of mouse ICAM-2 contains 28 amino acids (Xu et al., 1992), so only G-ICAM-2 containing the whole cytoplasmic domain was produced. We produced GST fusion proteins with the whole cytoplasmic domain of mouse E-cadherin (G-Ecad; Nagafuchi et al., 1987) and occludin (G-Oc; Furuse et al., 1994) as controls. Site-directed mutagenesis was performed by PCR, using appropriate mutagenic primers in pBluescript SK− vectors containing cDNAs encoding CD44, CD43, or ICAM-2 (see Fig. 8).
Figure 1.
Structure of GST fusion proteins (a) and E-cadherin chimeric proteins (b) of entire or truncated cytoplasmic domain of CD44, CD43, or ICAM-2. Details are described in Materials and Methods.
Figure 8.
Amino acid sequences of the juxta-membrane cytoplasmic domains of CD43 and CD44 and the entire cytoplasmic domain of ICAM-2 that are responsible for their ERM binding and ERM colocalization (Fig. 7). Positively charged amino acid residues are presented in bold letters. By in vitro mutagenesis, positively charged amino acid clusters, which were underlined in CD44, CD43, and ICAM-2, were substituted with noncharged amino acids to construct E-44/KKK:QIN, E-43/KRR:NGG, and E-ICAM-2/RRR:GGA, respectively. These sequence data are available from EMBL/GenBank/DDBJ under accession number Y00090 (rat CD43), X66081 (mouse CD44), and X6549/S46669 (mouse ICAM-2).
The GST fusion proteins were produced and purified basically according to the method of Smith and Johnson (1988) in Escherichia coli JM109 or HB101 cells. Synthesis of the GST fusion proteins was induced by incubating bacteria with 0.2 mM isopropyl β-d-thiogalactopyranoside for 2–5 h at 37°C. The cells were sedimented by centrifugation and the cell pellet was solubilized in buffer A (20 mM Tris buffer, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1.5% Sarkosyl, 1 mM PMSF, 20 μg/ml leupeptin) at 4°C according to the method of Frangioni and Neel (1993). Sarkosyl effectively decreased degradation of the fusion proteins during purification, which had not been technically circumvented in our previous study using E. coli (Hirao et al., 1996). After sonication, the cell debris was removed by centrifugation (10,000 g, 10 min, at 4°C) and the supernatant was mixed with an equal volume of buffer B (the same as buffer A except that 5% Triton X-100 was used instead of sarkosyl). The supernatant was mixed with glutathione-Sepharose 4B beads (Pharmacia Diagnostics AB) that had been washed with buffer C (1:1 mixture of buffers A and B) and then gently shaken for 10–30 min at 4°C. The beads were washed with buffer C to remove unbound bacterial proteins and stored on ice. The amount of GST fusion protein bound to the beads was estimated by SDS-PAGE.
In Vitro Binding Assay between ERM Proteins and GST Fusion Proteins
Mouse ezrin, radixin, and moesin were produced by recombinant baculovirus infection and purified as described (Hirao et al., 1996). For each reaction, 15–60 μl of glutathione-Sepharose bead slurry containing a GST fusion protein was suspended in 1 ml of buffer D (10 mM Hepes buffer, pH 7.5, 40 or 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 2 μg/ ml leupeptin) in a 1.5-ml tube, and recovered as a pellet by centrifugation (10,000 g, 1 min). After removing the supernatant, the pellet was resuspended in 1 ml of buffer D and this wash was repeated three times. Ezrin, radixin, or moesin was added to make 100–200 μl of bead suspension in buffer D containing 0.5–1 μg of ERM proteins. The beads were incubated for 30 min at room temperature with occasional mixing, and washed five times with buffer D by centrifugation. GST fusion protein was eluted with its associated protein using 150 μl of 50 mM Tris buffer (pH 8.0) containing 30 mM glutathione. The amount of GST fusion protein in each eluate was determined by SDS-PAGE. An appropriate amount of each eluate was again subjected to SDS-PAGE to contain the same amount of GST fusion protein. The amount of bound ERM protein was determined by immunoblotting with specific mAbs followed by densitometric scanning using a software NIH Image V1.54 and then relative amount of moesin bound per GST fusion protein (mol) was calculated.
SDS-PAGE and Immunoblotting
SDS-PAGE (12.5 or 10%) was performed according to the conventional method, and gels were stained with Coomassie brilliant blue R-250. For immunoblotting, proteins were electrophoretically transferred from gels onto nitrocellulose membranes. After incubation with first antibody, bound antibodies were visualized using biotinylated secondary antibody followed by avidin-conjugated alkaline phosphatase.
Mammalian Expression Vectors and Transfection
As shown in Fig. 1 b, a series of E-cadherin chimeric proteins with full-length or truncated cytoplasmic domains of mouse CD44, rat CD43, and mouse ICAM-2 were expressed in mouse L cells as described (Yonemura et al., 1993). These constructs corresponded to those of various GST fusion proteins (see Fig. 1 a). Appropriate restriction sites were introduced into the cDNA fragments obtained by PCR as described above, and subcloned into the pBATEM2 vector, which was designed for E-cadherin expression (Nagafuchi et al., 1987; Nose et al., 1988). All chimeric constructs consisted of the extracellular domain of mouse E-cadherin (from the amino terminus to BstPI site) and several amino acids of the extracellular domain/transmembrane domain/cytoplasmic domain of CD44, CD43, or ICAM-2.
All E-cadherin/CD44 chimeric molecules contained four amino acids of the extracellular domain of CD44. In E-44/20–70, N in the transmembrane domain located near the transmembrane–cytoplasmic junction was converted to T. All E-cadherin/CD43 chimeric molecules contained 23 amino acids of the extracellular domain of CD43. Both E-43/1–31 and E-43/1–47 contained additional PGIL at the carboxyl terminus, and E-43/ 1–9,49–124 contained RSA between amino acids 9 and 49. In our previous study, E-43, E-43/1–47, and E-43/1–9,49–124 were called CLS-1, CLS1-A, and CLS1-B, respectively (Yonemura et al., 1993). The E-cadherin/ ICAM-2 chimera (E-ICAM-2) contained 3 amino acids of the extracellular domain of ICAM-2. Site-directed mutagenesis was performed by PCR using appropriate mutagenic primers in pBluescript SK− vectors containing cDNAs encoding E-44, E-43, or E-ICAM-2 (see Fig. 8).
L cells were transfected with DNA using lipofectin or lipofectamine reagent (GIBCO BRL). Cells cultured on coverslips were washed twice with Opti-MEM (GIBCO BRL), and were incubated for 3–5 h with 1 ml Opti-MEM containing 1 μg of plasmid DNAs and 10 μl of the reagents, followed by the addition of 3 ml of normal medium containing FCS. Cells were then cultured for 2–3 d. L cells were also transfected by microinjection using a set of manipulators (MN-188 and MO-189; Narishige, Tokyo, Japan) connected to a microinjector 5242 (Eppendorf, Inc., Hamburg, Germany). Expression vectors in injection buffer (100 mM KCl, 10 mM Hepes buffer, pH 7.5) were injected into the nuclei of cells cultured on coverslips. Cells were examined 12–24 h after injection.
Immunofluorescence Microscopy
All procedures were performed at room temperature. Cells were fixed with 1–4% fresh formaldehyde in 0.1 M Hepes buffer (pH 7.5) for 10–15 min. After three washes with PBS containing 30 mM glycine (G-PBS), cells were soaked in blocking solution (G-PBS containing 2% normal goat serum) for 5 min and incubated with anti–E-cadherin mAb (ECCD-2) diluted with the blocking solution for 30 min. The cells were then washed three times with G-PBS, treated with 0.2% Triton X-100 in G-PBS for 10 min, and washed with G-PBS. The cells were soaked in blocking solution for 10 min, incubated with CR22 for 30 min, washed three times with G-PBS, and incubated with secondary antibodies. FITC-conjugated goat anti–rat Ig antibody (Biosource, Camarillo, CA) and rhodamine-conjugated goat anti–mouse IgG antibody (Chemicon International, Inc., Temecula, CA) were used as secondary antibodies. Cells were washed three times, and then mounted in 90% glycerol-PBS containing 0.1% para-phenylendiamine and 1% _n_-propylgalate. Specimens were observed using a Zeiss Axiophot photomicroscope (Carl Zeiss, Oberkochen, Germany). Images were taken on T-MAX 400 film (Eastman Kodak Co., Rochester, NY), or recorded with a cooled CCD camera (SenSys 0400, 768X512 pixels; Photometrics, Tucson, AZ) controlled by a Power Macintosh 7600/132 and the software package IPLab Spectrum V3.1 (Signal Analytics Corp., Vienna, VA).
Immunoprecipitation
Confluent monolayer cultures of stable L transfectants expressing E-43 or E-43/1–9,49–124 (Yonemura et al., 1993) on 10-cm dishes were used for immunoprecipitation. All procedures were carried out on ice. Cells were washed twice with a solution containing 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2 and 10 mM Hepes (pH 7.5), and then lysed in 1 ml of lysis buffer (150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes [pH 7.5], 0.1% Nonidet P-40, 1 mM DTT, 20 μg/ml leupeptin) for 30 min. The lysate was removed from the dish after fully dislodging any remaining cellular debris with a rubber policeman. After centrifugation at 12,000 g for 20 min, the supernatant was incubated for 1 h with 10 μl of protein G–Sepharose 4B beads (Zymed Labs, Inc., South San Francisco, CA) conjugated with anti– E-cadherin mAb, ECCD-2. The beads were collected and washed with lysis buffer five times by centrifugation at 1,250 g for 2 min. The immune complexes were eluted from the beads in 300 μl of 1 M CH3COOH for 10 min. The supernatant was freeze-dried and separated by SDS-PAGE followed by immunoblotting with ECCD-2 or TK89.
Results
In Vitro Binding of Moesin to GST Fusion Proteins with CD44, CD43, and ICAM-2
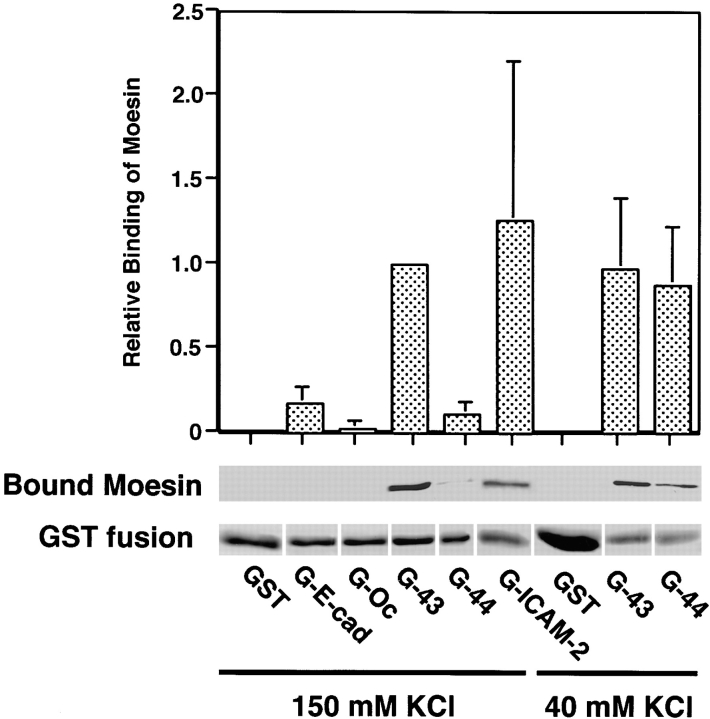
We established an in vitro binding assay to evaluate the interaction between recombinant ERM proteins and GST fusion protein with the cytoplasmic domain of CD44 (Hirao et al., 1996). Using this assay, we first compared the binding abilities of CD44, CD43, and ICAM-2 to recombinant moesin as a representative ERM protein. GST fusion proteins with the whole cytoplasmic domain of CD44 (G-44), CD43 (G-43), or ICAM-2 (G-ICAM-2) were purified on glutathione-Sepharose beads. As controls, GST fusion proteins with the whole cytoplasmic domain of E-cadherin (G-E-cad) and occludin (G-Oc) were also purified. These fusion protein-bound glutathione-Sepharose beads were incubated with recombinant moesin, washed, and eluted with glutathione. The eluate contained GST fusion protein and its associated protein. Moesin association with GST fusion proteins was evaluated by immunoblotting with antimoesin mAb followed by densitometry (Fig. 2). At low ionic strength (40 mM KCl), G-43 and G-44 bound to moesin with similar affinity. At physiological ionic strength (150 mM KCl), G-43 still bound to moesin, whereas the binding ability of G-44 to moesin was significantly decreased as previously reported (Hirao et al., 1996). G-ICAM-2 bound to moesin with affinity similar to G-43 at physiological ionic strength. In contrast, G-E-cad, G-Oc and GST showed no binding affinity to moesin. Considering that the dissociation constant between the cytoplasmic domain of CD44 and moesin at 40 mM KCl is ∼10 nM (Hirao et al., 1996), these findings showed that moesin directly and specifically binds to the cytoplasmic domains of CD43 and ICAM-2 at physiological ionic strength. We checked here that ezrin also bound to CD43 (data not shown), but other binding combinations remain to be examined.
Figure 2.
Association of moesin with the cytoplasmic domains of CD44, CD43, and ICAM-2. GST (GST) or GST fusion proteins with the entire cytoplasmic domain of E-cadherin (G-E-cad), occludin (G-Oc), CD43 (G-43), CD44 (G-44), or ICAM-2 (G-ICAM-2) were bound to Glutathione-Sepharose beads, and incubated with purified recombinant moesin at physiological (150 mM KCl) or low ionic strength (40 mM KCl). After washing, GST or GST fusion proteins were eluted together with their binding proteins from the beads with a buffer-containing glutathione. Proteins in the glutathione eluate were separated by SDS-PAGE followed by Coomassie brilliant blue staining to densitometrically estimate the amount of GST or GST fusion proteins in each eluate (GST fusion), or followed by immunoblotting with antimoesin mAb M22 to densitometrically estimate the amount of moesin bound to GST or GST fusion proteins (Bound Moesin). Relative binding ability of GST fusion proteins to moesin (Relative Binding of Moesin) was calculated by comparing the amount of bound moesin in each eluate containing a constant amount of nondegraded GST fusion protein with that in G-43 (150 mM KCl) eluate, with a careful attention to the effects of degradation products on the binding data. Values represent relative binding abilities averaged from three experiments ± SEM.
Moesin-binding Sites in the Cytoplasmic Domain of CD44 and CD43
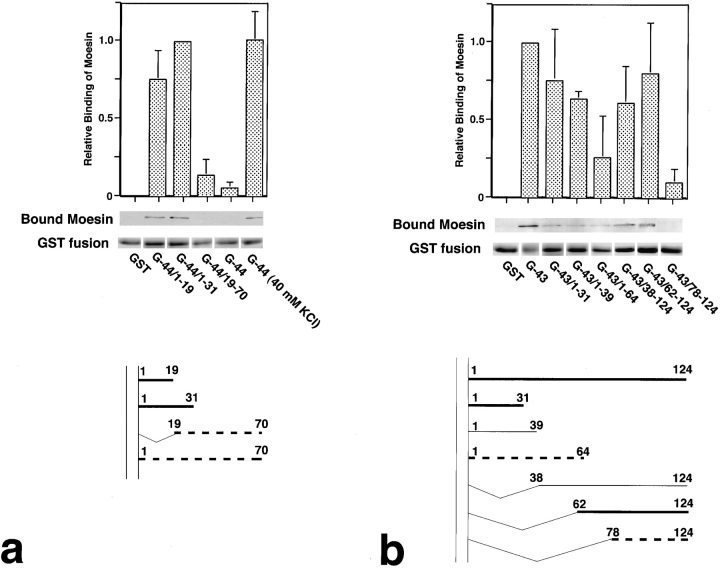
Because there is no significant homology among the cytoplasmic domains of CD44, CD43, and ICAM-2, we attempted to define the region responsible for moesin binding of each protein. Mouse CD44, rat CD43, and mouse ICAM-2 have 70, 124, and 28 amino acids in their cytoplasmic domains, respectively. We then constructed various deletion series of GST–CD44 and GST–CD43 fusion proteins (Fig. 1 a), and performed an in vitro binding assay using recombinant moesin at physiological ionic strength. As shown in Fig. 3 a, in the CD44 deletion series, G-44/1–19 and G-44/1–31 bound to moesin whereas G-44/19–70 did not. This indicates that the amino acid sequence of the juxta-membrane region (a.a.1–19) is responsible for moesin binding. The binding affinities of G-44/1–19 and G-44/1–31 to moesin at 150 mM KCl were similar to that of G-44 at 40 mM KCl (K d = ∼10 nM), suggesting an intramolecular suppressive interaction between the amino- and carboxyl-terminal parts of the cytoplasmic domain of CD44 under physiological conditions. We also found that G-44/1–19 and G-44/1–31 bound to ezrin and radixin with affinity similar to moesin under physiological conditions (data not shown).
Figure 3.
Comparison of the moesin-binding ability among various truncated cytoplasmic domains of CD44 (a) and CD43 (b). GST fusion proteins (Fig. 1 a) or GST were incubated with moesin at 150 mM KCl. Relative binding ability of each GST fusion protein to moesin was calculated as explained in Fig. 2. Relative-binding abilities of G-44/1–31 and G-43 were defined as 1 in a and b, respectively. In CD44, a.a.1–19 and a.a.1–31 (thick lines, bottom, a) bound to moesin with similar affinity to the whole cytoplasmic domain of CD44 at 40 mM KCl, whereas a.a.19–70 as well as a.a.1–70 showed very low affinity to moesin (broken lines, bottom, a), indicating that a.a.1–19 are responsible for moesin binding of CD44 (Fig. 2) and that a.a.19–70 are inhibitory for moesin binding of CD44. In CD43, a.a.1–31 and a.a.62– 124 strongly bound to moesin with similar affinity to the whole cytoplasmic domain of CD43 (thick lines, bottom, b), a.a.1–39 and a.a.38–124 were rather weakly associated with moesin (thin lines, bottom, b), and a.a.1–64 and a.a.78–124 showed very low affinity to moesin (broken lines, bottom, b). These findings indicate that at least in vitro both a.a.1–31 and a.a.62–78 appear to be responsible for moesin binding of CD43 (Fig. 2), and that the other region is inhibitory for moesin binding of CD43.
The CD43 deletion series yielded rather complex results (Fig. 3 b). G43/1–31 and G43/62–124 bound to moesin with affinities similar to the full-length cytoplasmic domain of CD43 (G-43). However, a construct longer than G-43/1–31 (G-43/1–39) weakly bound to moesin, and that longer than G-43/62–124 (G-43/38–124) also showed weak binding. Furthermore, G-43/1–64 and G-43/78–124 hardly bound to moesin. These observations indicate that there are two moesin-binding regions in the cytoplasmic domain of CD43 at least in vitro. One is located in the juxta-membrane domain (a.a.1–31) and the other is in the middle part of the cytoplasmic domain (a.a.62–78). The region between these two domains (a.a.32–61) appeared to be inhibitory for moesin binding. Similar results were obtained using ezrin (data not shown).
Colocalization of E-cadherin Chimeric Molecules Containing Cytoplasmic Domains of CD44, CD43, and ICAM-2 with ERM Proteins in Transfected Cells and Responsible Domains
To assess the physiological relevance of the above in vitro binding results, we constructed and introduced E-cadherin chimeric molecules consisting of the extracellular domain of E-cadherin and whole or various deletion constructs of the transmembrane/cytoplasmic domain of CD44, CD43, or ICAM-2 (Fig. 1 b) into mouse L fibroblasts that did not express any endogenous E-cadherin. Using an anti–E-cadherin mAb specific for the extracellular domain of E-cadherin, we then compared their subcellular distributions with those of ERM proteins by immunofluorescence microscopy. In parent L cells, ezrin, radixin, and moesin were precisely colocalized at microvilli, and the full-length E-cadherin introduced in L cells was not concentrated at microvilli (data not shown; see Yonemura et al., 1993). Because in L cells moesin was predominant among ERM proteins, the data obtained with antimoesin mAb (CR-22) are shown here.
As shown in Fig. 4 a, E-44 containing the full-length cytoplasmic domain of CD44 was co-concentrated with moesin at microvilli. Considering that endogenous CD44 was reported to be colocalized with moesin in microvilli of cultured fibroblasts (Tsukita et al., 1994), this finding indicates that the cytoplasmic, but not the extracellular domain of CD44 is responsible for its co-concentration with moesin in microvilli. We constructed various cytoplasmic domain deletion mutants of this chimeric molecule and introduced them into L cells (Fig. 1 b). E-44/1–19 (Fig. 4, c and d) as well as E-44/1–32 (data not shown) colocalized with moesin at microvilli, whereas E-44/20–70 was diffusely distributed over the cell surface and not concentrated at microvilli (Fig. 4, e and f). These observations indicated that the juxta-membrane region (a.a.1–19) of the cytoplasmic domain of CD44 is sufficient for E-cadherin– CD44 chimeric molecules to colocalize with moesin at microvilli (see Fig. 7).
Figure 4.
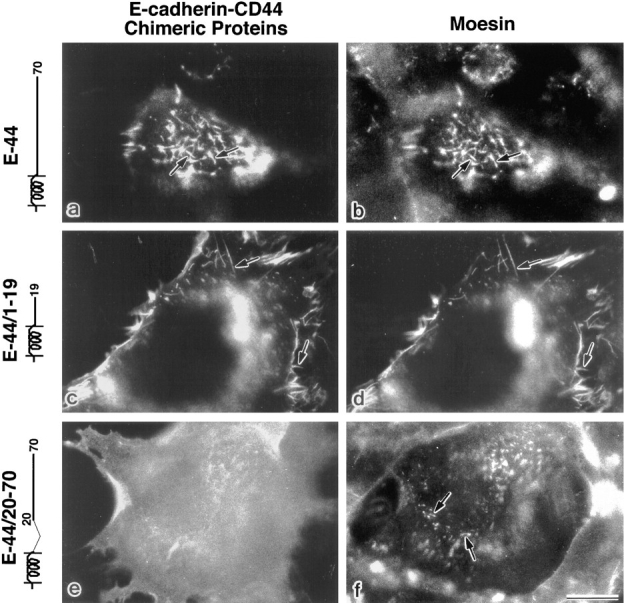
Immunofluorescence localization of E-cadherin chimeric molecules with entire and truncated cytoplasmic domains of CD44 (Fig. 1 b). L cells transiently expressing E-44 (a and b), E-44/1–19 (c and d) or E-44/ 20–70 (e and f) were doubly stained with anti–E-cadherin antibody (a, c, and e) and antimoesin antibody (b, d, and f). Both E-44 and E-44/1–19 were precisely co-concentrated with moesin at microvilli (arrows), whereas E-44/20–70 was diffusely distributed on cell surface, indicating that the ERM-colocalization signal resides in a.a.1–19 of the cytoplasmic domain of CD44. Bar, 20 μm.
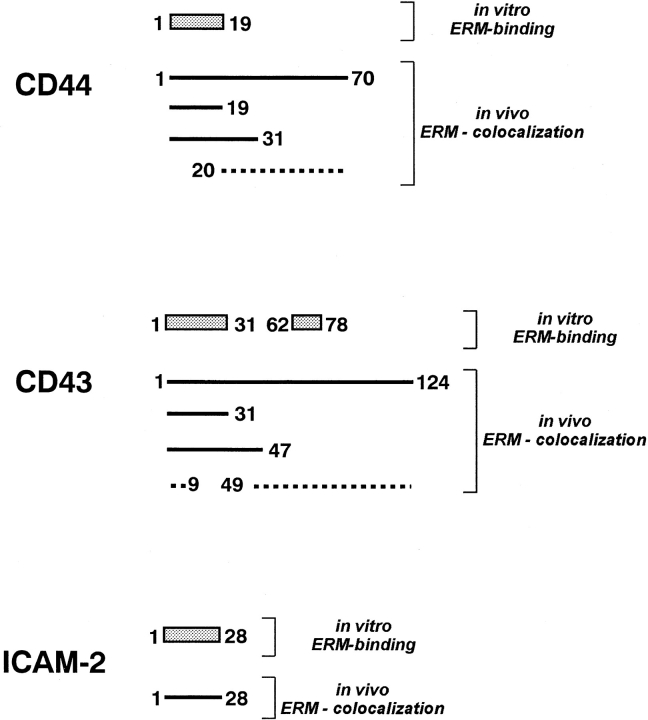
Figure 7.
Comparison of the results obtained from in vitro binding studies with transfection studies. Hatched squares represent the regions in cytoplasmic domains of CD44, CD43, and ICAM-2, which are responsible for their in vitro–direct binding to moesin (Fig. 3). In transfection experiments, E-cadherin chimeric molecules containing cytoplasmic regions represented as thick lines were co-concentrated with moesin at microvilli, whereas those containing regions represented by broken lines were diffusely distributed on the plasma membrane.
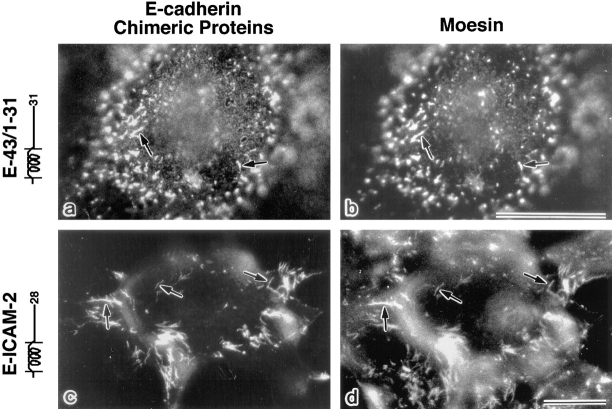
In our previous study using the same transfection system, we found that E-43, which contains the full-length cytoplasmic domain of CD43, was co-concentrated with moesin at microvilli, and that E-43/1–47 was also colocalized with moesin, whereas E-43/1–9,49–124 was not concentrated at microvilli (Yonemura et al., 1993; see Fig. 7). In this study, to further define the responsible domain, E-43/1–31 was introduced into L cells (Fig. 5, a and b). E-43/1–31 was highly concentrated at microvilli together with moesin, indicating that the juxta-membrane region (a.a.1–31) of the cytoplasmic domain of CD43 is responsible for colocalization of these E-cadherin–CD43 chimeric molecules with moesin.
Figure 5.
Immunofluorescence localization of E-cadherin chimeric molecules with the truncated cytoplasmic domain of CD43 (a and b; E-43/ 1–31) or the entire cytoplasmic domain of ICAM-2 (c and d; E-ICAM-2) (Fig. 1 b). L cells transiently expressing each protein were doubly stained with anti–E-cadherin antibody (a and c) and antimoesin antibody (b and d). Both chimeric proteins were precisely co-concentrated with moesin at microvilli (arrows). Bars, 20 μm.
We also produced an E-cadherin chimera with the whole cytoplasmic domain of ICAM-2 (E-ICAM-2) and transfected it into L cells. As shown in Fig. 5, c and d, this molecule was again highly concentrated at microvilli together with moesin.
Coimmunoprecipitation of ERM Proteins with E-43 from L Cell Transfectants
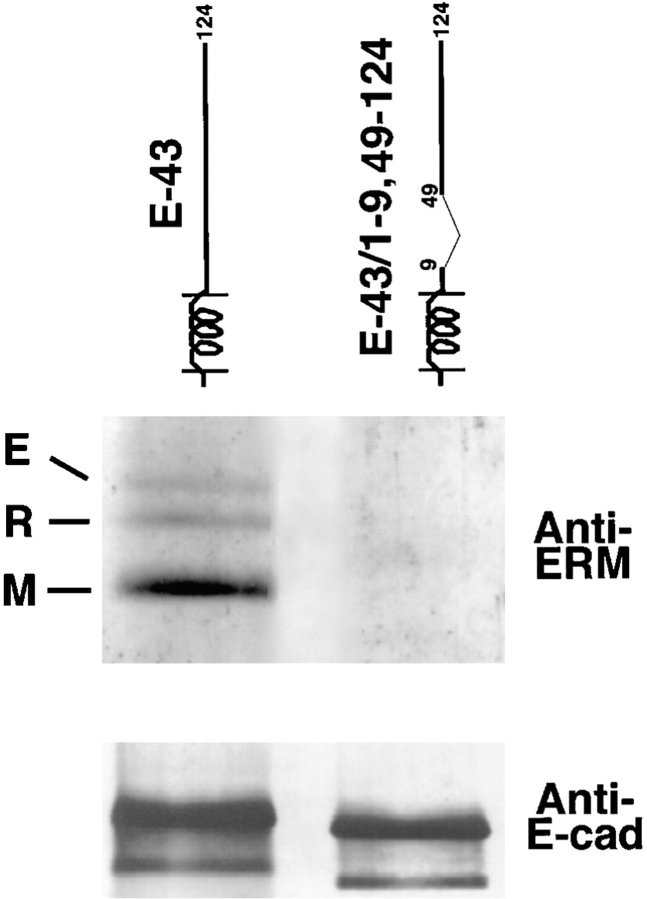
The association of ERM proteins with CD44 inside cells was detected by immunoprecipitation from BHK cells, but the detection of ERM protein–integral membrane protein complex by immunoprecipitation was difficult, because the molecular complex resists the detergent extraction due to its tight association with actin-based cytoskeletal components (Tsukita et al., 1994). However, probably because relatively large amounts of E-cadherin–CD43 chimeric molecules appeared on the cell surface of L cell transfectants, and probably because anti–E-cadherin mAb was very potent for immunoprecipitation, we were able to compare the amounts of ERM proteins in E-cadherin immunoprecipitates from E-43 stable transfectants with that from E-43/1–9,49–124 stable transfectants (in the former transfectants E-43 was co-concentrated with moesin at microvilli, whereas in the latter E-43/1–9,49–124 was not [see Fig.7]). As shown in Fig. 6, ERM proteins were coimmunoprecipitated with E-43, but not with E-43/1–9,49–124. This finding indicates not only that the juxta-membrane domain of CD43 is responsible for the CD43–ERM proteins interaction, but also that the L cell transfection system with E-cadherin chimeric molecules is useful for evaluation of ERM protein–integral membrane protein interaction. In E-ICAM-2 stable transfectants, the cell surface expression level of E-ICAM-2 was not sufficient for immunoprecipitation analyses.
Figure 6.
Coimmunoprecipitation of ERM proteins with E-cadherin chimeric protein with the whole cytoplasmic domain of CD43 (E-43). Cultured L cell transfectants expressing E-43 or E-43/1–9, 49–124 were solubilized with a lysis buffer containing 0.1% Nonidet P-40, and then immunoprecipitated with anti– E-cadherin mAb. The immunoprecipitate was separated by SDS-PAGE followed by immunoblotting with anti-ERM protein pAb (TK89; Anti-ERM) or anti–E-cadherin mAb (Anti–E-cad). TK89 recognized ezrin (E)/radixin (R) as well as moesin (M). ERM proteins were coimmunoprecipitated with E-43, but not with E-43/1–9,49–124.
Positively Charged Amino Acid Clusters Responsible for ERM Protein–Integral Membrane Protein Binding
As summarized in Fig. 7, in vitro binding analysis identified a.a.1–19 of the cytoplasmic domain of CD44, a.a.1–31, and 62–78 of the cytoplasmic domain of CD43, and the whole cytoplasmic domain (a.a.1–28) of ICAM-2 as moesin-binding sites. On the other hand, transfection experiments with E-cadherin chimeric molecules indicated that a.a.1–19 of CD44-, a.a.1–31 of CD43-, and a.a.1–28 of ICAM-2 cytoplasmic domains are responsible for co-concentration of these integral membrane proteins with ERM proteins at microvilli. Together with the results obtained from immunoprecipitation analysis, we concluded that the in vitro moesin–binding ability of a.a.62–78 of the CD43– cytoplasmic domain was not indispensable in the cells, and that the juxta-membrane 20–30-amino acid sequence of the cytoplasmic domains of CD44, CD43, and ICAM-2 can bind to ERM proteins and recruit these integral membrane proteins to microvilli together with ERM proteins.
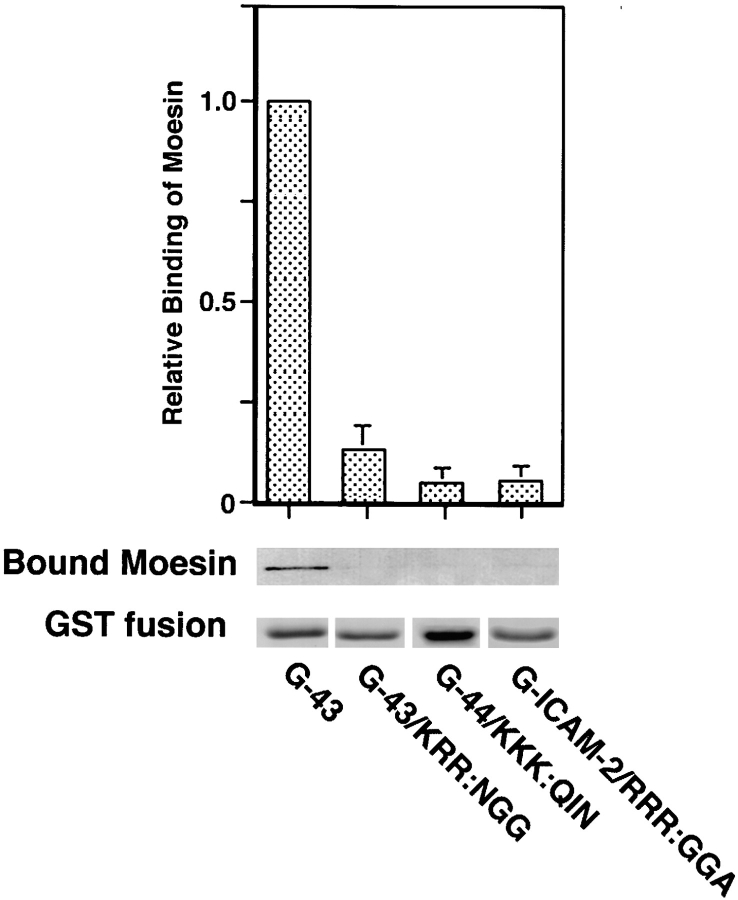
These three juxta-membrane domains showed no significant similarity in their amino acid sequences, but they were characterized by many positively charged amino acids, such as R and K (Fig. 8). To evaluate the importance of the positively charged amino acid clusters of CD44, CD43, and ICAM-2 in their binding to ERM proteins, we substituted the juxta-membrane KKK in G-44, KRR in G-43, and RRR in G-ICAM-2 with QIN, NGG, and GGA, respectively (Fig. 8). Using these site-directed mutants of GST fusion proteins (G-44/KKK:QIN, G-43/KRR:NGG, and G-ICAM-2/RRR:GGA, respectively) and recombinant moesin, we then performed an in vitro binding assay. As shown in Fig. 9, all of the mutant proteins lost their binding ability to moesin. Furthermore, when E-cadherin chimeric molecules with corresponding mutants (E-44/KKK: QIN, E-43/KRR:NGG, and E-ICAM-2/RRR:GGA) were constructed and introduced into L cells, all of these molecules were distributed diffusely on the cell surface (Fig. 10). They were not excluded from microvilli, but were not co-concentrated with moesin at microvilli. These findings indicated the importance of the juxta-membrane positively charged amino acid clusters in the ERM protein– integral membrane interaction.
Figure 9.
Moesin-binding abilities of site-directed mutants of the cytoplasmic domains of CD44, CD43, and ICAM-2. GST fusion proteins with the entire cytoplasmic domains of CD43 (G-43) and the site-directed mutants of CD43, CD44, and ICAM-2 (G-43/ KRR:NGG, G-44/KKK:QIN, and G-ICAM-2/RRR:GGA; Fig. 8) were incubated with moesin at 150 mM KCl (for G-43/KRR: NGG and G-ICAM-2/RRR:GGA) or at 40 mM KCl (for G-44/ KKK:QIN). Relative-binding ability of each GST fusion protein to moesin was calculated as explained in Fig. 2. Relative-binding ability of G-43 was defined as 1. As compared to G-43, the site-directed mutants appeared to lose their moesin-binding abilities.
Figure 10.
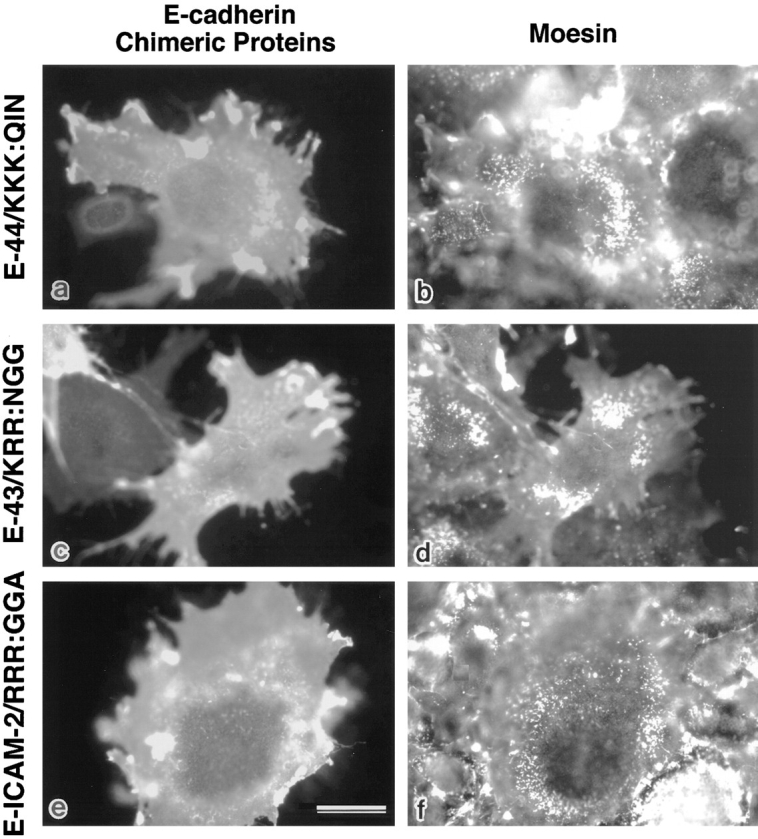
Immunofluorescence localization of site- directed mutants of E-cadherin chimeric molecules with entire cytoplasmic domains of CD44, CD43, and ICAM-2. KKK, KRR, and RRR in the juxta-membrane regions of CD44, CD43, and ICAM-2 were substituted with noncharged amino acids, respectively (E-44/KKK:QIN, E-43/ KRR:NGG, and E-ICAM-2/ RRR:GGA) (Fig. 8). L cells transiently expressing these mutants were doubly stained with anti–E-cadherin antibody (a, c, and e) and antimoesin antibody (b, d, and f). These mutants were diffusely distributed on the cell surface and were never concentrated at microvilli. Bar, 20 μm.
Discussion
Binding of ERM Proteins to CD43 and ICAM-2 As Well As CD44 through Their Juxta-Membrane Positively Charged Amino Acid Clusters
In this study, we first compared CD43 and ICAM-2 with CD44 in terms of moesin association in vitro. Close analyses using various deletion mutants revealed that moesin bound in vitro to juxta-membrane positively charged amino acid clusters of these membrane proteins. Judging from the affinity of moesin–CD44 binding (K d = ∼10 nM; Hirao et al., 1996), moesin binding to the clusters of CD43 and ICAM-2 appeared to be physiologically significant. Next, by transfecting various mutants of E-cadherin/ CD-44 (E-44), E-cadherin/CD-43 (E-43), and E-cadherin/ ICAM-2 (E-ICAM-2) chimeric molecules into L cells, we narrowed down the domains that were required for their colocalization with moesin at microvilli, and found that their juxta-membrane positively charged amino acid clusters were again responsible. These observations, together with the results of immunoprecipitation and site-directed mutagenesis studies, led us to conclude that moesin bound not only to the juxta-membrane region of CD44 but also to those of CD43 and ICAM-2 in vivo.
Most of the in vitro data presented here were from experiments using recombinant moesin, but we also confirmed that ezrin and radixin behaved in the same manner as moesin in several experiments. CD44 was reported to be associated with not only moesin but also ezrin and radixin both in vitro and in vivo (Tsukita et al., 1994; Hirao et al., 1996). Furthermore, in this study, ezrin and radixin, as well as moesin, were coimmunoprecipitated with E-43, and all ERM proteins were colocalized with E-44, E-43, and E-ICAM-2 in L cell transfectants. We therefore concluded that not only moesin, but also ezrin and radixin, bound to the juxta-membrane region of CD43, ICAM-2, as well as CD44.
Specificity of the Binding of ERM Proteins to the Juxta-Membrane Positively Charged Amino Acid Clusters
Most of the transmembrane proteins have positively charged amino acid residues in their juxta-membrane regions, and these residues are thought to form an anchor, arresting translocation across the bilayer during biosynthesis and assuring the correct topological orientation of membrane proteins (Boyde and Beckwith, 1990). For example, there are several positively charged amino acid residues in the juxta-transmembrane domains of E-cadherin and occludin, which did not bind to ERM proteins. As shown in Table I, we found that the balance of positively and negatively charged amino acid residues in the whole cytoplasmic domain was significantly different between ERM-binding proteins such as CD44, CD43, and ICAM-2, and ERM-nonbinding proteins such as E-cadherin and occludin. The calculated isoelectric points of the whole cytoplasmic domains of CD44, CD43, ICAM-2, E-cadherin, and occludin were 8.17, 9.24, 12.98, 3.89, and 5.85, respectively, indicating that, at neutral pH, CD44, CD43, and ICAM-2 have a net positive charge over their whole cytoplasmic domain and that E-cadherin and occludin have a net negative charge.
Table I.
Calculated Isoelectric Points of Cytoplasmic Domains
| Integral membrane protein | Calculated isoelectric point | ERM binding |
|---|---|---|
| CD44 | 8.17 | Weak |
| CD43 | 9.24 | Strong |
| ICAM-2 | 12.98 | Strong |
| E-cadherin | 3.89 | Undetectable |
| Occludin | 5.85 | Undetectable |
Several integral membrane proteins, L-selectin (Picker et al., 1991), ICAM-1 (Carpén et al., 1992), integrins α4β7 and α4β1 (Berlin et al., 1995), and P-selectin glycoprotein ligand-1 (Moore et al., 1995) were reported to be localized at microvilli. L-selectin and ICAM-1 have short cytoplasmic domains (∼10 and ∼30 amino acids, respectively) and their net charges are highly positive. It is postulated that α-actinin is a membrane–cytoskeleton linker for these membrane proteins. However, L-selectin lacking the α-actinin–binding site, which bears only a positively charged six–amino acid cytoplasmic domain (RRLKKG), is still localized at microvilli (Carpén et al., 1992; Pavalko et al., 1995), and ICAM-1 was reported to associate with ezrin (Helander et al., 1996). Both α4 integrin and P-selectin glycoprotein ligand-1 have positively charged amino acid clusters of ∼30 amino acids in their juxta-membrane region.
Taken together, for integral membrane proteins, the positively charged amino acid cluster in the juxta-membrane cytoplasmic domain might be a default signal for ERM binding and/or microvillar localization within cells. When the positively charged amino acid cluster is followed by a negatively charged amino acid cluster, i.e., the net charge of the whole cytoplasmic domain is negative or nearly neutral, the former cluster may be masked by the latter, losing its binding ability to ERM proteins. This hypothesis may explain the binding ability of integral membrane proteins to ERM proteins. However, the ERM-binding or ERM-colocalization ability of some of the truncated or site-directed mutants cannot be explained by this hypothesis. For example, although the isoelectric point of the cytoplasmic domain of E-43/1–64 was calculated as 10.73, this mutant does not bind to moesin (Fig. 3 b). The cytoplasmic domain of E-ICAM-2/RRR:GGA also has a net positive charge, but does not colocalize with moesin (Fig. 9). These findings suggest that the three-dimensional structure in or around the juxta-membrane positively charged amino acid clusters is also important for the specificity of ERM protein–integral membrane protein interaction.
Physiological Relevance of the Occurrence of Multiple Membrane Binding Partners for ERM Proteins
At present, it is not clear how many types of integral membrane proteins function as binding partners for ERM proteins in situ, but judging from the expression and distribution of CD44, CD43, and ICAM-2 in tissues, the occurrence of many other ERM-binding partners can be expected. In cells within tissues, as yet undetermined regulatory mechanisms may determine the combination of ERM proteins and their membrane-binding partners, resulting in the specific expression and distribution of ERM proteins in a cell type-specific manner (Franck et al., 1993; Amieva et al., 1994).
We found previously that at physiological ionic strength the association between CD44 and ERM proteins requires PIP2 in vitro, and that it is regulated by the Rho signaling pathway in vivo, which is thought to generally regulate actin-based cytoskeletal organization (Hirao et al., 1996). However, in marked contrast to CD44, even at physiological ionic strength, CD43 and ICAM-2 bound to ERM proteins in vitro with a relatively high affinity in the absence of PIP2. It is not clear at present whether the binding of ERM proteins to CD43 and ICAM-2 (and to as yet unidentified ERM membrane-binding partners) in vivo is also regulated by the Rho signaling pathway. Although this study was focused on the identification of membrane-binding partners other than CD44 and on their ERM protein-binding sites (positively charged amino acid clusters), the data obtained here would also provide some clues to understand the regulatory mechanism of ERM–membrane interaction in general. For example, the carboxyl terminus–truncated CD44 binds to ERM proteins in vitro even in the absence of PIP2 at physiological ionic strength. Studies are currently underway in our laboratory to clarify the regulatory mechanism of ERM–membrane interaction.
Footnotes
We thank all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. The anti– E-cadherin mAb, ECCD-2, and the plasmid pBATEM2 were gifts from M. Takeichi. S. Yonemura is grateful to Izumi and Takeru Yonemura for encouragement throughout this study.
This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science, and Culture of Japan.
Address all correspondence to Shigenobu Yonemura, Department of Cell Biology, Faculty of Medicine, Kyoto University, Sakyo-ku, Kyoto 606-01, Japan. Tel.: 81-75-753-4386. Fax: 81-75-753-4660. E-mail: yonemura@mfour.med.kyoto-u.ac.jp
1. Abbreviations used in this paper: a.a., amino acids; ECCD-2, anti-E-cadherin mAb; E-ICAM-2, E-cadherin/ICAM-2 chimera; ERM, ezrin/radixin/ moesin; G-PBS, PBS containing 30 mM glycine; GST, glutathione-S-transferase.
References
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129–140. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva MR, Wilgenbuns KK, Furthmayr H. Radixin is a component of hepatocyte microvilli in situ. Exp Cell Res. 1994;210:140–144. doi: 10.1006/excr.1994.1021. [DOI] [PubMed] [Google Scholar]
- Andréoli C, Martin M, Le-Borgne R, Reggio H, Mangeat P. Ezrin has properties to self-associate at the plasma membrane. J Cell Sci. 1994;107:2509–2521. doi: 10.1242/jcs.107.9.2509. [DOI] [PubMed] [Google Scholar]
- Arpin M, Algrain M, Louvard D. Membrane-actin microfilament connections: an increasing diversity of players related to band 4.1. Curr Opin Cell Biol. 1994;6:136–141. doi: 10.1016/0955-0674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- Bennett V. The spectrin-actin junction of erythrocyte membrane skeletons. Biochem Biophys Acta. 1989;988:107–121. doi: 10.1016/0304-4157(89)90006-3. [DOI] [PubMed] [Google Scholar]
- Berlin C, Bargatze RF, Campbell JJ, von Andrian UH, Szabo MC, Hasslen SR, Nelson RD, Berg EL, Erlandsen SL, Butcher EC. α4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell. 1995;80:413–422. doi: 10.1016/0092-8674(95)90491-3. [DOI] [PubMed] [Google Scholar]
- Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells, whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- Berryman M, Gary R, Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyde D, Beckwith J. The role of charged amino acids in the localization of secreted and membrane proteins. Cell. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol. 1983;97:425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Gary R, Berryman M. Soluble ezrin purified from placenta exists as stable monomers and elongated dimers with masked C-terminal ezrin-radixin-moesin association domains. Biochemistry. 1995;34:16830–16837. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- Carpén O, Pallai P, Staunton DE, Springer TA. Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and α-actinin. J Cell Biol. 1992;118:1223–1234. doi: 10.1083/jcb.118.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cohn JA, Mandel LJ. Dephosphorylation of ezrin as an early event in renal microvillar breakdown and anoxic injury. Proc Natl Acad Sci USA. 1995;92:7495–7499. doi: 10.1073/pnas.92.16.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J, Kan YW, Shohet SB, Mohandas N. Molecular cloning of protein 4.1, a major structural element of the human erythrocyte membrane skeleton. Proc Natl Acad Sci USA. 1986;83:9512–9516. doi: 10.1073/pnas.83.24.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens MHE, Stroeken PJM, Rodriguez NF, Erena, van der Valk MA, van Rijthoven EAM, Roos E. Targeted disruption of CD44 in MDAY-D2 lymphosarcoma cells has no effect on subcutaneous growth or metastatic capacity. J Cell Biol. 1995;131:1849–1855. doi: 10.1083/jcb.131.6.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl WR. Production of malignancy in vitro.IV. The mouse fibroblast cultures and changes seen in the living cells. J Natl Cancer Inst. 1943;4:165–212. [Google Scholar]
- Edwards KA, Montague RA, Shepard S, Edgar BA, Erikson RL, Kiehart DP. Identification of Drosophila cytoskeletal proteins by induction of abnormal cell shape in fission yeast. Proc Natl Acad Sci USA. 1994;91:4589–4593. doi: 10.1073/pnas.91.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck Z, Gary R, Bretscher A. Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J Cell Sci. 1993;105:219–231. doi: 10.1242/jcs.105.1.219. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Nell BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Funayama N, Nagafuchi A, Sato N, Sa, Tsukita, Sh, Tsukita Radixin is a novel member of the band 4.1 family. J Cell Biol. 1991;115:1039–1048. doi: 10.1083/jcb.115.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Heterotypic and homotypic association between ezrin and moesin, two putative membrane-cytoskeletal linking proteins. Proc Natl Acad Sci USA. 1993;90:10846–10850. doi: 10.1073/pnas.90.22.10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1 . EMBO (Eur Mol Biol Organ) J. 1989;8:4133–4142. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzel D, Reggio H, Bretscher A, Forte JG, Mangeat P. The secretion-stimulated 80K phosphoprotein of a parietal cells is ezrin, and has properties of a membrane cytoskeletal linker in the induced apical microvilli. EMBO (Eur Mol Biol Organ) J. 1991;10:2363–2373. doi: 10.1002/j.1460-2075.1991.tb07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Lesley J, Hyman R, Ishihara K, Kincade PW. Molecular isoforms of murine CD44 and evidence that the membrane proximal domain is not critical for hyaluronate recognition. J Cell Biol. 1992;119:1711–1719. doi: 10.1083/jcb.119.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander TS, Carpén O, Turunen O, Kovanen PE, Vaheri A, Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- Henry MD, Gonzalez C, Agosti, Solomon F. Molecular dissection of radixin: distinct and interdependent functions of the amino- and carboxyl-terminal domains. J Cell Biol. 1995;129:1007–1022. doi: 10.1083/jcb.129.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Sh, Tsukita, Sa, Tsukita Regulation mechanism of ERM protein/ plasma membrane association: possible involvement of phosphatidylinositol turnover and rho-dependent signaling pathway. J Cell Biol. 1996;135:37–52. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen N, Barclay AN, Willis AC, Williams AF. The sequence of rat leukosialin (W3/13 antigen) reveals a molecule with O-linked glycosylation of one third of its extracellular amino acids. EMBO (Eur Mol Biol Organ) J. 1987;6:4029–4034. doi: 10.1002/j.1460-2075.1987.tb02747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankes W, Furthmayr H. Moesin: a member of the protein 4.1-talin-ezrin family of proteins. Proc Natl Acad Sci USA. 1991;88:8297–8301. doi: 10.1073/pnas.88.19.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankes W, Griesmacher A, Grunwald J, Schwarts-Albiez R, Keller R. A heparin-binding protein involved in inhibition of smooth-muscle cell proliferation. Biochem J. 1988;251:831–842. doi: 10.1042/bj2510831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJG, Esch F, Furthmayr H, Hall A. Rho- and Rac- dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Andréoli C, Sahuquet A, Montcourrier P, Algrain M, Mangeat P. Ezrin NH2-terminal domain inhibits the cell extension activity of the COOH-terminal domain. J Cell Biol. 1995;128:1081–1093. doi: 10.1083/jcb.128.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL, Patel KD, Bruehl RE, Fugang L, Johnson DA, Lichenstein HS, Cummings RD, Bainton DF, McEver RP. P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J Cell Biol. 1995;128:661–671. doi: 10.1083/jcb.128.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A, Shirayoshi Y, Okazaki K, Yasuda K, Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature. 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Amieva MR, Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem. 1995;270:31377–31385. doi: 10.1074/jbc.270.52.31377. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Ozawa H. Immunolocalization of CD44 and ERM family in osteoblasts and osteoclasts in mouse tibiae. J Bone Miner Res. 1996;11:1715–1722. doi: 10.1002/jbmr.5650111115. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Pakkanen R, Hedman K, Turunen O, Wahlstrom T, Vaheri A. Microvillus-specific Mr 75,000 plasma membrane protein of human choriocarcinoma cells. J Histochem Cytochem. 1987;135:809–816. doi: 10.1177/35.8.3298422. [DOI] [PubMed] [Google Scholar]
- Pavalko FM, Walker DM, Graham L, Goheen M, Doerschuk CM, Kansas GS. The cytoplasmic domain of L-selectin interacts with cytoskeletal proteins via α-actinin: receptor positioning in microvilli does not require interaction with α-actinin. J Cell Biol. 1995;129:1155–1164. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestonjamasp K, Amieva MR, Strassel CP, Bauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker LJ, Warnock RA, Burns AR, Doerschuk CM, Berg EL, Butcher EC. The neutrophil selectin LECAM-1 presents carbohydrate ligands to the vascular selectins ELAM-1 and GMP-140. Cell. 1991;66:921–933. doi: 10.1016/0092-8674(91)90438-5. [DOI] [PubMed] [Google Scholar]
- Rees DJG, Ades SE, Singer SJ, Hynes RO. Sequence and domain structures of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Sato N, Yonemura S, Obinata T, Sa, Tsukita, Sh, Tsukita Radixin, a barbed end-capping actin-modulating protein, is concentrated at the cleavage furrow during cytokinesis. J Cell Biol. 1991;113:321–330. doi: 10.1083/jcb.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Funayama N, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita A gene family consisting of ezrin, radixin, and moesin. Its specific localization at actin filament/plasma membrane association sites. J Cell Sci. 1992;103:131–143. doi: 10.1242/jcs.103.1.131. [DOI] [PubMed] [Google Scholar]
- Shirayoshi Y, Nose A, Iwasaki K, Takeichi M. N-linked oligosaccharides are not involved in the function of a cell-cell binding glycoprotein E-cadherin. Cell Struct Funct. 1986;11:245–252. doi: 10.1247/csf.11.245. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene (Amst) 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Kawashima A, Nagafuchi A, Sh, Tsukita Structural diversity of band 4.1 superfamily members. J Cell Sci. 1994a;107:1921–1928. doi: 10.1242/jcs.107.7.1921. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J Cell Biol. 1994b;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-ezrin-radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppresser. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Hieda Y, Tsukita Sh. A new 82 kD-barbed end capping protein (radixin) localized in the cell-to-cell adherens junction: purification and characterization. J Cell Biol. 1989;108:2356–2382. doi: 10.1083/jcb.108.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sh, Tsukita Sa, Nagafuchi A, Yonemura S. Molecular linkage between cadherins and actin filaments in cell-to-cell adherens junctions. Curr Opin Cell Biol. 1992;4:834–839. doi: 10.1016/0955-0674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa,, Oishi K, Sato N, Sagara J, Kawai A, Tsukita Sh. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci (TIBS) 1997a;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997b;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- Turunen O, Wahlstrom T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen O, Winqvist R, Pakkane R, Grzeschik KH, Wahlstrom T, Vaheri A. Cytovillin, a microvillar Mr 75,000 protein. cDNA sequence, prokaryotic expression, and chromosomal localization. J Biol Chem. 1989;264:16727–16732. [PubMed] [Google Scholar]
- Urushidani T, Hanzel DK, Forte JG. Characterization of an 80-kDa phosphoprotein involved in parietal cell stimulation. Am J Physiol. 1989;256:G1070–G1081. doi: 10.1152/ajpgi.1989.256.6.G1070. [DOI] [PubMed] [Google Scholar]
- von Andrian UH, Hasslen SR, Nelson RD, Erlandsen SL, Butcher EC. A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell. 1995;82:989–999. doi: 10.1016/0092-8674(95)90278-3. [DOI] [PubMed] [Google Scholar]
- Xu H, Tong IL, De Fougerolles AR, Springer TA. Isolation, characterization, and expression of mouse ICAM-2 complementary and genomic DNA. J Immunol. 1992;149:2650–2655. [PubMed] [Google Scholar]
- Yonemura S, Nagafuchi A, Sato N, Tsukita Sh. Concentration of an integral membrane protein, CD43(leukosialin, sialophorin), in the cleavage furrow through the interaction of its cytoplasmic domain with actin-based cytoskeletons. J Cell Biol. 1993;120:437–449. doi: 10.1083/jcb.120.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]