A Class VI Unconventional Myosin Is Associated with a Homologue of a Microtubule-binding Protein, Cytoplasmic Linker Protein–170, in Neurons and at the Posterior Pole of Drosophila Embryos (original) (raw)
Abstract
Abstract. Coordination of cellular organization requires the interaction of the cytoskeletal filament systems. Recently, several lines of investigation have suggested that transport of cellular components along both microtubules and actin filaments is important for cellular organization and function. We report here on molecules that may mediate coordination between the actin and microtubule cytoskeletons. We have identified a 195-kD protein that coimmunoprecipitates with a class VI myosin, Drosophila 95F unconventional myosin. Cloning and sequencing of the gene encoding the 195-kD protein reveals that it is the first homologue identified of cytoplasmic linker protein (CLIP)–170, a protein that links endocytic vesicles to microtubules. We have named this protein D-CLIP-190 (the predicted molecular mass is 189 kD) based on its similarity to CLIP-170 and its ability to cosediment with microtubules. The similarity between D-CLIP-190 and CLIP-170 extends throughout the length of the proteins, and they have a number of predicted sequence and structural features in common. 95F myosin and D-CLIP-190 are coexpressed in a number of tissues during embryogenesis in Drosophila. In the axonal processes of neurons, they are colocalized in the same particulate structures, which resemble vesicles. They are also colocalized at the posterior pole of the early embryo, and this localization is dependent on the actin cytoskeleton. The association of a myosin and a homologue of a microtubule-binding protein in the nervous system and at the posterior pole, where both microtubule and actin-dependent processes are known to be important, leads us to speculate that these two proteins may functionally link the actin and microtubule cytoskeletons.
Global organization of the cell and the coordination of its physiology requires interaction between different cytoskeletal systems. During interphase, a typical eukaryotic cell has microtubules emanating from the centrosome located near the nucleus, which extend to the periphery of the cell, presumably interacting with the cortical actin filament meshwork. Microtubules during interphase are thought to be mainly required for the organization of the membrane systems (e.g., vesicular traffic and organelle movement). The actin-rich cortex is important for maintaining cell shape and for cellular movement.
There is increasing evidence of coordination between the actin and the microtubule cytoskeletons (Langford, 1995; Koonce, 1996). Data from a number of systems suggests that many cell types use a combination of microtubule and actin filament–based transport in vesicle and organelle trafficking. It is well established that microtubules are required for long distance transport of cellular components. In contrast, the actin cytoskeleton is thought to be required for more local traffic. The best evidence for transport along both cytoskeletal systems is in neurons. Vesicles appear to be transported along actin filaments in mammalian growth cones (Evans and Bridgman, 1995). Furthermore, gelsolin, which promotes depolymerization of actin filaments, has been shown to inhibit fast axonal transport in this system (Brady et al., 1984). In extruded squid axoplasm, Kuznetsov et al. (1992) observed what appeared to be the same vesicle moving along microtubules and then, subsequently, along microfilaments. Inhibitor studies provide evidence that mitochondria can move along both actin filaments and microtubules in neurons in vivo (Morris and Hollenbeck, 1995). These data support the idea that actin filament and microtubule-based transport cooperate to achieve proper organization of cellular components.
The same phenomenon may be occurring in other cell types. In yeast, the mutant phenotype of the MYO2 gene, which encodes an unconventional myosin, is suppressed by overexpression of a kinesin-related protein. These two proteins are colocalized in regions of active growth where a polarized arrangement of actin plays an important role (Lillie and Brown, 1992, 1994). Microtubules are not normally required for this growth. Thus, the basis for suppression is not completely understood. However, the phenotypic suppression suggests that perhaps microtubule-based transport can substitute for actin filament–based transport, under some conditions. In polarized epithelial cells, Fath et al. (1994) have isolated a population of vesicles containing both myosin and microtubule motors. They speculate that proper transport of vesicles relies on both microtubule and actin filament–based transport.
Previously, it has been shown that a class VI unconventional myosin, the Drosophila 95F unconventional myosin, transports particles along actin filaments during the syncytial blastoderm stage of Drosophila embryonic development (Mermall et al., 1994). 95F myosin activity is required for normal embryonic development (Mermall and Miller, 1995). 95F myosin is also associated with particulate structures in other cells of the embryo later in development where it may also be involved in actin-based transport. To investigate further the transport catalyzed by 95F myosin, we have begun studies to identify proteins associated with 95F myosin that might be cargoes or regulators. In this work, we have identified a protein that coimmunoprecipitates with 95F myosin. Sequence analysis reveals that this protein is the Drosophila homologue of cytoplasmic linker protein (CLIP)1–170. CLIP-170 is believed to function as a linker between endocytic vesicles and microtubules (Pierre et al., 1992). We have named this associated protein D-CLIP-190. Colocalization of 95F myosin and D-CLIP-190 at the subcellular level at several times in development and in cultured embryonic cells provides support for the in vivo association of these two proteins. The association of a myosin and a homologue of a microtubule-binding protein suggests that these two proteins may act to coordinate the interaction between actin filaments and microtubules.
Materials and Methods
Immunoprecipitations
Immunoprecipitations (IPs) were performed according to Govind et al. (1992) with minor modifications. The protocol is summarized below. Antibodies (900 μl of monoclonal supernatant or 50 μg of affinity-purified antibody) were incubated overnight at 4°C with 50 μl of protein A beads (Bio-Rad Laboratories, Hercules, CA). The beads were washed twice in 500 μl of IP buffer. Whole embryo extracts were made from 0–3-h embryos (25°C) in IP buffer (150 mM NaCl, 20 mM Hepes, pH 7.5, 250 mM sucrose, 1 mM Na molybdate, 1% NP-40) using 1 ml of embryos (∼1 g) in 4 ml of buffer. We routinely used IP buffer containing 1% NP-40 because more protein is immunoprecipitated than when 0.05% NP-40 is used; the identical profile of proteins is immunoprecipitated under either condition. The embryo homogenate was filtered through glass wool and spun at 5,000 g. The supernatant was diluted 1:3 in IP buffer and incubated overnight at 4°C with antibody protein A–beads. After this incubation the beads were thoroughly washed with IP buffer by pelleting at 100 g and resuspending in 500 μl of IP buffer five times. The beads were suspended in SDS-loading buffer and boiled. Protein samples were analyzed by SDS-PAGE.
Immunoprecipitations from later embryos in which the nervous system is developing were also performed. Embryos were collected for 4 h at 25°C and then aged at 18°C for 16 h. Immunoprecipitations were then performed as described above. The same profile of proteins was immunoprecipitated from later embryos as early embryos. Immunoprecipitations from early embryos were generally used because more of the 195-kD protein is immunoprecipitated. Immunoprecipitations were also performed with different salt conditions with qualitatively similar results. When higher than 150 mM NaCl was used in IPs, the amount of the 195-, 190-, and 170-kD proteins was reduced relative to the level of 95F myosin, whereas decreased concentrations of salt increased the amount of the 195-kD protein present (data not shown).
Anti-95F IPs were performed using monoclonal 3C7 culture supernatant (Fig. 1) or affinity-purified rabbit polyclonal antibody (data not shown). Immunoprecipitations with affinity-purified rabbit α195-kD (α6D1a) antibody (see Results; Fig. 1), mouse antisera raised against gel-isolated immunoprecipitated 195-kD protein (data not shown), or mouse antisera raised against 6D1a–glutathione-S-transferase (GST) fusion protein (data not shown) all immunoprecipitate the same profile of proteins. Control immunoprecipitations were performed in parallel, in which either protein A beads, or antibody or embryo extract were not included (data not shown). A dorsal monoclonal antibody generously supplied by R. Steward and affinity-purified rabbit HRP antibody (Sigma Chemical Co., St. Louis, MO) were also used as controls in immunoprecipitations (Fig. 1).
Figure 1.
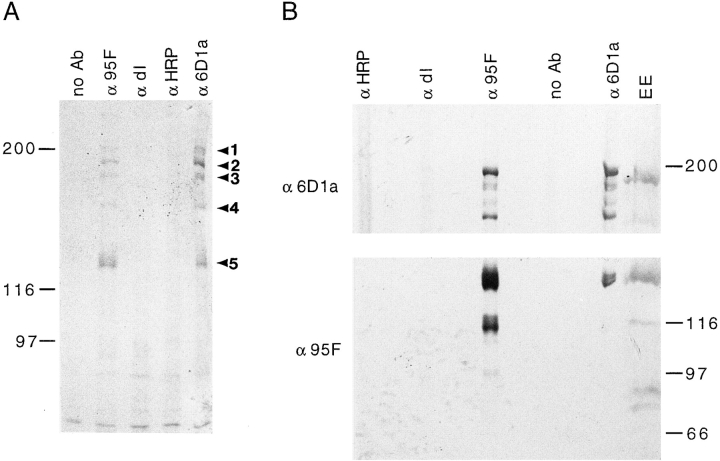
95F myosin and the 195-kD protein coimmunoprecipitate. (A) A Coomassie-stained gel of proteins present in IPs from 0–3-h embryos using the following conditions: no antibody included in IPs (no Ab), 95F unconventional myosin monoclonal antibody (α95F), dorsal antibody (αdl), HRP antibody (αHRP), and affinity-purified rabbit polyclonal α6D1a antibody raised against 195-kD cloned fusion protein (α6D1a). Arrowheads indicate the proteins described in the text and are labeled 1–5. A 200-kD protein (band 1), a 195-kD protein (band 2) and its breakdown products (the predominant proteins are 190 kD [band _3_] and 170 kD [band _4_]; an ∼180-kD protein is sometimes present) and 95F myosin (band 5) are immunoprecipitated with α95F myosin antibody and α6D1a antibody raised against 195-kD cloned fusion protein. These proteins were not immunoprecipitated with control antibodies (αHRP, αdl). (B) Identical samples to those shown in Fig. 1 A were transferred to nitrocellulose and the resulting blot was cut in half. The upper half was probed with mouse α6D1a antiserum raised against 195-kD cloned fusion protein (α6D1a) and the lower half with α95F myosin antibody (α95F) to confirm the identity of the proteins. 0–3-h embryo extract (EE) is also shown.
Western Analysis
Proteins were separated on 6 or 8% polyacrylamide gels. Afterwards, SDS-PAGE proteins were transferred to nitrocellulose in glycine buffer (150 mM glycine, 0.02% SDS, 25% methanol) overnight at 50 mA and 2 h at 320 mA. To determine whether similar amounts of protein were present in each lane, the blots were stained with Ponceau S red. After blocking with 1% BSA in TBST (TBS [10 mM Tris, pH 8, 150 mM NaCl]/ 0.05% Tween-20; Sigma Chemical Co.), blots were incubated for 1 h–overnight with primary antibody. Blots were rinsed three times and washed four times for 15 min in TBST. Goat αmouse secondary antibody conjugated to alkaline phosphatase (Boehringer Mannheim Biochemicals, Indianapolis, IN) was used at 1:5,000 to detect proteins. Alternatively, sheep αmouse secondary antibody conjugated to HRP (Amersham Corp., Arlington Heights, IL) was used at 1:2,000, and detection was performed using enhanced chemiluminescence (SuperSignal Substrate; Pierce, Rockford, IL). High molecular mass standards were from Bio-Rad (200, 116.2, 97.4, and 66.2 kD). The mouse α6D1a antisera was used at 1:2,500. The 95F myosin monoclonal antibody (3C7) culture supernatant was diluted 1:5. α-tubulin antibody (DM1A; gift of D. Kellogg and B. Alberts, University of California at San Francisco, San Francisco, CA) was diluted 1:10.
Cloning the Gene Encoding 195-kD Protein
Antisera to the 195-kD protein was obtained by injecting mice with protein isolated by immunoprecipitation. These IPs were subjected to SDS-PAGE, and the appropriate bands were isolated after Coomassie staining. The gel slices were processed according to Amero et al. (1987). Approximately 5–10 μg of each of the proteins was injected at monthly intervals into individual mice using standard protocols.
To obtain cDNA clones encoding the 195-kD protein, a Drosophila ovarian cDNA λgt11 expression library (Steinhaur et al., 1989) was screened according to standard methods (Synder et al., 1987), using anti– 195-kD antisera from mice. Nine clones were isolated and plaque purified. Southern analysis revealed that clones 8A2a and 6D1a cross-hybridize. Drosophila 0–4- and 8–12-h embryo cDNA libraries (Brown and Kafatos, 1988) were screened with the 6D1a clone. Cloning techniques were used according to Sambrook et al. (1989). Standard Southern and Northern analysis was used (Southern, 1975). Random primed probes were used for Southern and Northern experiments (Feinberg and Vogelstein, 1983).
Nitrocellulose Antibody Affinity Purification
The 6D1a–GST fusion protein or α95F myosin immunoprecipitation reactions were subjected to SDS-PAGE and transferred to nitrocellulose. After staining the nitrocellulose with Ponceau S to visualize the protein, the appropriate band was cut out and incubated with diluted antisera overnight at 4°C. The nitrocellulose strip was washed four times with TBST and rinsed one time with 150 mM NaCl, and the antibody was eluted sequentially with 0.1 M glycine, pH 2.9, 2.4, and 2.2. The elutions were neutralized with 2 M Tris, pH 8.4.
Antiserum raised against gel-purified 195-kD protein was affinity purified by absorption to protein (either the 195- or 170-kD protein isolated by immunoprecipitation) immobilized on nitrocellulose strips. Both the α195-kD antibodies and the α170-kD affinity-purified antibodies react with the 195-, 190-, and 170-kD proteins on immunoblots of 0–3 h embryo extract or α95F myosin IPs, suggesting that these proteins are related (data not shown).
Isolation of 6D1a–GST Fusion Protein
The 6D1a cDNA subcloned into pGEX-1λT (Pharmacia Biotech., Piscataway, NJ; Smith and Johnson, 1988) was freshly transformed into DH5αF′; colonies were picked and restreaked before fusion protein isolation. An overnight culture was diluted 1:10 in LB-Amp (50 μg/ml) and grown at 37°C for 1 h. IPTG was added to a final concentration of 1.25 mM, and the culture was further incubated at 37°C for 2.5 h. After harvesting, the cells were resuspended in 1/6 vol of TEGK buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 10% glycerol, 50 mM KCl, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A). Lysozyme was added to a final concentration of 0.1 mg/ml, and the cell suspension was incubated at 30°C for 15 min. The lysozyme treatment was repeated once. Cells were further lysed by sonication. DTT was added to 15 mM. Cell debris was pelleted at 9,000 g for 30 min. Glutathione beads were rehydrated and washed twice with TEGK buffer. 50 ml of cell supernatant was incubated with 1 ml of beads overnight at 4°C. The beads were washed three times with 10 ml of TEGK buffer containing 1 mM DTT. The GST fusion protein was eluted by incubating the beads on ice for 10 min with 5 mM glutathione in TEGK buffer. This procedure was repeated three to four times. Elutions were concentrated using a centricon-30 (Amicon Corp., Danvers, MA).
Antibodies
Antisera raised against D-CLIP-190 was obtained by injecting mice with 5–10 μg of the 6D1a–GST fusion protein at approximately monthly intervals using standard protocols. Rabbit polyclonal antisera was prepared by Cocalico Biologicals, Inc. (Reamstown, PA); rabbits were immunized with 100 μg of the 6D1a–GST fusion protein and boosted with 50 μg of the fusion protein. Rabbit antibodies were affinity purified against a soluble 6D1a-maltose–binding protein fusion (MBP). To make this fusion protein, the 6D1a cDNA was subcloned into the EcoRI site of pMal-c2 vector (New England Biolabs Inc., Beverly, MA). Bacterial cells transformed with this construct and expressing the 6D1a–MBP fusion protein were lysed, and the resulting supernatant was incubated with amylose resin according to the manufacturer's protocol (New England Biolabs). The 6D1a–MBP column was preeluted with 0.1 M glycine, pH 2.4, neutralized with 2 M Tris, pH 8.8, and washed with 1× PBS. The rabbit α6D1a–GST sera diluted in PBS was incubated overnight with the resin. The resin was washed with 1× PBS and the α6D1a antibody was eluted with 0.1 M glycine, pH 2.4. The antibody solution was neutralized with 2 M Tris, pH 8.8, dialyzed against 1× PBS, and concentrated using a centricon-30 (Amicon Corp.).
Sequencing
Both strands of the 10A cDNA clone were completely sequenced by making an M13 library and randomly sequencing to at least twofold coverage (Wilson et al., 1994). Single-strand templates were prepared and sequenced using a Prism fluorescent sequencing system (Perkin-Elmer Corp., Norwalk, CT). Sequence data was recorded and analyzed using an ABI 373–automated fluorescent DNA sequencer (Smith et al., 1986). The 10A cDNA encodes the last 4 kb of the transcript. The remainder of the cDNA sequence was obtained from clones 10C, 11A, and 11C by primer walking. Double-strand templates of 10C, 11A, and 11C cDNA clones were prepared and sequenced by the method of Sanger et al. (1977) using the Sequenase system (United States Biochemical Corp., Cleveland, OH) and custom primers (GIBCO BRL, Gaithersburg, MD) spaced 200–300 nucleotides apart. The sequence data from all three clones was compiled using the DNAStar program, SeqMan, to give at least twofold coverage on each strand. The last 1 kb of sequence of both the 10C and 11A clones was not determined as it overlaps that of the 10A clone.
Sequence databases were searched using BLASTX and BLASTP (Altschul et al., 1990; Gish and States, 1993). DNAStar programs EditSeq, MegAlign, and Align were used for sequence analysis. Genetics Computer Group (Madison, WI) program Bestfit was used to compare CLIP-170 to D-CLIP-190 (Devereux et al., 1984). Analysis of the α-helical coiled-coil regions was done using an algorithm developed by Lupas et al. (1991).
Microtubule Cosedimentation
Determination of the ability of D-CLIP-190 to cosediment with microtubules was performed according to Saxton (1994) with some modifications. The protocol is summarized below. Whole embryo extracts were made from 0–2-h embryos (25°C) in extraction buffer (0.1 M Pipes, pH 6.9, 0.9 M glycerol, 5 mM EGTA, 2.5 mM MgSO4). The extract was clarified by centrifugation at 15,000 g for 40 min at 4°C. The resulting supernatant was centrifuged at 50,000 g for 30 min at 4°C. To induce microtubule polymerization, 0.3 mM GTP and 20 μM taxol were added to this high speed supernatant. After agitation at room temperature for 20 min to allow microtubule polymerization and binding of microtubule-associated proteins, the microtubules were sedimented through a sucrose cushion (20% sucrose and 10 μM taxol in extraction buffer) by centrifugation at 23,000 g for 30 min at 4°C. The resulting supernatant and pellet were analyzed by SDS-PAGE and Western analysis. Embryo extract to which GTP and taxol had not been added was processed in parallel as a control.
Immunolocalization in Embryos
Immunolocalization was performed using a standard fluorescence microscope (Nikon, Melville, NY) or confocal imaging system (1024; Bio-Rad). Embryos were fixed with 6% formaldehyde in PEM (0.1 M Pipes, pH 6.9, 1 mM EGTA, and 1 mM MgCl2) with an equal volume of heptane. The vitelline membrane was removed with methanol/50 mM EGTA. Embryos were blocked with 1% BSA in PBST (1× PBS/0.05% tween 20) for 1–3 h. Embryos were incubated in α95F myosin monoclonal culture supernatant diluted 1:6 and α6D1a (αD-CLIP-190) rabbit affinity-purified antibody at 2 μg/ml. After overnight incubation at 4°C with primary antibody in 1% BSA in PBST, the samples were incubated for 1 h with fluorescent-labeled donkey αmouse and donkey αrabbit secondary antibodies (Chemicon International Inc., Waltham, MA). Embryos were stained with DAPI at 1 μg/ ml and mounted in 90% glycerol/PBS with 1 mg/ml _p_-phenylenediamine.
Cytochalasin D and Colchicine Treatment
Embryos were collected for 45 min, rinsed with Triton–salt buffer (0.4% NaCl, 0.1% Triton X-100), and dechorionated with 50% bleach. After thorough washing with distilled water and Triton–salt buffer, embryos were rinsed twice with 0.9% NaCl. Embryos were then treated with the desired concentration of drug in 2 ml 0.9% NaCl/2 ml octane. The NaCl solution was removed, and embryos were fixed in 12% formaldehyde/ PEM for 18–20 min. Embryos were then stored at 4°C or stained as described above.
When embryos were treated for 20–30 min with 10 μg/ml of cytochalasin D, the actin cytoskeleton appeared to be substantially depolymerized. Treated embryos stained with phalloidin, which binds filamentous actin, lacked normal actin structures found in untreated embryos such as the actin meshwork present in the cortex of early embryos and actin caps in later embryos. The nuclei were disorganized in treated embryos as expected because the axial expansion or spreading of nuclei is an actin-dependent process (von Dassow and Schubiger, 1994). When embryos were treated for 20–30 min with 10 or 20 μg/ml of colchicine, they lacked normal microtubule structures, such as the filamentous microtubule meshwork in the cortex of early embryos and normal spindles during mitosis. Embryos treated with cytochalasin D or colchicine under the same conditions were stained with αD-CLIP-190 and/or α95F myosin antibodies; embryos were also stained with DAPI to visualize the nuclei and assess whether the drugs were effective.
Results
Immunoprecipitation Identifies Several Proteins in Addition to 95F Myosin
To identify 95F myosin–associated proteins, we used α95F myosin antibodies in IPs of extracts from 0–3-h embryos or stage 14–17 embryos that are in the process of nervous system development (data not shown). 95F myosin (band 5) is immunoprecipitated as expected; in addition, several proteins of lesser abundance coimmunoprecipitate (Fig. 1, α95F myosin). The most prominent of these proteins are ∼200 kD (band 1), 195 kD (band 2), 190 kD (band 3), and 170 kD (band 4). The coimmunoprecipitated proteins are not detected in controls (Fig. 1, no antibody, αdorsal antibody, and αHRP antibody). The same set of proteins is also immunoprecipitated from later embryos (data not shown).
Antibodies to the 195-kD protein were raised against SDS gel–isolated protein. This antiserum reacts with the appropriately sized protein (band 2) on immunoblots of 0–3-h embryo extracts and immunoprecipitations performed with α95F myosin antibody (α95F myosin IPs) (Fig. 2 A). This antiserum also recognizes the 190- (band 3) and 170-kD (band 4) proteins (also see Methods and Materials). The amounts of the 190- and 170-kD proteins vary depending on the conditions under which the extract is prepared and in whole embryo extracts often only the 195-kD protein is detected. Thus, it appears these proteins (band 3 and 4) are breakdown products of the 195-kD protein (band 2). The antisera raised against the 195-kD protein also recognizes the 200-kD protein (band 1). This reaction is probably due to contamination of the 195-kD antigen with the 200-kD protein when isolating the protein from the gel rather than immunological relatedness (see below).
Figure 2.
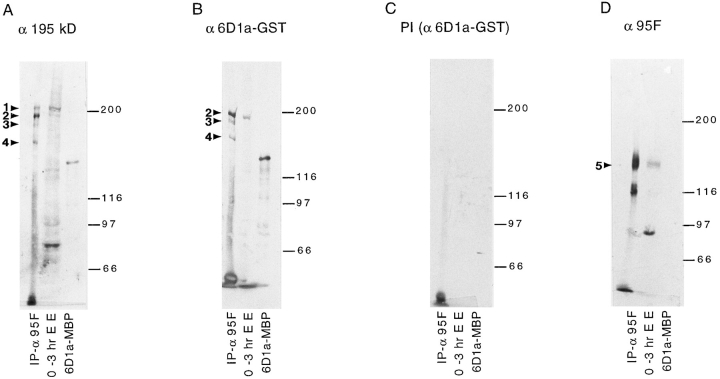
The 6D1a clone isolated using the α195-kD protein antibody corresponds to the gene that encodes the 195-kD protein. Duplicate immunoblots of identical samples are shown in each panel. On each blot are IPs from 0–3-h embryos using α95F myosin antibody (IP-α95F), 0–3-h embryo extract (0–3 hr E E) and bacterial extracts containing the 6D1a-maltose–binding protein fusion protein (6D1a– MBP). (A) Antiserum from a mouse immunized with the 195-kD protein gel purified after immunoprecipitation (α195 kD). (B) Antiserum from a mouse immunized with the 6D1a–GST fusion protein (α6D1a–GST). (C) Preimmune sera from mice immunized with the 6D1a–GST fusion protein PI(α6D1a-GST). (D) Anti–95F myosin monoclonal antibody (α95F). The proteins are labeled with arrowheads and numbers as in Fig. 1.
Cloning of the Gene Encoding the 195-kD Protein
To determine whether the 195-kD protein was associated with 95F myosin in vivo, we had hoped to use an antibody specific for the 195-kD protein to determine whether the expression pattern and distribution of the 195-kD protein overlapped that of 95F myosin. Unfortunately, the antibody raised against gel-isolated 195-kD protein was not useful for immunolocalization. To provide a source of antigen and begin molecular characterization, we cloned the gene encoding the 195-kD protein by screening an ovary expression library with the α195-kD protein antisera (Synder et al., 1987; Steinhaur et al., 1989). Several clones were isolated, including two (ov 6D1a and ov 8A2a) that overlapped based on Southern analysis. To determine which of the several clones might encode the 195-kD protein, but not other proteins recognized by the α195 kD serum, GST fusion proteins made with cDNAs obtained from the screen were used to affinity purify antibodies from the mouse polyclonal serum raised against the 195-kD protein and used to probe Western blots. These experiments suggested that the 6D1a clone encodes part of the 195-, 190-, and 170-kD proteins, but not the 200-kD protein.
We raised antibodies in mice against the 6D1a–GST fusion protein and used them to confirm that the 6D1a cDNA clone does indeed encode the 195-kD myosin–associated protein. Duplicate immunoblots of 0–3-h embryo extract (0–3 h EE), α95F myosin IPs (IP-α95F), and 6D1a-maltose–binding protein fusion (6D1a-MBP) were probed with several different antibodies/antisera. Anti-6D1a antiserum (α6D1a–GST; Fig. 2 B) recognizes the 6D1a–MBP fusion protein indicating that the serum contains antibodies that react with the 6D1a protein as expected. In addition, the serum recognizes a protein of 195 kD (band 2) in 0–3-h embryo extract and the 195-kD protein (band 2) and its breakdown products (band 3 and 4) in α95F myosin IPs. The pattern of bands (2–4) is identical to that seen when the α195-kD gel-isolated protein antiserum (α195 kD; Fig. 2 A) is used to probe a duplicate blot, except that the α195-kD protein antiserum recognizes the 200-kD protein (band 1) whereas α6D1a sera does not. The preimmune sera from the mouse in which this antibody was raised was used as a control (Fig. 2 C; PI(α6D1a-GST)). Thus, the 6D1a clone encodes at least part of the 195-kD protein. When α95F myosin monoclonal antibody is used to probe immunoblots of the same samples, no cross-reactivity is observed with the 200-kD protein, the 195-kD protein, or the 6D1a–MBP fusion protein (α95F; Fig. 2 D). This result suggests that the 195-kD protein coimmunoprecipitates with the 95F myosin antibody because of its association with 95F myosin and not because of cross-reactivity of the 95F myosin antibody.
Immunoprecipitation with Antibodies Raised against the 195-kD Protein Coimmunoprecipitate 95F Myosin
To determine if 95F myosin could be coimmunoprecipitated with the α195-kD protein antibody, affinity-purified rabbit antibody raised against the 195-kD cloned fusion protein (6D1a–GST; Fig. 1, α6D1a) or mouse polyclonal antisera raised against the 195-kD protein isolated by IP (data not shown) were used in IPs from early embryos. The proteins present in these IPs are essentially identical to those immunoprecipitated with α95F myosin antibodies. The 195-kD protein (band 2) and its breakdown products (band 3 and 4) are immunoprecipitated as expected. In addition, 95F myosin (band 5) is coimmunoprecipitated. Neither 95F myosin nor the 195-kD protein are immunoprecipitated in controls (Fig. 1, no antibody, αdorsal antibody, and αHRP antibody). The identities of the proteins in the Coomassie-stained gel (Fig. 1 A) were confirmed by immunoblot (Fig. 1 B). Thus, IPs with α95F myosin antibody and α195-kD protein antibody show that these two proteins are associated in embryo extracts. The 200-kD protein (band 1) seen previously in α95F myosin IPs is also present in α195-kD protein immunoprecipitations.
Sequence Analysis of cDNAs Encoding the 195-kD Protein
Based on Northern analysis with the 6D1a cDNA clone (2.5 kb), the transcript that encodes the 195-kD protein is expected to be ∼6.1 kb (data not shown). To obtain cDNA clones encoding the entire transcript, we screened Drosophila 0–4- and 8–12-h embryonic cDNA libraries (Brown and Kafatos, 1988) with the ov 6D1a cDNA clone (Fig. 3 A). Because a full-length cDNA was not obtained, four overlapping cDNA clones, all from the 8–12-h cDNA library, were sequenced using a combination of random automated fluorescent DNA sequencing and primer walking using standard dideoxy sequencing methods (Fig. 3 A; Materials and Methods).
Figure 3.

Sequence analysis of the 195-kD protein. (A) cDNAs encoding the 195-kD protein that were sequenced are depicted in schematic form. The ov 6D1a cDNA was isolated from an ovary expression library, and the partial cDNAs were isolated from an 8–12-h cDNA library. The Em 10C cDNA appears to extend to the 5′ end of the transcript whereas the Em 10A cDNA extends to the poly (A) tail. The sequence data are available from GenBank/ EMBL/DDBJ under the accession number AF041382. (B) An alignment of the entire amino acid sequence of D-CLIP-190 (uppercase) and CLIP-170 (lowercase) is shown. Identities (vertical lines), conservative substitutions (two dots), and less conservative substitutions (one dot) are determined by the Genetic Computer Group program Bestfit (Devereux et al., 1984). The first 48 amino acids of D-CLIP-190 do not align with CLIP-170. The amino terminus contains two repeats, rep1 and rep2, (black lines) that have been demonstrated to mediate binding of CLIP-170 to microtubules. The central region of both proteins is predicted to be an extended region of coiled coil (arrows indicate start and end points; Lupas et al., 1991). The predicted coiled-coil domain of D-CLIP-190 is interrupted after ∼100 amino acids by four prolines (asterisks), which are thought to disrupt α helices. The carboxy-terminal domain has two conserved sequences: a metal-binding motif (shaded) and a cysteine-rich sequence of 23 amino acids that does not conform to any previously recognized motif (dotted line).
The composite cDNA is 5,999 bases in length with a single long open reading frame (ORF) predicted to encode a protein of 1,690 amino acids. Because the largest transcript detected on Northern blots is ∼6.1 kb, this composite cDNA is likely to be full length. The calculated molecular mass of this protein is 189 kD, which is in good agreement with the size estimated from SDS polyacrylamide gels. The first ATG of this ORF is located at nucleotide 633 and is preceded by the sequence AAAA, which matches the Drosophila translational initiation consensus sequence (C/A, A, A, C/A; Cavener, 1987). After the large ORF is a 294 nucleotide 3′ untranslated region. At the end of the 10A cDNA are 16 adenine residues; 23 nucleotides 5′ of this stretch of adenines is the sequence AAUAAA, which is identical to the most commonly observed polyadenylation signal (Proudfoot and Brownlee, 1982). Thus, the composite cDNA extends to the poly(A) tail.
The 195-kD Myosin–associated Protein Is a CLIP-170 Homologue
When the sequence databases GenBank/EMBL/DDBJ, PDB, Swiss Prot, and PIR were searched using BLASTP (Altschul et al., 1990; Gish and States, 1993), the proteins that have the most significant similarity to the 195-kD protein are CLIP-170 (BLASTP score of 262 and a probability score of 6.1 × 10−106) and restin (BLASTP score of 262 and a probability score of 1.4 × 10−105). CLIP-170 was identified in HeLa cells as a protein that links endocytic vesicles to microtubules (Pierre et al., 1992). Restin, which was identified in human peripheral blood monocytes, has two isoforms, one identical to CLIP-170 and the other that differs from CLIP-170 by an insert of 35 amino acids (Bilbe et al., 1992). A search of the yeast sequence database with either CLIP-170 or the 195-kD protein did not reveal any proteins with such a high level of similarity at the amino acid level.
An alignment of the sequence of the 195-kD protein and CLIP-170 (Fig. 3) reveals that with the exception of 48 amino acids at the extreme amino terminus of the 195-kD protein, the two proteins can be aligned along their entire length. An amino-terminal domain of ∼350 amino acids and a carboxy-terminal domain of ∼90 amino acids are most similar (Fig. 4 A). These two domains are separated by a long region of predicted coiled coil (Lupas et al., 1991). Overall, the 195-kD and CLIP-170 are 34.5% identical. If conservative substitutions are included, the similarity increases to 53%. Thus, the 195-kD protein appears to be the Drosophila homologue of CLIP-170/restin. We have named the 195-kD protein, based on its predicted molecular weight, D-CLIP-190.
Figure 4.
Comparison of D-CLIP-190 and CLIP-170 predicted sequences. (A) The 195-kD protein (D-CLIP-190) is the Drosophila homologue of CLIP-170. A schematic drawing of the D-CLIP-190/CLIP-170 protein is shown. The protein can be divided into three regions: the amino-terminal domain, the coiled-coil domain, and the carboxy-terminal domain. The percent identity and similarity between D-CLIP-190 and CLIP-170 are indicated below each region. The main sequence and structural features of the protein are indicated. rep1 and rep2 are the two putative microtubule-binding motifs shared with CLIP-170. The coiled coil, the 23 amino acids–conserved motif (shaded box), and the metal-binding motif (thick black line) are also represented. (B) The MegAlign program (DNAStar), using the CLUSTAL method with the PAM250 residue weight table (Higgens et al., 1992), was used to align the putative microtubule-binding motifs from D-CLIP-190, CLIP-170 (Pierre et al., 1992), DP-150 (Holzbaur et al., 1991), Glued (Swaroop et al., 1987), BIK1 (Truehart et al., 1987), and yeast ORF YPL174c from the yeast sequence database (Genbank/EMBL/DDBJ accession number 1370367). There is a high degree of identity in this motif among the members represented. Notice that when comparing rep1 of D-CLIP-190 to that of CLIP-170 there is a higher level of conservation than when comparing rep1 to the motif from other proteins. The same is true for rep2. DP-150Glued is a component of the dynactin complex, an activator of dynein-mediated vesicle motility. DP-150Glued has one amino-terminal repeat as well as an extended region of coiled coil like CLIP-170/D-CLIP-190 (Swaroop et al., 1987; Truehart et al., 1987; Holzbaur et al., 1991). The yeast protein BIK1 is a microtubule-associated protein required for anaphase spindle movement (Berlin et al., 1990; Pellman et al., 1995). It also possesses a single repeat region and a region of coiled coil. Another yeast protein (ORF YPL174c; Genbank/EMBL/DDBJ accession number 1370367) with this motif was identified in a search of the yeast database. This protein has a putative microtubule-binding motif that contains the conserved sequence, GKN(D/S)G; however, it does not appear to have an extensive coiled-coil region (Lupas et al., 1991). (C) The metal-binding motifs from D-CLIP-190, CLIP-170 (Pierre et al., 1992), BIK1 (Truehart et al., 1987), and GAG (Berg, 1986) were aligned with the MegAlign program (DNAStar) using the CLUSTAL method with the PAM250 residue weight table (Higgens et al., 1992). The metal-binding motif consensus (MBM con) is also shown (Copeland et al., 1984; Berg, 1986). (D) A conserved motif of 23 amino acids in the carboxy terminus of D-CLIP-190 and CLIP-170 was aligned by Bestfit (Devereux et al., 1984). This highly conserved sequence, which is similar to the conserved metal-binding motif, contains several cysteines and a histidine (asterisks) whose spacing does not fit the consensus. Notably, a cysteine in CLIP-170 is not conserved in D-CLIP-190 (shaded).
Comparison of the Predicted Amino Acid Sequence of D-CLIP-190 and CLIP-170.
As expected for homologous proteins, the sequences of D-CLIP-190 and CLIP-170 predict a number of shared biochemical and structural features in addition to sequence motifs (see below and Figs. 3 B and 4). Both proteins can be divided into three regions: amino- and carboxy-terminal domains, separated by a region predicted to form an extended α-helical coiled coil (Fig. 4 A; Pierre et al., 1992). The amino-terminal region has two copies of a 57–amino acid motif or repeat. These motifs in CLIP-170 have been demonstrated to be capable of binding to microtubules. Comparison of the motifs reveals even higher identity (rep1 - 48%; rep 2 - 50%) and similarity (rep1 - 64%; rep 2 - 68%) than when the entire amino-terminal regions are compared (Fig. 4 A). The microtubule-binding motif has been identified in a number of other proteins that are known to interact with the microtubule cytoskeleton. Alignment of the D-CLIP-190 repeats with those of CLIP-170 and some of these other proteins is shown in Fig. 4 B.
The majority of the difference in the predicted size between the Drosophila and the human protein is in the second region, the coiled-coil domain. The coiled-coil region of CLIP-170 extends from amino acid 350 to 1310, a region of 960 amino acids. Regions of D-CLIP-190 that are predicted to form a coiled coil begin at amino acid 380 (Lupas et al., 1991). However, four prolines, which are predicted to break α helices, are closely spaced together at positions 469, 476, 477, and 480 (Fig. 3). Only one proline is present in the coiled-coil domain of CLIP-170 (position 492). After these prolines, D-CLIP-190 is predicted to form an extended coiled coil until amino acid 1600. The coiled-coil domain is 1,120 amino acids long in D-CLIP-190 compared to the 960–amino acid long region of CLIP-170; the region from position 380–480 in D-CLIP-190 may form an additional shorter domain of coiled coil. Not surprisingly, the level of similarity between CLIP-170 and D-CLIP-190 decreases in the coiled-coil domain (Fig. 4 A). However, a short sequence near the start of the coiled-coil regions of the two proteins is notably more similar than any other sequence in the coiled-coil domain (amino acids 370–393 in D-CLIP-190 and amino acids 354–377 in CLIP-170) (Fig. 3 B). In addition, near the end of the predicted coiled-coil regions is a stretch of sequence that is also more similar (amino acids 1573–1594 in D-CLIP-190 and amino acids 1277–1298 in CLIP-170).
The third region, the carboxy terminus, is ∼90 amino acids long and is the region most highly conserved between CLIP-170 and D-CLIP-190 (Figs. 3 B and 4 A). This region contains a putative metal-binding motif (CX2CX3GHX4C; Copeland et al., 1984; Berg, 1986; Pierre et al., 1992) of unknown function, which is also shared with BIK1 (Fig. 4 C). In addition to the metal-binding sequence, another stretch of 23 amino acids is highly conserved between CLIP-170 and D-CLIP-190; 19/23 amino acids are identical (Fig. 4 D). This sequence contains several cysteines and a histidine and is reminiscent of the metal-binding domain already noted. However, the spacing between the second cysteine and the histidine does not conform to the consensus for any metal-binding motifs that have been previously identified (Klug and Schwabe, 1995; Berg, 1986). Correct spacing of these critical amino acids is thought to be necessary for metal binding. Notably, one of the amino acids that is not conserved is a cysteine in CLIP-170 and a glutamic acid in D-CLIP-190. The similarity between the two proteins in this motif is much greater than that in the metal-binding region, indicating that this is likely to be a functionally important motif.
D-CLIP-190 Cosediments with Microtubules
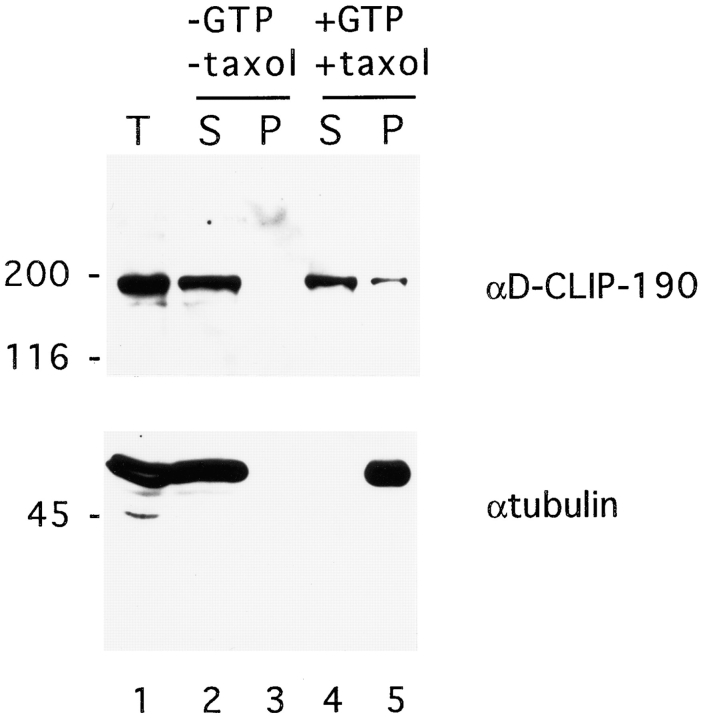
The similarity of D-CLIP-190 to CLIP-170 at the amino acid level and, in particular, in the putative microtubule– binding motifs predicts that D-CLIP-190 is capable of binding to microtubules. To determine if D-CLIP-190 can in fact associate with microtubules, the ability of D-CLIP-190 to cosediment with microtubules was assessed (Fig. 5). Microtubules were polymerized using endogenous tubulin by adding GTP and taxol to early Drosophila embryo extracts. A subset of proteins were cosedimented with microtubules in the presence of GTP and taxol (data not shown). In the absence of GTP and taxol neither microtubules nor these proteins were pelleted. A substantial fraction of D-CLIP-190 is present in the pellet in the presence of GTP and taxol (Fig. 5). Without the addition of GTP and taxol, D-CLIP-190 is not present in the pellet. Because the binding of CLIP-170 to microtubules is known to be regulated by phosphorylation (Rickard and Kreis, 1991), it is perhaps not surprising that not all of the D-CLIP-190 cosediments with microtubules. Thus, D-CLIP-190 can associate with microtubules in vitro.
Figure 5.
Cosedimentation of D-CLIP-190 with microtubules. Immunoblot of samples after polymerization and sedimentation of microtubules from embryo extract. Lane 1, embryo extract; lanes 2 and 4, supernatants; lanes 3 and 5, pellets without (lanes 2 and 3) or with (lanes 4 and 5) the addition of GTP and taxol. Supernatants and pellets from MT sedimentation experiments were transferred to nitrocellulose and the resulting blot was cut in half. The upper half was probed with αD-CLIP-190 antibody (mouse α6D1a) and the lower half with αtubulin antibody.
95F Myosin and D-CLIP-190 Are Coexpressed and Colocalized at the Subcellular Level in the Nervous System
If D-CLIP-190 is associated with 95F myosin in vivo, its distribution would be expected to overlap that of 95F myosin. To determine the expression pattern and subcellular localization of D-CLIP-190 at various stages of development, antisera raised against the cloned 195-kD protein (i.e., 6D1a–GST fusion) was used in immunolocalization studies. Overall, the distribution of D-CLIP-190 is very similar to that of 95F myosin at various stages of development, which is consistent with these two proteins being associated in vivo.
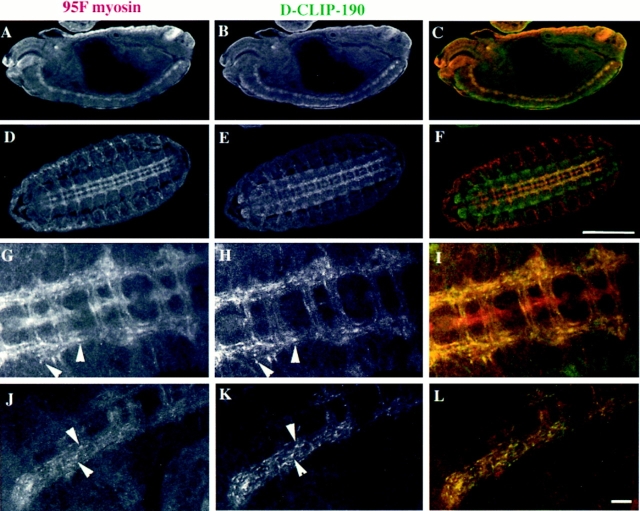
95F myosin is present in most if not all tissues throughout the lifetime of the fly. However, its level of expression in particular tissues is regulated, such that a subset of tissues express higher levels of protein at certain stages of embryonic development (K. Kellerman and K. Miller, unpublished observations). The overall immunostaining pattern of D-CLIP-190 and 95F myosin in stage 14–16 embryos is quite similar (Fig. 6). When viewed at low magnification, both proteins are enriched in the anterior structures of the head, the posterior spiracles, and the ventral nerve cord (Fig. 6, A–C).
Figure 6.
Colocalization of 95F myosin and D-CLIP-190 in the central nervous system. Confocal images of stage 14 (A–C) and stage 16 (D–L) embryos double labeled with α95F myosin (A, D, G, and J) and αD-CLIP-190 (B, E, H, and K) antibodies are shown. A lateral view is shown in A–C; ventral view is shown in D–F. High magnification views of the central nervous system in two different stage 16 embryos are shown in G–L. The embryo in G–I is oriented similarly to that shown in D–F. The embryo in J–L is oriented more laterally. An overlay of the localization of 95F myosin (red) and D-CLIP-190 (green) is shown in C, F, I, and L. Overlap in the distribution of the two proteins is yellow. Arrows indicate examples of punctate staining in which the two proteins colocalize. Bars: (F) 100 μm; (L), 10 μm.
The most striking example of their colocalization is the enrichment of both proteins in the central nervous system (CNS) located on the ventral side of the embryo. (Fig. 6, D–F; the ventral side faces the viewer). The cell bodies of the central nervous system form a broad, flat, compact region that surrounds the ladder-like connectives (longitudinal tracts) and commissures (transversal fibers that cross the ventral midline between the connectives; Hartenstein, 1993). The connectives and commissures are formed by axons from multiple neurons that are in large bundles as they run anterior, posterior, and across the midline to innervate their targets. Axons that leave the CNS do so at regular intervals, in a segmentally repeated manner. Both 95F myosin and D-CLIP-190 are enriched in the axonal processes that make up the connectives and commissures relative to overall staining levels in the embryo and in the nerve cell bodies. This enrichment is also apparent in processes that exit the CNS. In contrast, the two are differentially enriched in what appear to be glial cells in different positions (Fig. 6, D–F and G–I). 95F myosin, but not D-CLIP-190, is enriched in the midline glial cells that serve as guide cells for the formation of the commissures. Conversely, D-CLIP-190 is enriched in several cells that lie at the extreme periphery of the CNS in a segmentally repeated pattern that correlates with where the axons exit the CNS. At this position are glial cells that guide the axons as they exit the CNS. Thus, whereas in some cells or parts of cells the two proteins colocalize, in others they do not.
At higher magnification, the staining of the axons is nonuniform. Strikingly, the two proteins are present in the same particulate structures, possibly vesicles, in the neuronal processes. A ventral view of the axonal processes that lie in the longitudinal and transverse fiber tracts is presented in G–I. The anterior and posterior commissures of two segmental repeats, as well as the longitudinal fibers that connect them, are brightly stained with both antibodies. The individual axons that make up these tracts can occasionally be distinguished. Within these axons, punctate staining is observed with both antibodies. When the images are overlayed, it is clear that there is a one to one correspondence between the punctate spots that contain 95F myosin and those that contain D-CLIP-190. In Fig. 6, J–L, the anterior end and a stretch of about two segments length of one of the connectives is seen in longitudinal section. Again, striking punctate staining is seen along the length of each axon, with a clear colocalization of 95F myosin and D-CLIP-190 in the spots. In contrast, when embryos are double labeled with αD-CLIP-190 and αHRP, which stains neural cell membranes (Snow et al., 1987), the distribution of the two proteins appears rather different (data not shown). Whereas D-CLIP-190 staining is punctate, HRP staining appears more homogeneous. Thus, the punctate 95F myosin/D-CLIP-190 structures observed are distinct from the distribution of a uniformly distributed protein in axons of neurons of the CNS. Because the 95F myosin/D-CLIP-190 punctate structures are present in the axonal processes of neurons, it seems likely that they are vesicular or organellar structures.
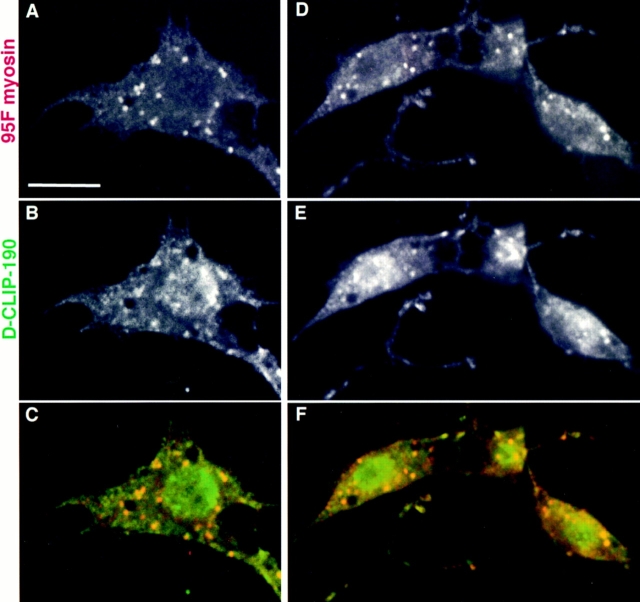
95F Myosin and D-CLIP-190 Are Colocalized at the Subcellular Level in Primary Embryonic Cultured Cells
Because subcellular localization is difficult to discern in whole embryos, we also have colocalized 95F myosin and D-CLIP-190 in primary cultures of embryonic cells. These cultures contain several different cell types, including myoblasts, which eventually fuse to form myotubes with sarcomeric structures, neurons that elaborate extensive processes, and hemocytes, a macrophage-like cell that has a flat, spread morphology (Cross and Sang, 1978). Cultures made from gastrulating embryos were fixed and stained with antibodies specific for the two proteins (Fig. 7). We see a number of interesting colocalizations in the different cell types. Neurons show 95F myosin and D-CLIP-190 particles like those seen in intact embryos (not shown). Colocalization in hemocytes is quite striking (A–C; well spread cell in D–F). There are a number of large inclusions in the cytoplasm, possibly vesicles or other organelles, that stain brightly for both proteins. In myoblasts, we also see these cytoplasmic organelles (less well spread cells in D–F) in which both proteins are present. Whereas 95F myosin staining appears to be primarily or exclusively confined to these organelles, D-CLIP-190 is clearly present in other areas of the cell. Particularly prominent is the D-CLIP-190 staining in the region of the nucleus. 95F myosin is not enriched in this area. We conclude that 95F myosin and D-CLIP-190 are colocalized in a subset of organelles in several cell types.
Figure 7.
Primary embryonic cultures stained with anti- D-CLIP-190 and 95F myosin antibodies. (A–C) A hemocyte showing colocalization of 95F myosin (A) and D-CLIP-190 (B) in large cytoplasmic organelles. The composite image in (C) shows D-CLIP-190 in green and 95F myosin in red. Regions of colocalization are yellow. (D–F) Two myoblasts and a hemocyte (well-spread cell) in which 95F myosin (D) and D-CLIP-190 (E) colocalize are shown. The composite image is shown in (F) with color scheme as in (C). Bar, 10 μm.
95F Myosin and D-CLIP-190 Are Colocalized at the Posterior Pole of the Early Embryo
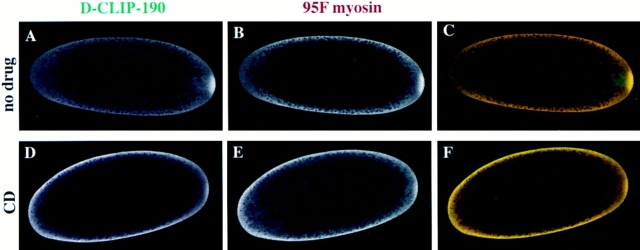
One of the most surprising features of the localization pattern of D-CLIP-190 is its distribution before migration of the nuclei to the cortex in precellularization embryos. In unfertilized eggs and during nuclear cycles 1–8 of embryogenesis, D-CLIP-190 protein is present at the posterior pole (Fig. 8 A). Consistent with the association of the two proteins, 95F myosin is also enriched at the posterior pole (Fig. 8 B). Because of the high level of protein present in the cortex of the embryo, the enrichment of 95F myosin at the posterior pole is not as striking as that of D-CLIP-190 protein; however, we consistently observe a higher level of protein at the posterior than in the adjacent cortex.
Figure 8.
The posterior pole localization of D-CLIP-190 and 95F myosin is dependent on the actin cytoskeleton. Confocal images of control embryos (no drug; A–C) and embryos treated with 10 μg/ml cytochalasin D for 30 min to depolymerize actin filaments (CD; D–F) are shown. These embryos were double labeled with αD-CLIP-190 (A and D) and α95F myosin (B and E) antibodies. A composite of the individual images is shown in C and F (D-CLIP-190, green; 95F myosin, red). Overlap of the two proteins is shown in yellow. D-CLIP-190 and 95F myosin proteins are enriched at the posterior pole. D-CLIP-190 appears to be present at a higher level and present in a broader distribution than 95F myosin at the posterior pole. Levels of 95F myosin are higher than D-CLIP-190 in the cortex. The cytochalasin D–treated embryo shown still has residual protein at the posterior pole (indicated as ± in Table I). The majority of αD-CLIP-190 and α95F myosin-labeled embryos have no observable posterior pole localization (Table I).
The Posterior Localization of D-CLIP-190 and 95F Myosin in the Early Embryo Is Dependent on the Actin but Not the Microtubule Cytoskeleton
The localization of D-CLIP-190 and 95F myosin at the posterior of this large syncytial cell gave us the opportunity to test whether the localization of these two proteins is dependent on the actin and microtubule cytoskeletons. As associated proteins, one might expect them to behave similarly in response to disruption of the cytoskeleton. Therefore, early embryos were treated with cytochalasin D to disrupt the actin cytoskeleton or colchicine to disrupt microtubules (Limbourg and Zalokar, 1973).
When embryos were treated with cytochalasin D, the actin cytoskeleton appeared to be substantially depolymerized (data not shown; see Materials and Methods). In addition, the nuclei were not distributed normally in treated embryos; this is expected if the cytochalasin D depolymerization was effective because the axial expansion or spreading of nuclei is an actin-dependent process (von Dassow and Schubiger, 1994). When such embryos were stained with αD-CLIP-190 or α95F myosin antibodies, the localization of these two proteins at the posterior pole was substantially disrupted (Table I, Fig. 8, D–F). Most cytochalasin D–treated embryos had little or no D-CLIP-190 or 95F myosin present at the posterior pole. A low level of D-CLIP-190 and 95F myosin may remain in some treated embryos because cytochalasin D does not depolymerize all actin filaments in some cases (Cooper, 1987). From these results we conclude that the posterior localization of both D-CLIP-190 and 95F myosin is dependent on the actin cytoskeleton.
Table I.
Posterior Localization of D-CLIP-190 and 95F Myosin Is Dependent on the Actin Cytoskeleton
| Treatment | D-CLIP-190 | 95F myosin | ||||||
|---|---|---|---|---|---|---|---|---|
| + | ± | − | n | + | ± | − | n | |
| Control | 70 | 19 | 10 | 259 | 63 | 29 | 8 | 143 |
| Cyto D | 6 | 20 | 73 | 300 | 2 | 4 | 93 | 45 |
| Colchicine | 71 | 22 | 7 | 117 | 70 | 22.5 | 7.5 | 40 |
| Both | 0 | 21 | 79 | 84 | ND |
Embryos were also treated with colchicine to assess the effect of depolymerizing the microtubule cytoskeleton on localization at the posterior pole of D-CLIP-190 and 95F myosin. Microtubules appeared to be substantially depolymerized by colchicine treatment (data not shown; see Materials and Methods). The distribution of 95F myosin appeared to be unaffected by disrupting the microtubule cytoskeleton (Table I). D-CLIP-190 also appears unaffected, although at higher concentrations of colchicine (20 μg/ml), the protein does appear somewhat more diffuse. Thus, the posterior localization of D-CLIP-190 and 95F myosin does not appear to be dependent on the microtubule cytoskeleton. Embryos were also treated with both cytochalasin D and colchicine. Because this treatment did not completely abolish localization in the majority of embryos, we conclude that D-CLIP-190 is not substantially more affected when compared to treatment with cytochalasin D alone (Table I).
Discussion
We have identified a 195-kD protein, D-CLIP-190, that coimmunoprecipitates with a class VI myosin, Drosophila 95F unconventional myosin. The same profile of proteins are immunoprecipitated from embryo extracts with α95F myosin and αD-CLIP-190 antibodies. The fact that these two proteins are coimmunoprecipitated with antibodies against either protein argues that their association in vitro is specific. Cloning and sequencing of the gene encoding the 195-kD protein revealed that it is a homologue of the human protein, CLIP-170, which is involved in microtubule–vesicle interactions (Pierre et al., 1992). In addition to coimmunoprecipitation data, immunolocalization studies support the contention that these two proteins are associated. D-CLIP-190 is coexpressed and colocalized with 95F myosin at several times in development and in a number of tissues. In the central nervous system, 95F myosin and D-CLIP-190 are coexpressed in a subset of cells and associated with the same particulate structures/vesicles in axonal processes of neurons. They are also colocalized in the same subcellular structures in primary embryonic cultured cells. Interestingly, the proteins are colocalized at the posterior of the early embryo. The association of this CLIP-170 homologue and an unconventional myosin suggests that these proteins may be important in coordination of certain microtubule and actin filament–based processes in cells.
D-CLIP-190 Is a Homologue of CLIP-170
Comparison of the amino acid sequence of D-CLIP-190 and CLIP-170 reveals that the two proteins are predicted to have the same overall structure and to share sequence motifs throughout their length. A predicted long α-helical coiled coil separates the relatively highly conserved amino- and carboxy-terminal domains of the proteins. CLIP-170 was originally isolated as a protein that mediates binding of endocytic vesicles to microtubules in vitro (Rickard and Kreis, 1990; Scheel and Kreis, 1991). When the CLIP-170 cDNA is transformed into HeLa cells, CLIP-170 binds along the length of microtubules; however, normally it localizes to structures at the plus ends of peripheral microtubules. The amino-terminal domain of CLIP-170 possesses two copies of a motif that has been demonstrated to mediate binding to microtubules both in vitro and in vivo (Pierre et al., 1992, 1994). Binding to microtubules in vitro has been shown by assaying the binding to purified microtubules of recombinant CLIP-170, including versions in which the microtubule-binding domains have been mutated. Because of the high degree of similarity between the two proteins in these motifs, it is likely that D-CLIP-190 is also capable of binding microtubules. Consistent with this idea, D-CLIP-190 cosediments with microtubules. Interestingly, the carboxy-terminal region of CLIP-170 is thought to be required for its association with cytoplasmic structures that accumulate at the plus ends of peripheral microtubules (Pierre et al., 1994). It is possible that the two conserved sequences in this region of CLIP-170/D-CLIP-190 are involved in mediating binding to other proteins on vesicles or other cellular structures.
The Posterior Localization of D-CLIP-190 and 95F Myosin Are Similarly Affected upon Disruption of the Cytoskeleton
The localization of D-CLIP-190 and 95F myosin at the posterior pole of the precellularization embryo is dependent on the actin cytoskeleton but not the microtubule cytoskeleton. The similarity in the behavior of D-CLIP-190 and 95F myosin when the cytoskeleton is disrupted provides additional support that these two proteins are associated in vivo. However, assuming that 95F myosin binds actin filaments and D-CLIP-190 binds microtubules at the posterior pole of the embryo, it is rather curious that disruption of microtubules does not substantially disrupt the localization of at least D-CLIP-190. This result could be due to incomplete depolymerization of microtubules, such that the two proteins remain bound at the posterior via colchicine-resistant microtubules. However, the observed behavior could also be explained by the fact that human CLIP-170 appears to associate with microtubules only transiently (Rickard and Kreis, 1996). When microtubules are disrupted, D-CLIP-190 may still be part of a complex with 95F myosin and bound to the actin cytoskeleton and, thus, still present at the posterior pole. When the actin cytoskeleton is disrupted, however, the complex may not remain at the posterior pole because D-CLIP-190 is not stably bound to microtubules; therefore, the complex may then diffuse from the posterior pole.
Both the microtubule and actin cytoskeleton are thought to be involved in localization of determinants (Yisraeli et al., 1990; Erdélyi et al., 1995; Pokrywka, 1995; Guo and Kemphues, 1996; Tetzlaff et al., 1996). In Drosophila the cytoskeleton plays important roles during oogenesis and early embryogenesis when axial polarity is being established. A group of genes has been characterized that are required for posterior polarity and germ cell formation, both of which require the localization of determinants at the posterior pole (St. Johnston, 1993). Many of the gene products encoded by these posterior group genes such as oskar are present at the posterior of the oocyte and early embryo. During oogenesis, microtubules are required for localization of several mRNAs including oskar (Theurkauf et al., 1993; Pokrywka, 1995). The actin cytoskeleton may also be involved in localization because oskar mRNA is not localized in normal amounts to the posterior pole in oocytes and early embryos from females mutant for cytoplasmic tropomyosin (Erdélyi et al., 1995; Tetzlaff et al., 1996). In addition, when the actin cytoskeleton is disrupted with cytochalasin D, pole plasm components are not stably maintained at the posterior of the early embryo (Lantz, V.A., S. Clemens, and K. Miller, manuscript submitted for publication). These results suggest that RNA localization at the posterior of the Drosophila embryo may require coordination of the actin and MT cytoskeletons.
This potential coordination of actin- and microtubule-based processes is not unique to Drosophila. In Xenopus, microtubules are responsible for Vg1 mRNA transport whereas actin filaments are required for its maintenance at the vegetal pole (Yisraeli et al., 1990). The localization of 95F myosin and D-CLIP-190 at the posterior pole raises the possibility that they could be involved in stable association of pole plasm components at the posterior pole through their association with actin filaments. Further studies will be required to provide a functional link between these two proteins and posterior patterning.
A Family of Linker Proteins May Mediate Interactions between Different Cellular Components
CLIP-170 and its Drosophila counterpart appear to be a member of, at present, a small family of proteins that includes DP-150Glued and BIK1 (Rickard and Kreis, 1996). The other members of the family share only limited regions of sequence similarity and a subset of structural features with D-CLIP-190/CLIP-170. DP-150Glued is known to be part of a larger complex, the dynactin complex, which stimulates dynein-mediated motility of vesicles in vitro (for review see Schroer, 1996). Biochemical evidence suggests that DP-150Glued binds microtubules via the single microtubule-binding motif in the amino-terminal region and is a key protein in linking components of the complex together.
D-CLIP-190/CLIP-170 may act in a manner analogous to DP-150Glued, as a linker protein between microtubules and in this case the actin cytoskeleton via 95F myosin. Because CLIP-170 is located at the peripheral end of microtubules near the actin cortex, it is possible that this protein and a class VI myosin like 95F myosin may provide a direct link between microtubules and the actin cortex. This complex could function to anchor microtubules in the actin cortex (Koonce, 1996). Such binding may be important for microtubule cytoskeleton organization and microtubule stability. Alternatively, it is possible that the 95F myosin/D-CLIP-190 complex functions at a transition point for the transfer of vesicles or other cytoplasmic structures transported along microtubules to actin filaments in the cortex or vice versa. At the posterior pole of the oocyte, such a link could be required to allow transfer of posterior proteins/mRNAs from microtubules along which they were transported to actin filaments where they can be anchored at the posterior. Maintaining this link, which initially forms during oogenesis through early embryogenesis, could be important for the stable localization of posterior pole plasm components.
Because these two proteins colocalize in structures that most likely include other proteins and perhaps lipids, it is not clear whether their association is direct. The ability to coimmunoprecipitate 95F myosin and D-CLIP-190 in 1% NP-40 would tend to suggest that these two proteins are directly associated. Further biochemical studies suggesting direct binding would support either of the above models.
Coordination of Microtubule and Actin-based Transport
Because 95F myosin has been implicated in transport and the vertebrate homologue of D-CLIP-190, CLIP-170, is suspected of being involved in transport (Rickard and Kreis, 1996), the colocalization in axons, where vesicle/organelle transport along both actin and microtubules has been observed, is particularly intriguing. The particulate distribution of both 95F myosin and D-CLIP-190 in nerve processes in the embryo and also in cultured cells from Drosophila embryos is consistent with these structures being vesicles. CLIP-170 has been suggested to participate in loading endocytic vesicles on to the plus ends of microtubules (Rickard and Kreis, 1996). No previous data exist that suggest an interaction of CLIP-170 with the actin cytoskeleton. However, the plus ends of peripheral microtubules to which the CLIP-170–associated vesicles appear to bind are adjacent to the actin filament–rich cortex. One function for D-CLIP-190 and 95F myosin in the same structures may be to coordinate the transport of specific cargoes along both microtubules and actin filaments to facilitate their proper localization in the cell.
There is some evidence of a role for unconventional myosins in vesicle traffic (Fath and Burgess, 1994; Langford, 1995). In the nervous system, in particular, in a variety of organisms, there is increasing evidence for vesicular and organellar transport along both microtubules and microfilaments (Brady et al., 1984; Lillie and Brown, 1992; Kuznetsov et al., 1992; Fath et al., 1994; Morris and Hollenbeck, 1995; Evans and Bridgman, 1995). More recently, in yeast, class I myosins have been implicated in endocytosis (Geli and Riezman, 1996). MYO2 (Myosin V) mutants have defects that have been interpreted as recycling defects (Govindan et al., 1995). Furthermore, expression of activated RhoD, a small GTPase, causes reorganization of the actin cytoskeleton and affects endosome motility and distribution (Murphy et al., 1996). The D-CLIP-190/95F myosin complex could serve to permit movement of vesicles from one cytoskeletal filament type to the other during secretion, transport, and/or endocytosis.
One model for how transport may occur in neurons is that long range movement occurs along microtubules whereas actin-based transport is required for traversing the actin-rich cortex along the axon periphery and at the nerve terminal. Thus, a myosin, like 95F myosin, and a microtubule motor, like a kinesin-related protein or dynein, would be present on the same vesicle. When a vesicle is being transported along the axon, it would primarily use its microtubule motor(s), potentially being attached or regulated through association with D-CLIP-190. Actin-based transport would only come into play when the vesicle encounters the relatively microtubule-free, actin-rich peripheral cytoplasm. Once a vesicle reached the terminal, actin-based transport events, mediated by 95F myosin, would then be required to bring it to its final target. Actin-based transport might also be important in local recycling of vesicles at the nerve terminal.
We have shown that D-CLIP-190, a microtubule-binding protein and CLIP-170 homologue, and the 95F (class VI) unconventional myosin are present in the same complex. Our studies reveal a potentially important link between the actin and microtubule cytoskeletal systems. They support other studies that have provided evidence for coordination between actin and microtubules-based cellular processes.
Footnotes
We are especially grateful for the help of J. Verbsky and C. Cheney (both from Washington University, St. Louis, MO) with automated fluorescent DNA sequencing. In addition, we thank the Department of Genetics at Washington University School of Medicine for use of the ABI Prism sequencing system. We thank J. Diani and the animal facility in the Biology Department at Washington University for injecting and bleeding the mice. We thank R. Steward for the dorsal antibody. We thank J. Cooper, J. McNally, C. Cheney, C. Wagner, R. Hopmann, J. Hicks, B. Clifford (all from Washington University), and T. Schroer (Johns Hopkins University, Baltimore, MD) for helpful suggestions on the manuscript.
This work was supported by an American Cancer Society postdoctoral fellowship to V. Lantz and a National Institutes of Health grant to K.G. Miller.
Address all correspondence to Valerie A. Lantz, Department of Biology, Washington University, St. Louis, MO 63130. Tel.: (314) 935-7339. Fax: (314) 935-5125. E-mail: Lantz@biodec.WUSTL.edu
1. Abbreviations used in this paper: CLIP, cytoplasmic linker protein; CNS, central nervous system; GST, glutathione-S-transferase; IP, immunoprecipitation; MBP, maltose-binding protein fusion; ORF, open reading frame.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amero, S.A., T.C. James, and S.C.R. Elgin. 1987. Raising antibodies to protein bands in gels. In Methods in Molecular Biology. Vol. 3. J.M. Walker, editor. Humana Press, Inc., Clifton, NJ. 355–362.
- Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berlin V, Styles CA, Fink GR. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiaecolocalizes with tubulin. J Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbe G, Delabie J, Bruggen J, Richener H, Asselbergs FAM, Cerletti N, Sorg C, Odink K, Tarcsay L, Wiesendanger W, DeWolf-Peeters C, Shipman R. Restin: a novel intermediate filament-associated protein highly expressed in the Reed-Sternberg cells of Hodgkin's disease. EMBO (Eur Mol Biol Organ) J. 1992;11:2103–2113. doi: 10.1002/j.1460-2075.1992.tb05269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST, Lasek RJ, Allen RD, Yin HL, Stossel TP. Gelsolin inhibition of fast axonal transport indicates a requirement for actin microfilaments. Nature. 1984;310:56–58. doi: 10.1038/310056a0. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland TD, Morgan MA, Oroszlan S. Complete amino acid sequence of the basic nucleic acid binding protein of Feline Leukemia Virus. Virol. 1984;133:137–145. doi: 10.1016/0042-6822(84)90432-x. [DOI] [PubMed] [Google Scholar]
- Cross DP, Sang JH. Cell culture of individual Drosophila embryos. J Embryol Exp Morphol. 1978;45:161–172. [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdélyi M, Michon A-M, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskarmRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Evans LL, Bridgman PC. Particles move along actin filament bundles in nerve growth cones. Proc Natl Acad Sci USA. 1995;92:10954–10958. doi: 10.1073/pnas.92.24.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Burgess DR. Membrane motility mediated by unconventional myosin. Curr Opin Cell Biol. 1994;6:131–135. doi: 10.1016/0955-0674(94)90126-0. [DOI] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR. Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol. 1994;126:661–675. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6–13 and 137:266–267. [DOI] [PubMed]
- Geli M, Riezman H. Role of type I myosins in receptor-mediated endocytosis in yeast. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Govind S, Whalen AM, Steward R. In vivoself-association of the Drosophila rel-protein dorsal. Proc Natl Acad Sci USA. 1992;89:7861–7865. doi: 10.1073/pnas.89.17.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Hartenstein, V. 1993. Atlas of Drosophila Development. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Higgens DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comp Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Holzbaur ELF, Hammarback JA, Paschal BM, Kravit NG, Pfister KK, Vallee RB. Homology of a 150K cytoplasmic dynein-associated polypeptide with the Drosophila gene Glued. . Nature. 1991;351:579–583. doi: 10.1038/351579a0. [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JWR. Zinc fingers. FASEB (Fed Am Soc Exp Biol) J. 1995;9:597–604. [PubMed] [Google Scholar]
- Koonce MP. Making a connection: the “other” microtubule end. Cell Motil Cytoskeleton. 1996;35:85–93. doi: 10.1002/(SICI)1097-0169(1996)35:2<85::AID-CM1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Langford GM. Actin- and microtubule-dependent organelle motors: interrelationships between the two motility systems. Curr Opin Cell Biol. 1995;7:82–88. doi: 10.1016/0955-0674(95)80048-4. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. . J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbourg B, Zalokar M. Permeabilization of Drosophila egg. Dev Biol. 1973;35:382–387. doi: 10.1016/0012-1606(73)90034-1. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mermall V, McNally JG, Miller KG. Transport of cytoplasmic particles by an unconventional myosin in living Drosophila embryos. Nature. 1994;369:560–562. doi: 10.1038/369560a0. [DOI] [PubMed] [Google Scholar]
- Mermall V, Miller KG. The 95F unconventional myosin is required for proper organization of the Drosophilasyncytial blastoderm. J Cell Biol. 1995;129:1575–1588. doi: 10.1083/jcb.129.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F–actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lutcke A, Parton RG, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu H, Fink GR. Two microtubule–associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. . J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Pierre P, Pepperkok R, Kreis TE. Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci. 1994;107:1909–1920. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson E. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1982;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. Identification of a novel nucleotide-sensitive microtubule binding protein in HeLa cells. J Cell Biol. 1990;110:1623–1633. doi: 10.1083/jcb.110.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. Binding of pp170 to microtubules is regulated by phosphorylation. J Biol Chem. 1991;266:17597–17605. [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. CLIPs for organelle-microtubule interactions. Trends Cell Biol. 1996;6:178–183. doi: 10.1016/0962-8924(96)10017-9. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM. Isolation and analysis of microtubule motor proteins. Methods Cell Biol. 1994;44:279–288. doi: 10.1016/s0091-679x(08)60919-x. [DOI] [PubMed] [Google Scholar]
- Scheel J, Kreis TE. Motor independent binding of endocytic carrier vesicles to microtubules in vitro. J Biol Chem. 1991;266:18141–18148. [PubMed] [Google Scholar]
- Schroer TA. Structure and function of dynactin. Semin Cell Dev Biol. 1996;7:321–328. [Google Scholar]
- Smith DB, Johnson KS. Single step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Smith LM, Sanders JC, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SBH, Hood LE. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- Snow PM, Patel NH, Harrelson AL, Goodman C. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper. J Neurosci. 1987;712:4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- St. Johnston, D. 1993. Pole plasm and the posterior group genes. In The Development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Press, Cold Spring Harbor, NY. 325–364.
- Steinhaur WR, Walsh RC, Kalfyan LJ. Sequence and structure of the Drosophila melanogaster ovarian tumor gene and generation of an antibody specific for ovarian tumor protein. Mol Cell Biol. 1989;9:5726–5732. doi: 10.1128/mcb.9.12.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Swaroop M, Garen A. Sequence analysis of the complete cDNA and encoded polypeptide for the Gluedgene of Drosophila melanogaster. Proc Natl Acad Sci USA. 1987;84:6501–6505. doi: 10.1073/pnas.84.18.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synder M, Elledge S, Sweetser D, Young RA, Davis RW. Lambda gt11: gene isolation with antibody probes and other applications. Methods Enzymol. 1987;154:107–128. doi: 10.1016/0076-6879(87)54073-3. [DOI] [PubMed] [Google Scholar]
- Tetzlaff MT, Jäckle H, Pankratz MJ. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskarmRNA required for germ cell formation. EMBO (Eur Mol Biol Organ) J. 1996;15:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Alberts BM, Jan YN, Jongens TA. A central role for microtubules in the differentiation of Drosophila oocytes. Development (Camb) 1993;118:1169–1180. doi: 10.1242/dev.118.4.1169. [DOI] [PubMed] [Google Scholar]
- Truehart J, Boeke JD, Fink G. Two genes required for cell fusion during cell conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Schubiger G. How an actin network might cause fountain streaming and nuclear migration in the syncytial Drosophilaembryo. J Cell Biol. 1994;127:1637–1653. doi: 10.1083/jcb.127.6.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development (Camb) 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]