Homer1a is a core brain molecular correlate of sleep loss (original) (raw)
Abstract
Sleep is regulated by a homeostatic process that determines its need and by a circadian process that determines its timing. By using sleep deprivation and transcriptome profiling in inbred mouse strains, we show that genetic background affects susceptibility to sleep loss at the transcriptional level in a tissue-dependent manner. In the brain, Homer1a expression best reflects the response to sleep loss. Time-course gene expression analysis suggests that 2,032 brain transcripts are under circadian control. However, only 391 remain rhythmic when mice are sleep-deprived at four time points around the clock, suggesting that most diurnal changes in gene transcription are, in fact, sleep–wake-dependent. By generating a transgenic mouse line, we show that in _Homer1_-expressing cells specifically, apart from Homer1a, three other activity-induced genes (Ptgs2, Jph3, and Nptx2) are overexpressed after sleep loss. All four genes play a role in recovery from glutamate-induced neuronal hyperactivity. The consistent activation of Homer1a suggests a role for sleep in intracellular calcium homeostasis for protecting and recovering from the neuronal activation imposed by wakefulness.
Keywords: homeostasis, microarray, mRNA tagging, sleep deprivation, sleep function
Two main processes regulate sleep. A homeostatic process regulates sleep need and intensity according to the time spent awake or asleep. A circadian process regulates the appropriate timing of sleep and wakefulness across the 24-h day. A highly reliable index of the homeostatic process is provided by the amplitude and prevalence of delta (1- to 4-Hz) oscillations in the electroencephalogram (EEG) of nonrapid eye movement (NREM) sleep (hereafter, “delta power”). Delta power is high at sleep onset and decreases during sleep, in parallel with sleep depth. Sleep deprivations and naps induce a predictable increase or decrease, respectively, in delta power during subsequent sleep. The interaction between homeostatic and circadian processes is mathematically described in the two-process model of sleep regulation, which provides a framework for prediction and interpretation of a large body of experimental data (1).
Among hypotheses concerning the physiological function of waking-induced changes in sleep, the most compelling suggests that sleep plays a key role in synaptic plasticity (2, 3). More specifically, EEG delta power during NREM sleep has been shown to play a critical role in learning-induced plasticity (4–6). In general, the prediction is that local neural activation due to specific behavioral (cognitive) demands imposes a burden on the brain which necessitates sleep and which is reflected by the EEG delta power.
On the basis of mathematical modeling and experimental data, we have shown that sleep need, as indexed by the EEG delta power, is under genetic control (7), which is of direct relevance for explaining the interindividual vulnerability to sleep loss in human subjects (8, 9). However, deciphering the molecular bases of sleep need is rendered difficult because the contributions of the homeostatic and circadian processes are difficult to separate and because the impact of genetic background on brain gene expression is poorly understood. From a series of gene-profiling experiments, we here report a comprehensive transcriptome analysis that specifically takes these interacting factors into account. We show that short-term sleep loss induces changes in brain gene expression for a few genes only. These genes are all part of a highly specific pathway involved in neuronal protection and recovery after waking-induced glutamate overstimulation.
Results
We have previously reported (7) that the dynamics of sleep need varies strongly among inbred mouse strains, with AKR/J (AK) mice showing a dramatic increase in delta power after 6-h sleep deprivation, whereas DBA/2J (D2) mice have a blunted response. Through quantitative linkage analysis, a significant quantitative trait locus (QTL) was identified on mouse chromosome 13 (Dps1: delta power in slow-wave sleep 1) that explains >50% of variance in delta power after sleep deprivation in BXD recombinant inbred lines (RIs) derived from inbred mouse strains C57BL/6J (B6) and D2 (7). The best polymorphic marker associated was D13Mit126 at 46.5 cM (95% CI = 25 cM), suggesting a large QTL region (≈38 Mb). However, based on the most recent high-resolution, single-nucleotide polymorphism (SNP) genetic map of BXD RIs (10) (http://gscan.well.ox.ac.uk/gs/strains.cgi), the smallest differential region corresponds to an 11-Mb sequence flanked by SNPs rs3669221 and rs397202. According to the Ensemble database (Mus musculus release 46), this region contains 33 known genes, 15 potential unknown coding sequences, and three pseudogenes. Among these, the short splice variant of the Homer1 (Homer1a) gene is the only transcript in the region that was previously reported to be among the up-regulated genes after sleep deprivation (11–14). However, the specificity of this finding, compared with other gene expression changes after sleep loss, has not yet been established.
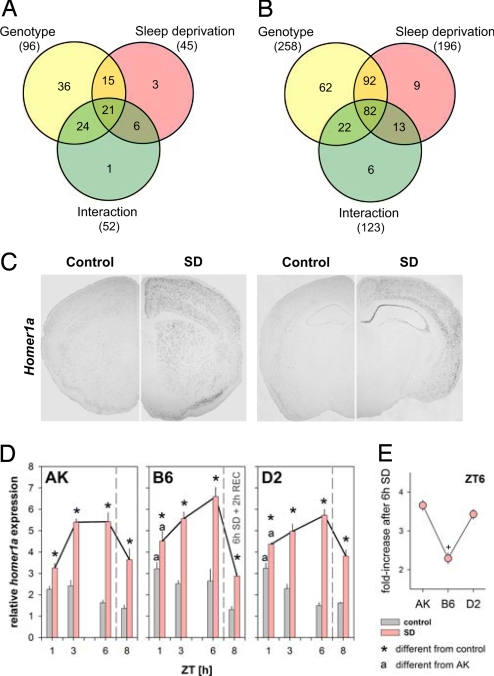
To confirm and verify whether differences in sleep need between mouse strains are regulated at the transcriptional level, mice of three genotypes (AK, B6, and D2) were deprived of sleep by gentle handling [see supporting information (SI) Materials and Methods] for 6 h, starting at light onset, and killed at the same time of day [Zeitgeber time (ZT)6] with their non-sleep-deprived controls. Because it is believed that sleep fulfills a brain-specific function, we also sampled the liver as a peripheral reference organ. Microarray results were analyzed by a two-way ANOVA, with strain and condition as main factors. Venn diagrams summarizing the data are shown in Fig. 1 A and B and suggest that very few genes show consistent changes in expression across genetic backgrounds. We use the terms “consistent” and “reliable” hereafter only for transcripts either increased or decreased in all three inbred strains and across experimental conditions.
Fig. 1.
Transcriptome analysis of the brain and liver after sleep deprivation in three inbred mouse strains indicates that Homer 1a is specifically upregulated in the brain. (A and B) Venn diagrams of a two-way ANOVA microarray data analysis of total RNA extracted from brain (A) and liver (B) in AK, B6, and D2 inbred mouse strains at ZT6 after a 6-h sleep deprivation. Results are reported as a function of genetic background (genotype), experimental condition (sleep deprivation), and their interaction. (C) Homer1a is overexpressed after sleep deprivation (SD) in the cortex and caudate putamen (Left) and in the hippocampus (Right). (D) Mean (±1 SEM) forebrain mRNA levels of Homer1a for AK, B6, and D2 mice after SDs of 1, 3, and 6 h (pink bars), with their time-matched controls (gray bars). All SDs started at light onset (ZT0). Effects of recovery sleep on expression were assessed by allowing 2 h of recovery after 6-h SD (ZT8). Homer1a expression was affected by SD, genotype, and time of day (P < 0.05; three-way ANOVA). (E) Fold change increase of Homer1a expression level after 6 h of SD varies among strains. Ratios between SD and control mRNA levels at ZT 6 were calculated as described in SI Materials and Methods, with their standard errors. The effect of SD on Homer1a expression is statistically different between B6 and the two other strains (+, P < 0.006; two-way ANOVA).
Sleep deprivation induced changes in only 42 brain probe sets in B6, 92 in D2, and 188 in AK, among which 52 sets were affected by genotype. To our surprise, sleep loss induced almost three times more transcriptional changes in liver compared with whole brain (Fig. 1A). As in brain, among all transcripts with significant changes in liver, ≈50% changed as a function of genetic background. Thus, for genes differentially expressed in each tissue of the three strains in response to sleep deprivation, we have focused on those showing a significant two-way ANOVA interaction (see SI Tables 1 and 2). As reported in refs. 11, 15, and 16, sleep deprivation most significantly up-regulated the expression of heat shock protein genes in both brain and liver, strongly suggesting that this group of genes is part of a general stress–response pathway induced by enforced wakefulness in most organs, and, therefore, is not specific to the brain. The most significant decrease was found for the cold-induced RNA binding protein (Cirbp) in both tissues, again suggesting a general rather than a brain-specific implication (SI Tables 1 and 2). In brain, the expression of Homer1a was most affected by both genotype and sleep loss. In situ hybridization analysis confirmed the strong induction of Homer1a transcript after sleep deprivation and indicated a restricted up-regulation in cortex, striatum, and hippocampus (Fig. 1C). Homer1 encodes several transcripts by alternative splicing, among which long forms are constitutively expressed while two short forms, Homer1a and Ania-3, are activity-induced. These postsynpatic density proteins bind calcium-signaling molecules and have been implicated in synaptic plasticity.
In contrast to most other activity-induced genes, Homer1a overexpression seemed highly specific. For instance, brain-derived neurotrophic factor (Bdnf), a plasticity-related gene, showed a significant increase in AK and D2 only; the immediate early gene Fos did not show a significant increase in D2; and Arc, Egr1, and Egr3 were induced by sleep deprivation mainly in AK (SI Table 1). Thus, comparisons among genotypes, and between brain and liver, identified Homer1a as the most specific transcriptional index of the whole brain in response to sleep loss.
We then analyzed the time course of Homer1a induction by real-time quantitative RT-PCR in a dose–response experiment. Mice of all three strains were sleep-deprived for 1, 3, and 6 h and killed together with their time-matched controls. An additional group, which was also sleep-deprived for 6 h, was killed 2 h into recovery sleep. As shown in Fig. 1D, Homer1a is rapidly and strongly induced by sleep deprivation in a dose-dependent manner. Parallel to this increase, Homer1a expression decreases with increasing accumulated sleep in non-sleep-deprived animals, and its relative level remains significantly higher than in the time-matched controls after only 2 h of recovery sleep, indicating that the time course of Homer1a expression closely parallels sleep need. However, the fold change of Homer1a expression after 6-h sleep deprivation was similar between D2 and AK, which represent the two extreme strains in their response to sleep deprivation, and was significantly higher than in B6 mice (Fig. 1E). In accord with our findings, sequence comparisons in public databases for the Homer1 region indicated that D2 and AK share the same SNP haplotype, which is different from that of B6, although no coding sequence differences between any of the three strains can be identified. This finding suggested that, even though Homer1a is the best candidate for the Dps1 QTL segregating with response to sleep loss between D2 and B6 mice, other genes may affect sleep need in other inbred strains, or posttranscriptional or translational changes of Homer1a may differ between D2 and AK mice.
Time-of-Day Effects of Sleep Loss on Brain Transcriptional Changes.
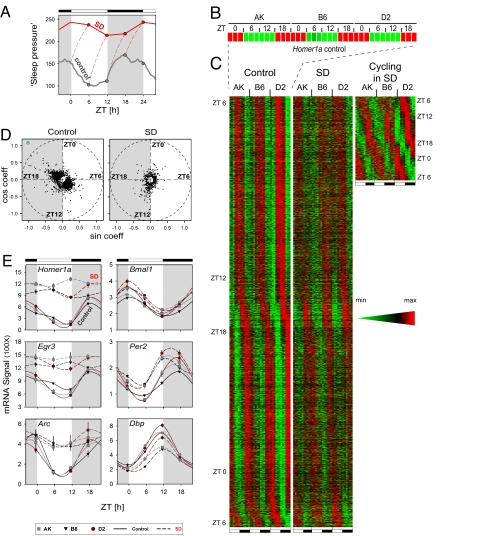
As shown in Fig. 1D, the level of Homer1a expression under baseline conditions shows significant variation that might be either due to a direct circadian effect or driven by the diurnal sleep–wake distribution, as we suggest. To separate time-of-day and homeostatic effects on brain gene expression, we have simulated the time course of sleep need (quantified as EEG delta power) according to published assumptions and parameters (7) under entrained baseline conditions and after 6-h sleep deprivation at four time points around the 24-h day. If a gene is implicated in the processes underlying sleep need, then changes in its expression can be expected to parallel the predicted time course of sleep need (Fig. 2A). Under control conditions, sleep need is high at sleep onset, which coincides with light onset (ZT0), and during the latter part of the active period (ZT18), whereas sleep need is lowest between ZT6 and ZT12 (i.e., dark onset) (Fig. 2A). Under sleep deprivation conditions, these pronounced sleep–wake-dependent changes are expected to be strongly reduced. Thus, we sleep-deprived mice of the three strains for 6 h, starting at ZT0, ZT6, ZT12, or ZT18, and killed them together with their home-cage controls (n = 9 per strain per time per condition; i.e., total 216). A linear statistical model was first used (see SI Materials and Methods) and identified 2,540 probe sets that were significantly changed at any time point (false-positive rate; false discovery rate <5%). These probe sets were then assessed for time-of-day variation separately in the baseline and sleep-deprivation conditions. Under the baseline condition, ≈8% (2,032; see SI Table 3) of probe sets detected in the brain showed a significant time-of-day pattern of expression (Fig. 2 C and D), with two major, opposite phases of peak expression (between ZT6 and ZT12 and between ZT18 and ZT0, respectively; Fig. 2D). Under the sleep-deprivation condition, only 391 (SI Table 4) of the 2,032 sets at baseline were still significantly affected by time of day, indicating that most others changed according to the sleep–wake distribution, rather than as a result of (or in addition to) a direct circadian effect. Among the 391 transcripts that maintained a significant cycling pattern after sleep deprivation, a large majority reached maximum levels of expression between ZT0 and ZT6 and between ZT12 and ZT18 (Fig. 2 C and D). Among all rhythmic transcripts under control conditions, Homer1a clearly showed the largest amplitude of variation (Fig. 2 B, D, and E), beyond that of all canonical circadian genes. Time course of expression of six representative genes under the two conditions is depicted in Fig. 2E. As predicted by the simulation analysis (Fig. 2A), activity-regulated genes such as Homer1a, Arc, and Egr3 show a high amplitude variation (up to 6-fold for Homer1a), closely following the diurnal sleep–wake distribution, and no, or greatly reduced, variation after sleep deprivation. Although the pattern of expression of circadian genes remains little affected by sleep deprivation, their relative levels can be significantly affected (unchanged for Bmal1, increased for Per2, and decreased for Dbp; Fig. 2E), as we also reported in refs. 17 and 18.
Fig. 2.
Around-the-clock transcriptome analysis of the brain after sleep deprivation indicates that most changes are affected by behavioral states. (A) Simulation of sleep need in the around-the-clock sleep-deprivation experiment. Sleep-need dynamics in the control (gray) were modeled mathematically according to Franken et al. (7): lights on at ZT0, lights off at ZT12. The increase in sleep-need during the four 6-h sleep deprivations (SD) was determined by assuming a saturating exponential increase during wakefulness (red dashed lines). The red solid line connects values of sleep need reached at the end of the SDs. (B and C) SD affects the cycling pattern of gene expression in the brain. Rhythmic transcripts were selected, as outlined in SI Materials and Methods, and their temporal expression patterns were aligned according to phase. (B) Enlargement of heat map for Homer1a gene in control condition. For AK, B6, and D2, the four time points (ZT0, 6, 12, and 18) are represented in triplicate. Green and red represent minimal and maximal expression levels, respectively. (C Left) The 2,032 genes cycling in controls are depicted. The peak time of expression is indicated at left. (Center) The same genes are represented after SD. Note that the rhythmic accumulation of most transcripts is severely blunted after SD. (Right) The 391 probe sets for which cycling expression profiles are not affected by SD (with the peak of expression indicated at right). (D) Cycling genes shown in C are plotted according to their amplitude and phase. Homer1a, outlined in green, is the most rhythmic transcript under control conditions. (E) Temporal expression profiles of representative homeostatic (Left) and circadian (Right) genes in whole brain. Normalized expression signals from microarray experiments are plotted as a function of time for control (solid lines) and SD (dashed lines) mice of each strain. Each point is the mean ± SEM of three pools of three animals.
Comparison of sleep-deprived and control mice at all four time points again revealed that <2% (343 probe sets) of the expressed genes in the brain are up-regulated (>70%, or 249 probe sets) or down-regulated (<30%, or 94 probe sets) by sleep loss (SI Table 5). Significant interaction between condition and time of day was detected for 585 probe sets. Again, the most significantly overexpressed gene after sleep deprivation was Homer1a, followed by those belonging to stress–response and synaptic-plasticity gene groups. The major functional gene groups that reduced their expression after sleep deprivation concern protein synthesis, membrane trafficking, and protein transport (SI Table 5) (12). Real-time quantitative RT-PCR verification for 41 candidate genes (found here and by others) at ZT6 confirmed our microarray findings at this reference time point (SI Table 6).
Cell-Specific Transcriptional Changes Due To Sleep Loss.
Previous studies (12, 19, 20) investigated transcriptomes of different brain structures, rather than the whole brain. In contrast to most other organs, the brain is a highly heterogeneous tissue, with specialized regions and nuclei having very different functional roles. Whole-brain transcriptome, therefore, suffers several potential limitations, including dilution of low copy-number transcripts and missing those with opposing patterns of transcriptional changes among brain regions. On the other hand transcriptome analysis of selected regions also has drawbacks because of our limited knowledge of exactly where functionally significant molecular changes can be expected and because, even in well defined regions, cell types vary greatly. To overcome these limitations, we chose to analyze changes in mRNA profiles of those neurons that are selectively and specifically activated by sleep deprivation. To this end, we used a modified mRNA tagging technique originally established in Caenorhabditis elegans and Drosophila (21–23).
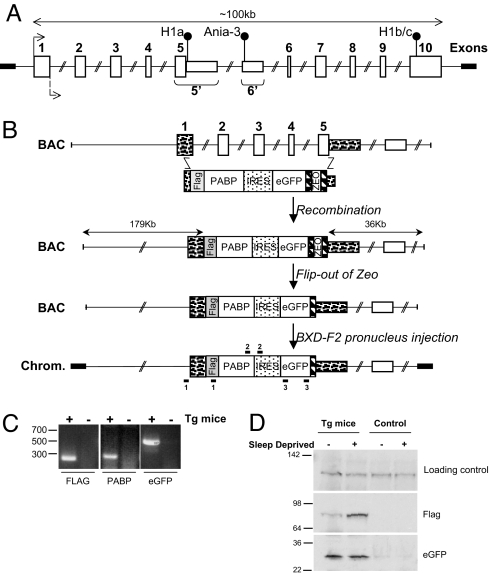
We have shown here that Homer1a is the transcript most consistently increased by enforced wakefulness. The Homer1 gene is therefore a good marker for neuronal populations activated by sleep loss but not restricted to a single structure. We thus generated BAC-based transgenic mice by replacing the first five exons of Homer1 (corresponding to activity-induced Homer1a transcript; Fig. 3A) by a FLAG-tagged poly(A) binding protein 1 (PABP) followed by internal ribosome entry site (IRES)-eGFP (Fig. 3B). Because PABP binds poly(A) tails of mRNAs, affinity-purification of FLAG-tagged PABP proteins from whole-brain lysates is expected to coprecipitate all mRNAs from neurons expressing Homer1a. Seven transgenic lines were obtained, and analysis indicated that at least three lines expressed the transgene at very similar amounts, at both mRNA and protein levels (Fig. 3 C and D). Also, the induction of the transgene by 6-h sleep deprivation was very similar to that of endogenous Homer1a, indicating that our BAC construction contains the regulatory elements for the correct functional expression of Homer1a (Fig. 3D).
Fig. 3.
A transgenic mouse model to analyze neuron-specific gene expression. (A) Schematic representation of the genomic structure of Homer1. The putative transcriptional initiation site is depicted by a bent arrow at the beginning of exon 1; the translational stops for short activity-induced Homer1a (H1a) and Ania-3, as well as for long constitutively expressed Homer1b/c (H1b/c) are indicated by black circles. Intron 5 is here divided into four segments (4.4 kb of Homer1a 3′ UTR, 5.7 kb up to Ania-3, 1.4 kb of _Ania-3_-specific sequence, and 18.8 kb to exon 6). The _Homer1a_-specific exon 5′ extends exon 5 by the intron 5 sequence. The _Ania-3_-specific exon 6′ sits within intron 5 (adapted from ref. 25). (B) General strategy for generating _Homer1a_-PABP transgenic mice. The relative position of the Homer1a gene in the fully sequenced 227,644-bp RP23–262I3 BAC clone (GenBank accession no. AC120347), and the different steps used to introduce a PCR-amplified construction containing PABP cDNA, followed by IRES-eGFP and the zeocine (ZEO) selection cassette flanked by FRT sites (hashed boxes) by BAC recombination in the EL250 bacterial stain (see SI Materials and Methods). (C) Transgenic mice (Tg) were identified by RT-PCR with the primer pairs depicted in B for the presence of the FLAG (primers 1/1), PABP (primers 2/2), and eGFP (primers 3/3). (D) Western blot verification of transgene expression in transgenic mice line 36 (Tg) indicated the presence of the FLAG and eGFP and the up-regulation of the FLAG after sleep deprivation. The loading control is a nonspecific band generated by the GFP antibody.
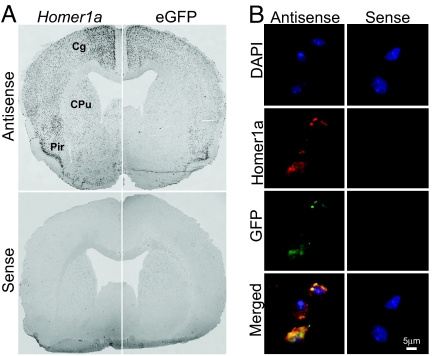
The expression of this construct was also verified by in situ hybridization (Fig. 4A). Although eGFP inserted after the IRES did not result in reliable signal under epifluorescence, riboprobes against Homer1a and eGFP clearly indicated that both are similarly coexpressed in the same brain regions (Fig. 4A). Double fluorescent in situ hybridization (FISH) with riboprobes against Homer1a and eGFP revealed that, at least in two transgenic lines, eGFP was almost exclusively expressed in _Homer1a_-expressing neurons (Fig. 4B), which represented ≈40% of the neurons in the cingulate cortex and 30% in the dorsal striatum (not counted in the hippocampus). All mRNA immunoprecipitations and microarray data presented below are from a single transgenic line (line 36). Sleep recordings in _Homer1_-PABP transgenic mice indicated that they react to sleep deprivation similar to their wild-type littermates (SI Fig. 5). The specificity of this mRNA pull-down method was also verified by quantitative RT-PCR with probes specific for eGFP, FLAG, Homer1a, and Hcrt (a gene expressed only in the lateral hypothalamus, where Homer1a expression is very low). The results indicated that eGFP, FLAG, and the endogenous Homer1a were enriched and induced by sleep deprivation, whereas only trace amounts of Hcrt could be detected (SI Fig. 6).
Fig. 4.
Colocalization of Homer 1a and FLAG-tagged PABP eGFP transcripts. (A) In situ hybridization with Homer1a and eGFP antisense riboprobes indicated that both are expressed in similar brain structures. Cg, cingulate cortex; CPu, caudate putamen; Pir, piriform cortex. (B) Confocal images of FISH analysis. FISH experiments were performed with both Homer1a (red) and eGFP (green) riboprobes at the same time and revealed that almost all positive neurons were double-labeled, indicating the colocalization of the endogenous Homer1a and the transgene. Negative control conditions were obtained with both sense riboprobes.
Immunoprecipitated mRNAs were prepared from _Homer1_-PABP transgenic mice with (n = 6) or without (n = 6) a 6-h sleep deprivation at light onset for gene expression profiling. For comparison, RNA extractions were also made from supernatants (n = 4) after immunoprecipitation and from transgenic whole brains (n = 8). To test for transcriptional changes after sleep deprivation in Homer1_-expressing cells, we proceeded in two steps: (i) we identified probe sets enriched in the pull-down extracts and (ii) among those probe sets, we compared sleep deprivation with control condition in both pull-down (6 vs. 6-chip comparison) and whole-brain (4 vs. 4-chip comparison) extracts. We found that 4,728 probe sets were significantly enriched at 5% false discovery rate when pull-downs were compared with both supernatant and whole-brain extracts (SI Materials and Methods). Again, very few genes were identified (SI Table 7), among which the most significant ones—_Homer1a, Egr2 (NGFI-B), and Fosl2 (Fos-like antigen 2)—were similarly induced after sleep loss in the pull-down and the whole-brain extracts, suggesting that gene expression changes in the pull-down samples recapitulate the most significant changes at the whole-brain level. In addition, several unique immediate-early genes were specifically identified in pull-down samples that might be coinduced with Homer1a, namely prostaglandin–endoperoxide synthase 2 (Ptgs2), junctophilin 3 (Jph3), and neuronal pentraxin 2 (Nptx2). Interestingly, both Jph3 and Nptx2 are activity-induced through either ryanodine receptor-mediated intracellular calcium mobilization (Jph3) or activity-induced AMPA receptor synaptic clustering (Nptx2). Among the very few down-regulated transcripts (SI Table 7), we have identified another activity-induced gene, 4-nitrophenylphosphatase domain and nonneuronal SNAP25-like protein homolog 1 (Nipsnap1), suggesting that plasticity genes can be up- or down-regulated by sleep deprivation.
Discussion
The results presented here demonstrate that 6 h of sleep deprivation, which importantly impacts sleep physiology and behavior, results in only minimal changes in brain transcriptional adaptation. As reported for changes in delta power (7), we also showed here that sleep loss-induced transcriptional changes are largely affected by genetic background. As opposed to other studies that did not take genetic background into account and did not contrast their findings to peripheral tissue (12, 15, 20), our results indicate that only a few genes reliably change expression after sleep deprivation. The surprising finding that sleep loss induced a larger number of changes in liver than in brain suggests either that sleep deprivation might have a specific impact on the liver or that the brain might be protected against major transcriptional changes.
Another important aspect of the present study is the interaction between the homeostatic and circadian processes. Although we did not sleep-deprive the animals under constant conditions, and therefore the direct and indirect effects of light, for instance, on gene expression could not be accounted for, we have shown that a large majority (>80%) of changes in gene expression were driven by the prior sleep–wake history.
We also adopted, and further developed, a reliable mRNA tagging technique to investigate gene expression changes in neurons. This technique can be used to evaluate different neuronal subpopulations without the burden of sampling several structures or using labor-intensive laser microdissection to isolate neurons. The results of this technique confirmed that sleep loss-induced transcriptional changes occur for very few genes, among which Homer1a remains the most specific.
In addition to Homer1a, we identified overexpression of other genes involved in synaptic plasticity, but only Egr2 and Homer1a were found to consistently change across experiments. Others reported overexpression for a number of plasticity-related genes, and these observations are commonly used in support of a functional role for sleep in plasticity. Because the expression of most plasticity-related genes was not reliably changed, our findings do not support such a general conclusion and instead suggest that the molecular mechanisms might not be identical for most plasticity-regulated genes. In this context it is important to note that Nipsnap1, one of the proposed plasticity genes (24), is actually down-regulated after sleep deprivation in our mRNA tagging experiment.
Three different genes in mammals encode Homer proteins. Homer1 encodes constitutively expressed long-form proteins, whereas short-form Homer1a is activity-induced (25). Homer1 long-form proteins dimerize and interact with metabotropic glutamate receptors and increase calcium from intracellular stores. Short-form proteins, which lack the dimerization domain, function as natural activity-dependent dominant negative forms that regulate the scaffolding and signaling capabilities of the long forms and reduce glutamate-induced intracellular calcium release (26, 27). We have recorded sleep, and the response to a 6-h sleep deprivation, in Homer1 (all forms) mutant mice but found a very similar pattern compared with their wild-type littermates (data not shown). This finding could be expected due to the fact that, because Homer1a functions as a dominant negative form of the long forms, constitutive loss of all isoforms might not result in any specific sleep phenotype.
Conceptually, spontaneous or enforced wakefulness represents a stressor activating a series of stress–response mechanisms of the organism, which, at the transcriptional level, could be translated into the induction of genes such as heat shock proteins in most tissues. However, unlike in other organs, brain-specific stress–response pathways are primarily triggered by glutamate. Glutamate is the major excitatory neurotransmitter in the central nervous system and acts through either ionotropic or metabotropic receptors (mGluRs). Long Homer1 tetramers bind group I mGluRs and inositol 1,4,5-triphosphate receptors, thus enabling efficient calcium release from intracellular stores, whereas monomeric Homer1a competitively disrupts synaptic glutamatergic signaling complexes to reduce glutamate-induced intracellular calcium release (26, 27). Homer1 also activates ryanodine receptors and l-type calcium channels (28, 29). Interestingly, Jph3, which was identified by our mRNA-tagging strategy as being up-regulated by sleep deprivation, has been shown to play a major role in ryanodine receptor-mediated, calcium-induced opening of small-conductance, calcium-activated potassium (SK) channels (28, 30). SK channels are responsible for the generation of slow afterhyperpolarizations in neurons of the nucleus reticularis thalami and thus contribute to the EEG slow waves characteristic of NREM sleep (31).
According to Tononi and Cirelli (3), plastic processes occurring during wakefulness result in increased synaptic strength, whereas the role of sleep is to downscale synaptic strength to a basal level. Homer1a transcription is rapidly up-regulated in neurons in response to synaptic activity induced by long-term potentiation, seizure, inflammation, stimulant drugs, or even selectively in the hippocampus of rodents by exploratory behavior (32, 33). In this context, Homer1a, by buffering intracellular calcium and disassembling synaptic glutamatergic signaling complexes, could play a pivotal role in synaptic downscaling. Because Homer1a and the other genes identified here (Egr2, Fosl2, Ptgs2, Jph3, and Nptx2) are all induced by stressful conditions such as seizure, stroke and hypoxia, and inflammation, an alternative, complementary view could be that they play a primary brain-protective or recovery role. This view is also of relevance for the etiology of neuropsychiatric disorders because it is increasingly recognized that stress is implicated in many of such disorders (34). Both environmental and pharmacological stressors up-regulate Homer1a mRNA in key structures involved in higher brain functions (35): the same structures in which Homer1a is up-regulated after sleep deprivation. It has also been shown that overexpression of Homer1a after inflammation, seizure, and psychostimulant or antipsychotic drug use plays a major role in neuroprotection (28, 35). It is tempting to relate the dramatic improvement in depression in humans after sleep-deprivation (36) to the sleep deprivation-induced up-regulation of Homer1a. In conclusion, converging evidence strongly implicates Homer1a as a brain-coping marker against stressors, and our findings suggest that Homer1a might represent the molecular link between sleep, cognition, and neuropsychiatric disorders.
Materials and Methods
Animal Handling.
All experiments were performed in accordance with the protocols approved by the Ethical Committee of the State of Vaud Veterinary Office, Switzerland. Sleep-deprivation and sleep-recording procedures are described in SI Materials and Methods.
cRNA Preparation, cDNA Microarray Hybridization, and Real-Time RT-PCR.
For the first experiment, we isolated total RNA from whole brain and liver by using the RNAXEL kit (Eurobio), treated the RNA with DNase, and cleaned it using RNeasy columns (Qiagen). Equal quantities of total RNA from three individual mice of each strain were pooled in triplicate (nine mice of each strain in each condition). A hybridization mixture containing 15 μg of biotinylated cRNA was hybridized to GeneChip Mouse Expression Set 430. Chips were washed, scanned, and analyzed with Affymetrix GeneSpring software.
For the around-the-clock microarray experiment, RNA from whole brain was isolated and purified with the RNeasy Lipid Tissue Midi kit (Quiagen) and DNase-treated. All RNA quantities were assessed with a NanoDrop ND-1000 spectrophotometer, and the quality of RNA was controlled on Agilent 2100 bioanalyzer chips. Equal amounts of total RNA were pooled from three mice within each of the 24 experimental groups (three strains, two conditions, 4 ZT = 24; in triplicate: 24 · 3 = 72 chips). Three micrograms of each of these pools were used to perform the chip array experiment, according to the Affymetrix Gene Expression procedure. Twelve micrograms of biotinylated cRNA from each sample were fragmented and hybridized to GeneChip Mouse 430 2.0 arrays, according to standard procedures. Microarray analyses and qPCR verifications were performed as reported in SI Materials and Methods.
Immunoblot and in Situ Hybridization.
Total protein extract was prepared with RIPA lysis buffer. Protein concentration was calculated by using the bicinchoninic acid assay (Pierce) with BSA as a standard. Eighty micrograms of each fraction were analyzed by SDS/PAGE, followed by Western blotting using antibodies as follows: mouse anti-tubulin 1/1,000 (Santa Cruz), goat anti-Homer1a 1/200 (Santa Cruz), mouse anti-Flag M2 1/300 (affinity-purified; Sigma), and rabbit anti-GFP 1/2,500 (AbCam). Secondary antibodies were all coupled with HRP, except for the anti-goat antibody, which was IRDye800-conjugated for Lycor analysis.
In situ hybridizations with coronal cryosections of 12 μm were performed according to Allen Brain Atlas protocols (enzymatic BCIP/NBT revelation) (37). All reagents and solutions were purchased and prepared based on Eurexpress II in situ hybridization consortium instructions. GFP and Homer1a riboprobes were synthesized by in vitro transcription on a linearized pGEM-Easy vector (Promega) containing the corresponding sequences. The cDNA insert of this plasmid was generated by RT-PCR from mouse brain RNA, using the following primers: Homer1a forward, 5′-GCTGTCAGAAGCTTAGGATGTG-3′; Homer1a reverse, 5′-AAAGTGCAGAAAGTCCAGCAGC-3′; GFP forward, 5′-GAGCTGGACGGCGACGTAAACG-3′; and GFP reverse, 5′- AGGACCATGTGATCGCGCTTCTC-3′.
FISH was performed as described in ref. 38, using anti-DIG-POD 1/600 (Roche), anti-FLU-AP 1/100 (Roche), and SA-Alexa 488 (Molecular Probes) and counterstained with DAPI (Sigma).
Transgenic and mRNA Tagging.
Transgenic mice were generated as described in Fig. 3. See SI Materials and Methods for details.
Supplementary Material
Supporting Information
Acknowledgments
We thank K. Harshman, A. Paillusson, and M. Bueno for assistance in microarray and real-time RT-PCR analyses at the Lausanne DNA Array Facility; P. Descombes, M. Docquier, D. Chollet, and C. Delucinge for assistance in microarray and real-time RT-PCR analyses at the Geneva Genomics Platform, National Center for Competence in Research Frontiers in Genetics; S. Excoffier for help in the transgenic construction; P. Seeburg, M. Schwarz (Max Planck Institute, Heidelberg, Germany), and P. Worley (Johns Hopkins School of Medicine, Baltimore, MD) for providing Homer1 mutant mice; and A. Vassali for constructive discussions. This work was supported by the Swiss National Science Foundation and the State of Vaud (M.T.) and in part by National Institutes of Mental Health Grant MH67752 (to P.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9444).
References
- 1.Dijk DJ, Franken P. In: Principles and Practice of Sleep Medicine. Kryeger TH, Roth TH, Dement W, editors. Philadelphia: Saunders; 2005. pp. 418–434. [Google Scholar]
- 2.Krueger JM, Obal F. J Sleep Res. 1993;2:63–69. doi: 10.1111/j.1365-2869.1993.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 3.Tononi G, Cirelli C. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Huber R, Ghilardi MF, Massimini M, Ferrarelli F, Riedner BA, Peterson MJ, Tononi G. Nat Neurosci. 2006;9:1169–1176. doi: 10.1038/nn1758. [DOI] [PubMed] [Google Scholar]
- 5.Marshall L, Helgadottir H, Molle M, Born J. Nature. 2006;444:610–613. doi: 10.1038/nature05278. [DOI] [PubMed] [Google Scholar]
- 6.Molle M, Marshall L, Gais S, Born J. Proc Natl Acad Sci USA. 2004;101:13963–13968. doi: 10.1073/pnas.0402820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franken P, Chollet D, Tafti M. J Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Dongen HP, Baynard MD, Maislin G, Dinges DF. Sleep. 2004;27:423–433. [PubMed] [Google Scholar]
- 9.Tucker AM, Dinges DF, Van Dongen HP. J Sleep Res. 2007;16:170–180. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 10.Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirelli C, Gutierrez CM, Tononi G. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 12.Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack A. Physiol Genomics. 2007;31:441–457. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 13.Nelson SE, Duricka DL, Campbell K, Churchill L, Krueger JM. Neurosci Lett. 2004;367:105–108. doi: 10.1016/j.neulet.2004.05.089. [DOI] [PubMed] [Google Scholar]
- 14.Huber R, Tononi G, Cirelli C. Sleep. 2007;30:129–139. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- 15.Terao A, Steininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 16.Wisor JP, Morairty SR, Huynh NT, Steininger TL, Kilduff TS. Neuroscience. 2006;141:371–378. doi: 10.1016/j.neuroscience.2006.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franken P, Thomason R, Heller HC, O'Hara BF. BMC Neurosci. 2007;8:87. doi: 10.1186/1471-2202-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisor JP, O'Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, Edgar DM, Franken P. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirelli C, Faraguna U, Tononi G. J Neurochem. 2006;98:1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- 20.Terao A, Greco MA, Davis RW, Heller HC, Kilduff TS. Neuroscience. 2003;120:1115–1124. doi: 10.1016/s0306-4522(03)00395-6. [DOI] [PubMed] [Google Scholar]
- 21.Roy PJ, Stuart JM, Lund J, Kim SK. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 22.Kunitomo H, Uesugi H, Kohara Y, Iino Y. Genome Biol. 2005;6:R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Edenberg HJ, Davis RL. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh K, Takeuchi M, Oda Y, Deguchi-Tawarada M, Sakamoto Y, Matsubara K, Nagasu T, Takai Y. Genes Cells. 2002;7:187–197. doi: 10.1046/j.1356-9597.2001.00505.x. [DOI] [PubMed] [Google Scholar]
- 25.Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worley PF, Zeng W, Huang G, Kim JY, Shin DM, Kim MS, Yuan JP, Kiselyov K, Muallem S. Cell Calcium. 2007;42:363–371. doi: 10.1016/j.ceca.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao B, Tu JC, Worley PF. Curr Opin Neurobiol. 2000;10:370–374. doi: 10.1016/s0959-4388(00)00087-8. [DOI] [PubMed] [Google Scholar]
- 28.Kakizawa S, Kishimoto Y, Hashimoto K, Miyazaki T, Furutani K, Shimizu H, Fukaya M, Nishi M, Sakagami H, Ikeda A, et al. EMBO J. 2007;26:1924–1933. doi: 10.1038/sj.emboj.7601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto K, Sakagami Y, Sugiura S, Inokuchi K, Shimohama S, Kato N. Eur J Neurosci. 2005;22:1338–1348. doi: 10.1111/j.1460-9568.2005.04278.x. [DOI] [PubMed] [Google Scholar]
- 30.Moriguchi S, Nishi M, Komazaki S, Sakagami H, Miyazaki T, Masumiya H, Saito SY, Watanabe M, Kondo H, Yawo H, et al. Proc Natl Acad Sci USA. 2006;103:10811–10816. doi: 10.1073/pnas.0509863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blethyn KL, Hughes SW, Toth TI, Cope DW, Crunelli V. J Neurosci. 2006;26:2474–2486. doi: 10.1523/JNEUROSCI.3607-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tappe A, Klugmann M, Luo C, Hirlinger D, Agarwal N, Benrath J, Ehrengruber MU, During MJ, Kuner R. Nat Med. 2006;12:677–681. doi: 10.1038/nm1406. [DOI] [PubMed] [Google Scholar]
- 33.Kato A, Ozawa F, Saitoh Y, Fukazawa Y, Sugiyama H, Inokuchi K. J Biol Chem. 1998;273:23969–23975. doi: 10.1074/jbc.273.37.23969. [DOI] [PubMed] [Google Scholar]
- 34.Southwick SM, Vythilingam M, Charney DS. Annu Rev Clin Psychol. 2005;1:255–291. doi: 10.1146/annurev.clinpsy.1.102803.143948. [DOI] [PubMed] [Google Scholar]
- 35.Szumlinski KK, Kalivas PW, Worley PF. Curr Opin Neurobiol. 2006;16:251–257. doi: 10.1016/j.conb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Giedke H, Schwarzler F. Sleep Med Rev. 2002;6:361–377. [PubMed] [Google Scholar]
- 37.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 38.Ishii T, Omura M, Mombaerts P. J Neurocytol. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information