Roles of LAP2 Proteins in Nuclear Assembly and DNA Replication: Truncated LAP2β Proteins Alter Lamina Assembly, Envelope Formation, Nuclear Size, and DNA Replication Efficiency in Xenopus laevis Extracts (original) (raw)
Abstract
Humans express three major splicing isoforms of LAP2, a lamin- and chromatin-binding nuclear protein. LAP2β and γ are integral membrane proteins, whereas α is intranuclear. When truncated recombinant human LAP2β proteins were added to cell-free Xenopus laevis nuclear assembly reactions at high concentrations, a domain common to all LAP2 isoforms (residues 1–187) inhibited membrane binding to chromatin, whereas the chromatin- and lamin-binding region (residues 1–408) inhibited chromatin expansion. At lower concentrations of the common domain, membranes attached to chromatin with a unique scalloped morphology, but these nuclei neither accumulated lamins nor replicated. At lower concentrations of the chromatin- and lamin-binding region, nuclear envelopes and lamins assembled, but nuclei failed to enlarge and replicated on average 2.5-fold better than controls. This enhancement was not due to rereplication, as shown by density substitution experiments, suggesting the hypothesis that LAP2β is a downstream effector of lamina assembly in promoting replication competence. Overall, our findings suggest that LAP2 proteins mediate membrane–chromatin attachment and lamina assembly, and may promote replication by influencing chromatin structure.
Keywords: nuclear envelope, chromatin structure, prereplication complex, emerin, MAN
The nuclear envelope generates a unique structural and functional environment for chromosomes inside the nucleus. Molecules move through the nuclear envelope via nuclear pore complexes (NPCs)1 which regulate nucleocytoplasmic transport while permitting the passive diffusion of small molecules (<40 kD) and ions (reviewed by Ohno et al., 1998). The inner nuclear membrane and NPCs are anchored to the lamina, which is a polymeric network of nuclear-specific intermediate filament proteins named lamins (reviewed by Hutchison et al., 1994; Gant and Wilson, 1997). There are two major types of lamins, B type and A/C type, which are encoded by different genes. B lamins remain membrane-associated throughout the cell cycle, predominantly through their association with lamin-binding membrane proteins (see below), whereas lamins A/C become soluble and are dispersed throughout the cytosol during mitosis. During interphase, B type lamins have also been detected immunologically inside the nucleus at sites of DNA replication (Moir et al., 1994). Biochemical studies suggest that the tail domains of lamins can bind to DNA and core histones (Burke, 1990; Glass et al., 1993; Taniura et al., 1995; reviewed in Gant and Wilson, 1997).
The lamina is a key structural element of the nucleus. When nuclei are assembled in cell-free extracts immunodepleted of soluble lamins, the lamin-depleted nuclei fail to undergo DNA replication (Newport et al., 1990; Meier et al., 1991; Jenkins et al., 1993), suggesting that lamina assembly is linked to the assembly or function of replication complexes. However, this link is likely to be indirect. When nuclei are exposed to dominant negative mutant lamin A proteins, DNA replication sites (Mills et al., 1989; Hozak et al., 1994) become physically and functionally disrupted (Spann et al., 1997). However, dominant lamin B mutants have different effects: nuclei with a preexisting lamina can remain replication-competent even when their lamina is gradually dissolved by the mutant lamin B proteins (Ellis et al., 1997). In both cases, the mutant lamins are thought to sequester depolymerized wild-type lamins and prevent them from recycling (Schmidt et al., 1994). It was not clear why DNA replication would depend, either initially or in an ongoing capacity, on the integrity of the lamina. Furthermore, lamin assembly may not be essential. New results show that plasmid DNA can be fully replicated in vitro if it is incubated first in Xenopus laevis cytosol and subsequently in concentrated nucleosol, suggesting that high concentrations of factors inside the nucleus, rather than nuclear structure per se, are essential for replication competence (Walter et al., 1998).
The inner nuclear membrane contains several unrelated resident proteins that bind to lamins (reviewed by Gerace and Foisner, 1994; Gant and Wilson, 1997), including the lamin B receptor (LBR, also known as p58; Worman et al., 1988, 1990), three isoforms of the lamina-associated polypeptide-1 (LAP1; Martin et al., 1995), and several isoforms of LAP2 (Foisner and Gerace, 1993; Harris et al., 1994, 1995; Berger et al., 1996). Because LBR, the C isoform of LAP1, and α and β isoforms of LAP2 are phosphorylated during mitosis, these proteins are postulated to play structural roles that must be modified for nuclei to disassemble at mitosis (Simos and Georgatos, 1992; Foisner and Gerace, 1993; Ye and Worman, 1994; Martin et al., 1995; Dechat et al., 1998). LAP1, LAP2β, and LBR do not appear to associate with each other. Instead, LBR and LAP1 form separate complexes, each of which has a distinct protein kinase (Simos and Georgatos, 1992; Nikolakaki et al., 1996; Maison et al., 1997). It is not known if LAP2 proteins associate with a kinase. Two proteins related to LAP2, named emerin (Bione et al., 1994; Manilal et al., 1996; Nagano et al., 1996) and MAN (Paulin-Levasseur et al., 1996; H. Worman, personal communication), also reside at the inner nuclear membrane. Loss of emerin causes Emery-Dreifuss muscular dystrophy, a rare form of muscular dystrophy in humans (see Bione et al., 1994; Gant and Wilson, 1997), an effect which has not yet been explained at the functional level.
LBR and LAP2 both bind to chromatin in vitro, and therefore are both in a position to directly mediate chromosome attachment to the inner nuclear membrane. The chromatin partner for LBR is Hp1, a chromodomain protein associated with repressive (transcriptionally silent) chromatin structure (Ye and Worman, 1996; Ye et al., 1997; reviewed by Lamond and Earnshaw, 1998). Based on immunoprecipitation and liposome reconstitution experiments, LBR appears to play a major role in targeting membranes to chromatin (Pyrpasopoulou et al., 1996). In contrast, immunodepletion of LAP2 had little effect on membrane targeting to chromatin in vitro (Pyrpasopoulou et al., 1996).
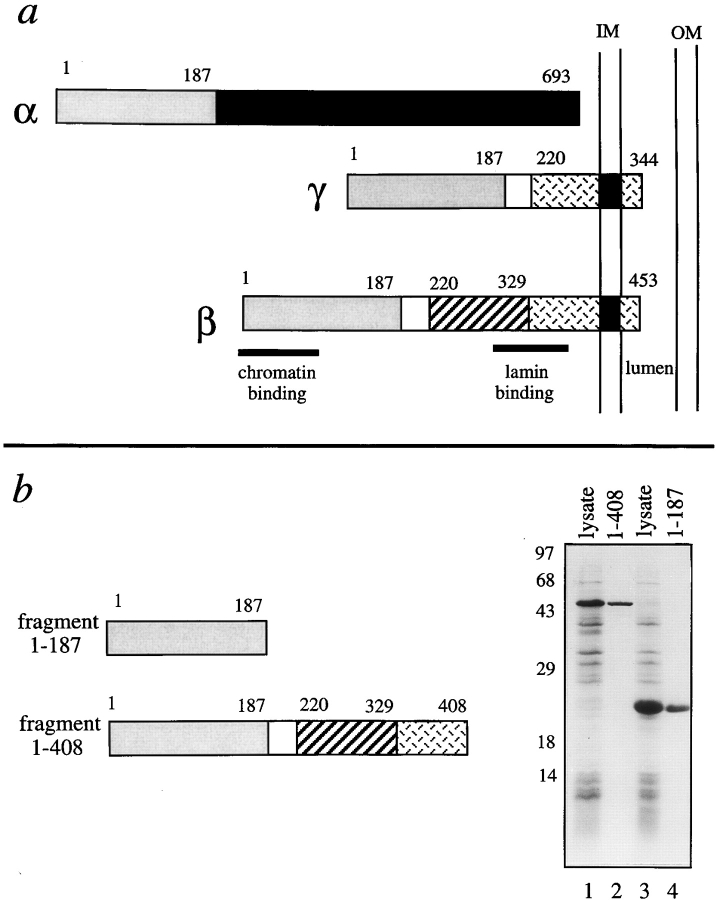
We focused on the role of LAP2 in nuclear assembly, structure, and function. LAP2, which was originally discovered, cloned, and characterized in rat, binds in vitro to lamin B1, and to mitotic HeLa chromosomes with an affinity of 40–80 nM (Foisner and Gerace, 1993; Furukawa et al., 1995). Cloning of human and mouse LAP2 cDNAs showed that there are three major alternatively spliced isoforms, named LAP2α (75 kD), β (51 kD), and γ (39 kD; Fig. 1 a; Harris et al., 1994, 1995), and four minor isoforms (Berger et al., 1996). LAP2 is highly conserved among mammals; for example, human LAP2β is 91% identical to rat LAP2. All LAP2 isoforms share a common NH2-terminal domain, which is encoded by three exons in humans. Beyond this NH2-terminal region (residues 1–187), LAP2α differs from all other LAP2 isoforms; α has no transmembrane anchor, whereas LAP2β and γ (and presumably most other β-related minor isoforms) are anchored to the inner nuclear membrane by a single predicted transmembrane span near the COOH terminus. The α isoform is located inside the nucleus, where it associates tightly with intranuclear structures and cofractionates with the detergent-resistent lamina–matrix fraction (Dechat et al., 1998). The existence of both membrane-bound and soluble isoforms of LAP2 suggests that these isoforms serve distinct functions. Proliferating cells consistently express LAP2β, whereas γ and α (but not β) are expressed at high levels in nonproliferating cerebellar tissue (Ishijima et al., 1996). Expression of LAP2 isoforms is also regulated during spermatogenesis; in mature rat sperm the only detectable LAP2 isoform is α (Alsheimer et al., 1998). We have now cloned and sequenced three LAP2 cDNAs from a Xenopus oocyte library, demonstrating the presence of multiple LAP2 isoforms in Xenopus eggs.
Figure 1.
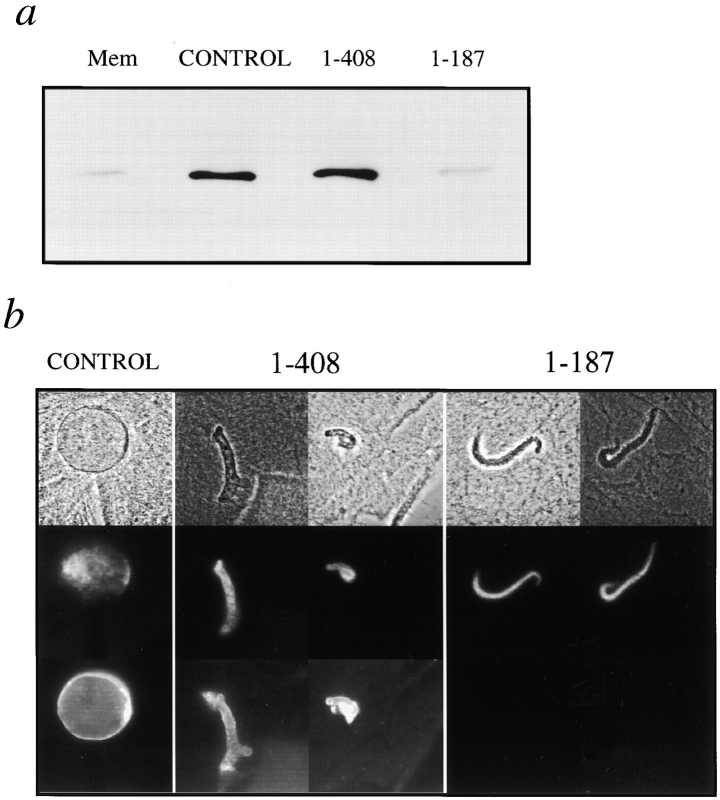
Human LAP2 proteins. (a) Diagrams of human LAP2α, β, and γ. All three LAP2 isoforms share the same first 187 residues. Regions of rat LAP2β that are reported to bind to chromatin (residues 1–85) and to lamins (residues 298–370) in vitro are indicated (see Furukawa et al., 1998). (b) Recombinant human LAP2 fragments used in these studies, shown schematically, and before and after purification. Lysates of bacteria expressing fragment 1–408 (lane 1), fragment 1–187 (lane 3), or 5 μg of each purified fragment (1–408, lane 2; 1–187, lane 4) were subjected to SDS-PAGE (12% gel) and proteins were visualized by Coomassie blue stain.
To investigate the function of LAP2, we tested the effect of two recombinant polypeptides derived from human LAP2β, on nuclear assembly in cell-free extracts of Xenopus eggs. Xenopus egg extracts are an efficient system for assembling replication-competent nuclei in vitro (Wilson and Wiese, 1996). Xenopus extract components also interact efficiently with nuclear proteins from other species; for example, Xenopus membranes readily incorporate into rat liver nuclei, Xenopus extracts support nucleocytoplasmic transport into all exogenous nuclei tested (e.g., Newmeyer et al., 1986), and Xenopus extracts can assemble nuclei around isolated mitotically condensed mammalian chromosomes (e.g., Lawlis et al., 1996). Our results, presented here, show that human LAP2 fragments are functional in the Xenopus extracts, and inhibit nuclear assembly in distinct ways. Our most unexpected finding was that LAP2β fragment 1–408 can influence the efficiency of DNA replication.
Materials and Methods
Reagents and Solutions
Membrane wash buffer (MWB) consisted of 250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2, 50 mM Hepes, pH 8.0, 1 mM dithiothreitol, 0.5 mM ATP, 1 μg/ml aprotinin, and 1 μg/ml leupeptin. Sonication buffer consisted of 50 mM NaPO4, pH 8.0, and 300 mM NaCl. Protease inhibitors benzamidine (5 mM final concentration; Sigma Chemical Co.), PMSF (0.5 mM final concentration; Sigma Chemical Co.), and pepstatin A (1 μg/ml final concentration; Sigma Chemical Co.) were included in the sonication buffer during sonication, but were not added in subsequent steps. Stop buffer consisted of 80 mM Tris-HCl, pH 8.0, 8 mM EDTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, and 0.2% Bromophenol blue. TBS consisted of 100 mM Tris-HCl, pH 7.5, plus 0.9% (wt/vol) NaCl. WGA (Sigma Chemical Co.) was kept frozen as a 10 mg/ml stock at −80°C. Aphidicolin (Sigma Chemical Co.; catalog number A-0781) was kept as a 2.5 mg/ml stock in DMSO at −20°C.
In Vitro Nuclear Assembly Reactions and Import Assays
Membrane and cytosol fractions were prepared from unactivated Xenopus eggs as previously described (Newmeyer and Wilson, 1991; Boman et al., 1992). Demembranated Xenopus sperm chromatin was also prepared as previously described (Lohka and Masui, 1983; Newmeyer and Wilson, 1991). Chromatin, at a final concentration of ∼40,000 sperm/μl, was stored at −80°C. For nuclear assembly reactions, 2 μl membranes (∼30 mg protein/ml), 20 μl cytosol (∼25–30 mg protein/ml, supplemented with 10 mM phosphocreatine, 1 mM ATP, and 50 μg/ml creatine phosphokinase as an ATP regenerating system), and 1 μl demembranated sperm chromatin were mixed on ice and transferred to 22–24°C to initiate nuclear assembly. All reactions were done using components that had been frozen and thawed once. For reactions that contained recombinant LAP2 fragments, 1 μl of LAP2 protein (in MWB) was added to mixed cytosol and membranes to yield the indicated final concentration of LAP2 fragment. Chromatin was added, and reactions were mixed again and transferred to 22– 24°C to initiate assembly.
To assay for nuclear import, rhodamine-labeled nucleoplasmin was prepared according to Newmeyer et al. (1986), and added to nuclei after 2 h of assembly in the presence or absence of LAP2 fragment 1–408. Nuclei were imaged by epifluorescence microscopy 30 min later (time = 2.5 h). As a negative control for import, WGA (final concentration, 1 mg/ ml) was added 5 min before adding fluorescent nucleoplasmin. WGA inhibits active transport by binding to _O_-GlcNAc-modified nucleoporins at the NPC (see Finlay and Forbes, 1990, and references therein).
Preparation of Recombinant LAP2 Proteins
Escherichia coli cells, strain BL21(DE3)pLysS (Novagen, Inc.), were transformed with the pET-23a expression vector (Novagen, Inc.) containing inserts coding for either residues 1–408 or residues 1–187 of human LAP2β, or residues 1–164 of Xenopus LAP2 (see below). We followed the convention of numbering amino acids that excludes the initiating methionine, consistent with previous papers (Foisner and Gerace, 1993; Harris et al., 1994). The pET-23a expression vector adds a His tag (Leu-Glu-His6) to the COOH terminus of the expressed protein. Thus, LAP2β fragment 1–408 is a 417–amino acid, 46.49-kD protein, and LAP2 fragment 1–187 is a 196–amino acid, 21.716-kD protein. To produce each recombinant protein, an overnight culture of a single colony was diluted 1:60 in fresh media. Upon reaching an OD600 of ∼0.6, protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h, and the bacteria were pelleted by centrifugation (6,000 g for 15 min at 4°C). The pellet was frozen in liquid N2 and stored at −80°C. To purify each recombinant protein, the pellet was resuspended in sonication buffer, subjected to pulse sonification, and centrifuged (20,000 g for 20 min at 4°C). The supernatant was applied to a Ni-NTA–agarose column (Qiagen, Inc.), which was washed successively with 10 column volumes each of sonication buffer and sonication buffer plus 10 mM imidazole. Recombinant His-tagged proteins were eluted with sonication buffer containing 100 mM imidazole. The proteins were concentrated and desalted using Centricon-30 units (Amicon, Inc.). Small aliquots were frozen in liquid N2 and stored at −80°C; the thawed proteins were stable at 4°C for at least a week. Mock purifications were done in parallel using uninduced bacteria to provide a control for potential nonspecific effects due to either imidazole or bacterial proteins. However, in no case did the effect of mock-purified proteins differ from that of buffer alone.
Light Microscopy and Photography
Aliquots of assembly reactions were fixed in MWB containing 3.7% formaldehyde, 20 μg/ml Hoechst 33342 (a DNA stain; Calbiochem Corp.). The samples were observed using a Nikon Microphot fluorescence microscope and photographed with Kodak Tri-X Pan 400 film. In some cases samples were imaged using a Photometrics SenSys cooled CCD camera, and images were processed and printed using IPLabSpectrum software.
Transmission Electron Microscopy
Samples for electron microscopy were fixed for 30 min on ice in 1.5% (vol/ vol) glutaraldehyde and 1% (vol/vol) paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4. Samples were pelleted for 1 min in an Eppendorf centrifuge at 4°C, and the chromatin/nuclear pellet was rinsed in cacodylate buffer. Pellets were postfixed for 30 min at 4°C in 1% reduced osmium tetroxide, dehydrated, and embedded in Spurr's medium. Samples were sectioned (90-nm sections) and poststained in uranyl acetate followed by lead citrate. Electron micrographs of thin sections were taken on a TEM10 microscope (Carl Zeiss, Inc.) at 60 or 80 kV.
In Vitro Replication Assays
DNA replication was assayed by incorporation of α[32P]dCTP (see Powers et al., 1995). In brief, 1 μl of α[32P]dCTP (Redivue, 3,000 Ci/mmol; Nycomed Amersham) was added to 24-μl nuclear assembly reactions (20 μl cytosol, 2 μl membranes, 1 μl chromatin [stock concentration ∼40,000/μl], plus 1 μl buffer or recombinant LAP2β polypeptides). As a negative control, aphidicolin was added at a final concentration of 50 μM; this agent inhibits the activity of DNA polymerase α. Alternatively, independent negative control reactions were made 1 mM in GTPγS to inhibit nuclear membrane formation (Boman et al., 1992) and, indirectly, DNA replication. After 3 h, samples were combined with an equal volume of stop buffer, proteinase K (Boehringer Mannheim GmbH) was added to 1 mg/ ml final concentration, and samples were incubated at 37°C for 2 h. To detect incorporated α[32P]dCTP, the protease-digested samples were mixed thoroughly by pipetting to ensure homogeneity, and 5-μl aliquots were electrophoresed through 0.8% agarose gels. Note that all reactions contained equal numbers of nuclei, and samples were processed without pelleting steps that might cause loss of material. Gel loading was monitored by ethidium stain contained in the gel. Dried gels were exposed to x-ray film, and signals quantitated with scanning densitometry using the Microcomputing Imaging Device (Imaging Research Inc.). In one experiment, the signal was quantitated by both phosphorimaging and densitometry, with the same results.
To measure the time course of replication, 10-μl aliquots were removed every 15 min from 200-μl reactions (190 μl crude extract, 10 μl chromatin, 5 μl [32P]dCTP, plus 6 μl of buffer or purified LAP2 fragment 1–408), digested with proteinase K, and the DNA was separated on agarose gels and quantitated by PhosphorImager as described above.
Bromodeoxyuridine (BrdU) Density Substitution
Density substitution experiments were done essentially as described by Hua and Newport (1998), using both freshly prepared crude nuclear assembly extracts (10,000 g cytoplasmic fraction), and high speed fractionated, frozen, reconstituted extracts, with the same results. Nuclei were assembled for 90–120 min in the presence or absence of 3.3 μM fragment 1–408. Crude reactions contained 60 μl cytoplasm with 2,000 sperm per μl, plus 0.5 mM BrdU (Sigma Chemical Co.), 0.5 mM MgCl2, and 0.2 μCi of α[32P]dCTP per μl of extract. Fractionated/reconstituted reactions contained 60 μl cytosol, 12 μl membranes, and a total of 100,000 sperm chromatin, plus 0.2 μCi/μl α[32P]dCTP and 0.5 mM BrdU. Reactions were stopped by adding 1 ml ice cold buffer A (50 mM KCl, 50 mM Hepes-KOH, pH 7.4, 5 mM MgCl2, and 1 mM DTT), incubated on ice for 5 min, centrifuged 5 min at 16,000 g in a microfuge, resuspended in 100 μl buffer A, made 0.5% in SDS and 0.4 mg/ml in proteinase K, and digested for 2 h at 37°C. DNA was then extracted three times with phenol-chloroform, once with chloroform, and ethanol precipitated using 0.3 M sodium acetate. Each ethanol pellet was resuspended in 100 μl TE, mixed with 12.7 ml of 1.75 g/ml CsCl, loaded into a Beckman 16 × 76 mm Quick-Seal Tube, and centrifuged 45 h at 30,000 rpm at 20°C in a Beckman Ti-70.1 rotor. 46– 50 fractions (250–300 μl each) were collected by needle puncture from the bottom of each tube. To quantitate radioactivity, an aliquot of each fraction was counted by liquid scintillation (Beckman LS-7000). The refractive index of each fraction was measured using a refractometer (Bausch & Lomb Inc.), and converted to density using the equation: d (in units of g/ml) = 0.99823 × d 20 20, where d 20 20is the specific gravity of solution at 20°C. Values for d 20 20for cesium chloride at each index of refraction (n) are provided in the CRC Handbook (Weast, 1967).
Immunoblotting
To detect lamin accumulation by immunoblotting, nuclei were assembled for 3 h in the presence or absence of human LAP2β fragments, diluted with 100 μl MWB, and then pelleted at top speed in an Eppifuge for 1 min and washed with 200 μl MWB. The washed nuclear pellet was resuspended in SDS-sample buffer, subjected to SDS-PAGE (12% gel), and proteins transferred to Immobilon PVDF membrane (Millipore Corp.). The Immobilon was blocked with 5% dry milk in TBS 0.1% Tween-20 (TBS-Tw) for 30 min, rinsed briefly in TBS-Tw, and incubated overnight at 4°C with either of two mAbs: antibody 46F7 (1:750 dilution in TBS-Tw; a kind gift from Prof. Georg Krohne; Lourim and Krohne, 1993), which is specific for Xenopus lamin B3 (formerly known as lamin Liii), the major lamin found in Xenopus eggs (Lourim et al., 1996), or a monoclonal directed against human lamin B (Calbiochem; final concentration 100 μg/ml in TBS-Tw). Blots were rinsed six times (5 min each) with TBS-Tw and then incubated for 1 h at 22–24°C with HRP-conjugated anti–mouse secondary antibody in TBS-Tw (Nycomed Amersham). The blots were washed again (six times for 5 min) and developed using enhanced chemiluminescence (ECL) reagents (Nycomed Amersham). Both antibodies gave identical results, detecting a single band of ∼70 kD that was present in Xenopus cytosol and (much less abundantly) in Xenopus membrane fractions.
Indirect Immunofluorescence of In Vitro Assembled Nuclei
To visualize nuclear lamins by immunofluorescence, nuclei were assembled in the presence or absence of LAP2β fragments for 3 h. A 2.5-μl aliquot of each assembly reaction was placed on a slide and covered with the siliconized side of an 18 mm square coverslip. The slide was plunged into liquid N2 for 10 s. Subsequent steps were performed at 22–24°C. The coverslip was quickly peeled off, and the sample was fixed/dehydrated in 100% methanol for 1 h. The slide was then rehydrated by incubation for 5 min each in 70, 50, and 30% methanol, then in PBS. After washing twice in PBS/0.1% Triton, and blocking for 5 min in PBS/0.1% Triton/2% BSA, the sample was incubated with 100 μg/ml mouse anti–human lamin B mAb (Calbiochem-Novabiochem Corp.) in PBS/0.1% Triton/2% BSA for 1 h. After extensive washes with PBS/2% BSA, the sample was incubated for 30 min with Texas red–conjugated goat anti–mouse antibody (Organon Teknika), washed twice with PBS/2% BSA, and incubated with the DNA dye Hoechst 33342 for 5 min. After three more washes with PBS/ 2% BSA, the sample was overlaid with 5 μl glycerol, covered with an 18 mm square coverslip, and viewed by phase-contrast and immunofluorescence on a Nikon Microphot fluorescent microscope.
Cloning Xenopus LAP2 Isoforms
A Xenopus stage VI oocyte cDNA library in the UniZap vector (Stratagene Cloning Systems) was screened using full-length human LAP2β DNA as a probe. The probe was radiolabeled with α[32P]dCTP using the Multiprime DNA labeling system (Nycomed Amersham). The Xenopus cDNA library was a kind gift from D. Patterton and A. Wolffe (National Institutes of Health, Bethesda, MD). For the primary screen of 106 plaques, we used moderate stringency hybridization (30% formamide, 5× SSC, 42°C; washes in 2× SSC, 42°C), and obtained ∼300 positives. 20 strong positives were rescreened, and 12 remained positive. Single positive plaques from the tertiary screen were picked and converted to phagemids according to the manufacturer's protocol. Insert DNA was analyzed using restriction enzymes, revealing four distinct DNA inserts, which were sequenced. Sequences were aligned using ClustalW Multiple Sequence Align (http://dot.imgen.bcm.tmc.edu:9331/multi-align/Options/clustalw. html) and BOXSHADE (http://ulrec3.unil.ch/software/BOX_form.html) software; DNA sequence information was further analyzed using DNA Strider. Clone 1 was a variant of clone 2, with several frameshifting point mutations, and was not studied further. GenBank accession numbers for Xenopus LAP2 clones are: clone 2 (AF048815), clone 3 (AF048816), and clone 4 (AF048817).
To construct Xenopus LAP2β fragment 1–164, a 5′ primer was designed to code for an Nde1 site followed by the first five amino acids 5′(GGGCATATGCCCGAGTTTCTG)3′, and a 3′ primer designed to encode the six terminal amino acids followed by an Xho1 site 5′(CCCCTCGAGTTCTTTATCACTGTAATG)3′. These primers were used in a PCR reaction to obtain a 510-bp fragment using clone 2 as template. This PCR product was digested with Nde1 and Xho1 (Life Technologies, Inc.; GIBCO BRL) and ligated into corresponding sites in pET23a (Novagen, Inc.). In this case, the vector is predicted to express Xenopus LAP2β amino acids 1–164 followed by the six-His tag (173 amino acids; predicted mass of 19,749 D). The Xenopus fragment was expressed and purified as described above for the human LAP2 fragments.
Results
LAP2β Fragment 1–408 Inhibits Nuclear Envelope Expansion; Fragment 1–187 Inhibits Enclosure
To explore LAP2β function, we purified two His-tagged recombinant polypeptides derived from human LAP2β (Fig. 1 b). Fragment 1–408 (∼46.5 kD) included the entire nucleoplasmic portion of LAP2β, and is predicted to have both lamin-binding and chromatin-binding properties (Furukawa et al. 1998). Fragment 1–187 (∼21.7 kD) corresponds to the three conserved NH2-terminal exons present in all LAP2 isoforms and includes residues 1–85, which are sufficient for binding to chromatin (Furukawa et al., 1998). We used fragment 1–187 for two reasons. First, conserved exons frequently code for conserved structural domains within a protein, and second, fragment 1–187 was expected to compete with all endogenous LAP2 isoforms for binding to partners other than lamins.
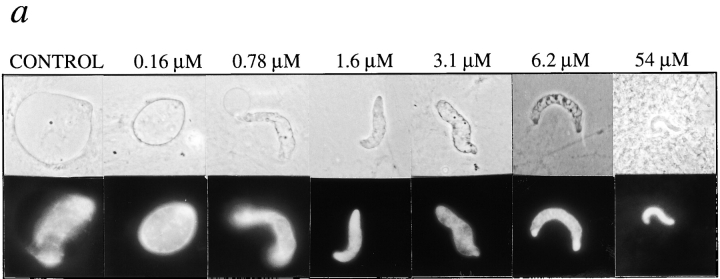
To determine its effects on nuclear assembly, fragment 1–408 was purified (Fig. 1 b) and added to Xenopus nuclear assembly reactions at concentrations ranging from 0.16 to 54 μM (Fig. 2 a). Fragment 1–408 had no detectable effect on vesicle binding, envelope enclosure, or nuclear import (see below), but inhibited envelope growth at concentrations as low as 1–3 μM (Fig. 2 a). The nuclei became enclosed by an intact nuclear envelope at the same time as control nuclei (∼30 min) but did not increase in size for at least 4.5 h. The final size of the arrested nuclei correlated inversely with the amount of fragment 1–408 in the reaction: at higher concentrations, the nuclei were smaller. At concentrations of 1.5–3 μM, fragment 1–408 reproducibly inhibited nuclear growth in all assembly extracts tested (three independent preparations), and virtually all nuclei were similar to those seen in Fig. 2 a. Inhibition was not due to residual imidizole, since control nuclei, which were assembled with proteins purified from uninduced bacteria, assembled and grew normally (Fig. 2 a, control).
Figure 2.
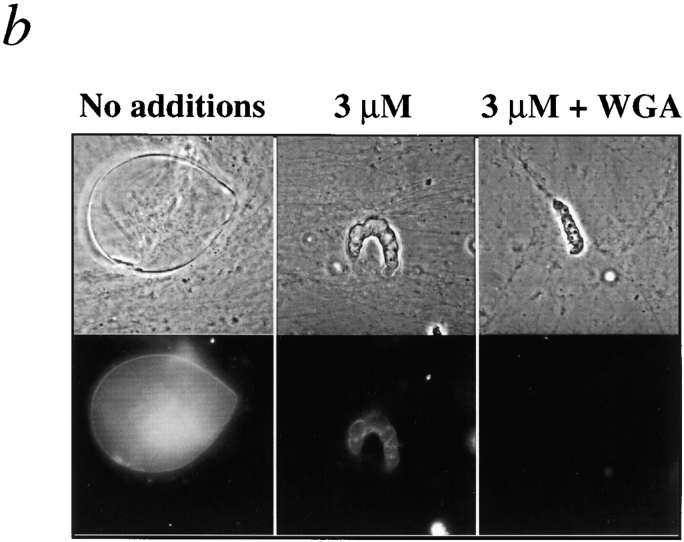
Inhibition of nuclear growth by human LAP2β fragment 1–408. (a) Nuclei assembled for 1.5 h in reactions containing the indicated concentration of fragment 1–408. The small phase-dense black dots (center of control nucleus) are coiled bodies, which form only in transport-competent nuclei. Upper panels show phase-contrast images of representative nuclei; lower panels show corresponding DNA, as visualized by Hoechst staining. (b) LAP2-arrested nuclei import a fluorescent karyophilic substrate. Nuclei were assembled for 2 h in the presence or absence of 3 μM LAP2 fragment 1–408, supplemented with rhodamine-conjugated nucleoplasmin, and viewed by epifluorescence 30 min later. As a negative control, WGA, which inhibits nuclear import, was added to samples 5 min before adding nucleoplasmin (right panels). Upper panels show phase-contrast images of representative nuclei; lower panels show the corresponding nuclei by rhodamine fluorescence, indicative of transport activity.
Nuclei Growth-arrested by Fragment 1–408 Are Active for Nuclear Import
Three lines of evidence showed that nuclei growth-arrested by fragment 1–408 were not defective for nuclear import. First, they contained prenucleolar coiled bodies (Fig. 2 a), the formation of which is dependent on nuclear import (Bell et al., 1992; Bauer et al., 1994). Second, they were active for DNA replication (see below), which also requires nuclear import. Third, we directly tested for import activity by first assembling nuclei for 2 h in the presence or absence of fragment 1–408, then adding a fluorescent karyophilic protein (rhodamine-conjugated nucleoplasmin), and imaging 30 min later. Nucleoplasmin accumulated at the nuclear rim and interior of the positive controls, and 1–408-arrested nuclei (Fig. 2 b, left and middle). Negative control nuclei, pretreated with the transport inhibitor WGA for 5 min before adding the nucleoplasmin, failed to accumulate the transport substrate (Fig. 2 b, right). Because fragment 1–408 had no detectable effect on nuclear import in three independent assays, we concluded that the nuclear expansion defect was probably a direct effect of fragment 1–408 on other nuclear structures or pathways.
Inhibition by LAP2 Fragment 1–187: Scalloped, Nonenclosed Nuclear Envelopes
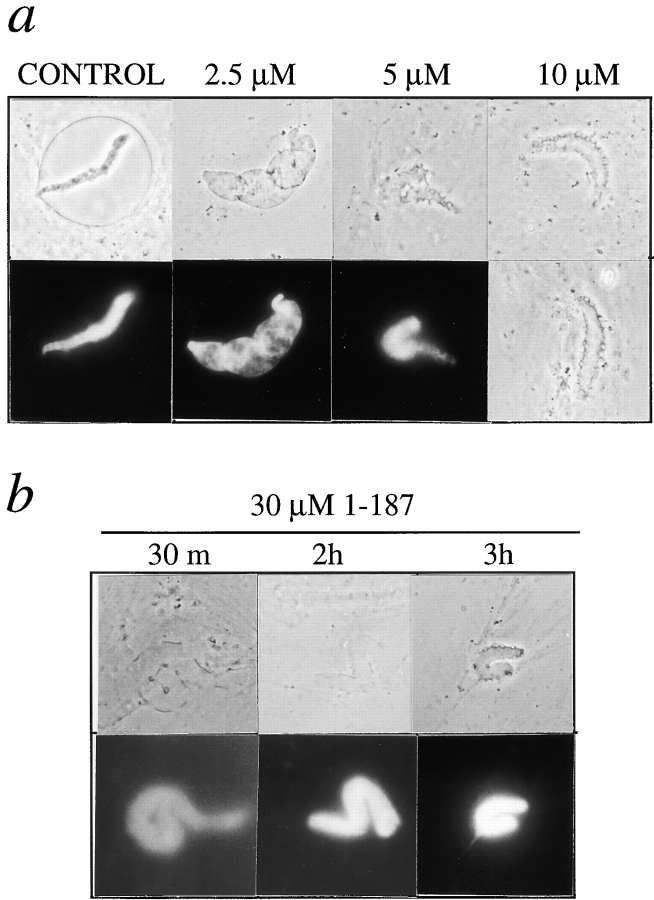
Nuclei assembled for 3 h in the presence of purified fragment 1–187 appeared very different from 1–408-arrested nuclei, as seen by comparing Fig. 2 a (panel labeled 3.1 μM) with Fig. 3 a (upper right). Nuclei inhibited by fragment 1–187 remained small, and did not acquire a typical enclosed nuclear envelope. These effects were titratable over the low micromolar range. Nuclei assembled in the presence of 2–3 μM fragment 1–187 were smaller than positive controls. At 5–10 μM, the nuclear membranes consistently had an unusual scalloped morphology: some regions seemed flattened against the chromatin, whereas other regions appeared as oversized, unflattened vesicles. This scalloped appearance did not change for ≥5 h. The effects of fragment 1–187 were reproduced in three independent extracts. At a higher concentration (30 μM), fragment 1–187 delayed the attachment of membranes to chromatin (Fig. 3 b): after 1–2 h of assembly, only a few patches of nuclear envelope were flattened onto chromatin. However, by 3 h the nuclear membranes had assembled enough to appear scalloped, like those assembled in lower concentrations (5–10 μM) of 1–187. These results showed that the putative chromatin-binding fragment of LAP2 inhibited membrane attachment to chromatin at a concentration of 30 μM. However, at lower concentrations the NH2-terminal fragment had quite different effects, interfering with both the enclosure and morphology of the nuclear envelope.
Figure 3.
Inhibition of nuclear assembly by human LAP2 fragment 1–187. (a) Nuclei assembled for 3 h in reactions containing the indicated concentration of fragment 1–187. Upper panels show phase-contrast images; lower panels show DNA as visualized by Hoechst, except the right-most panel which shows a second phase-contrast image of a nucleus inhibited by 10 μM fragment 1–187. (b) Nuclei were assembled in a reaction containing 30 μM fragment 1–187 for the indicated times. Membrane association with chromatin is delayed, but after 3 h the nuclei appear similar to those arrested by 5–10 μM fragment 1–187.
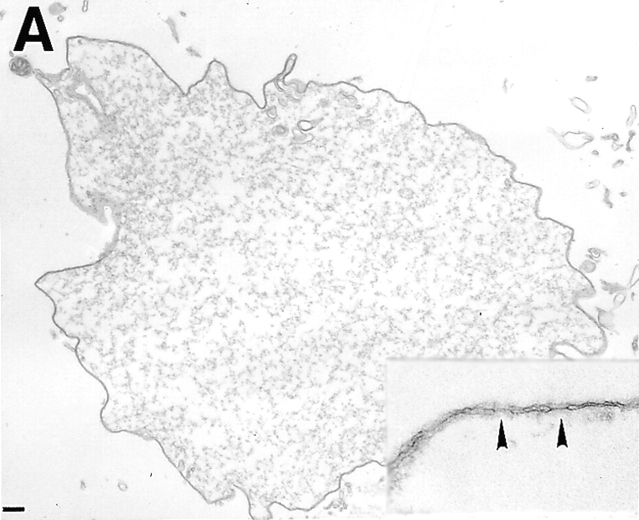
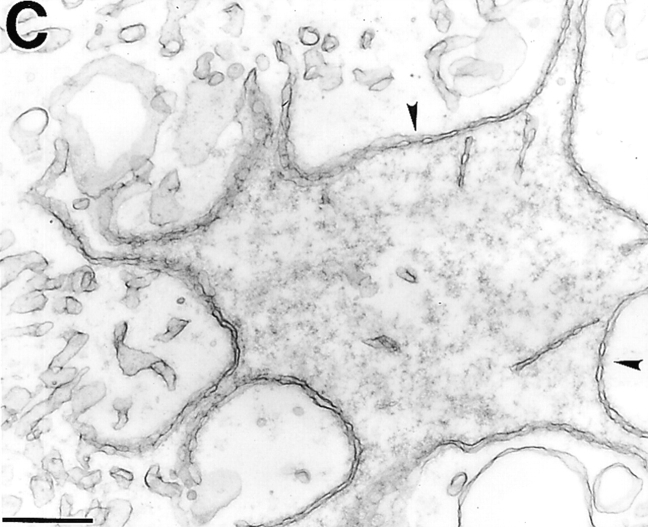
Ultrastructure of Arrested Nuclei
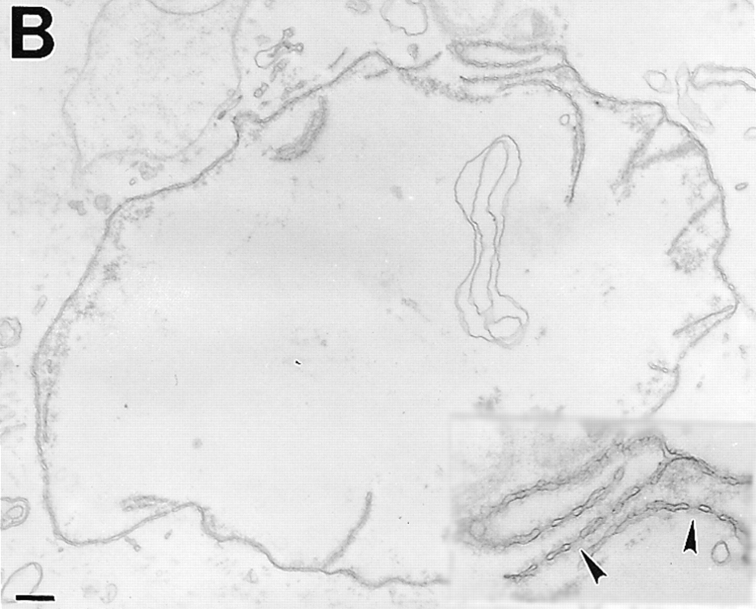
To study their morphology in greater detail, nuclei were examined by transmission electron microscopy (TEM; Fig. 4). As expected, control nuclei were enclosed by two nuclear membranes and studded with NPCs (Fig. 4 a; inset arrows point to NPCs). Nuclei inhibited by fragment 1–408 had an enclosed nuclear envelope (Fig. 4 b), confirming our phase-contrast observations (Fig. 2 a). Although the particular cross-sections shown in Fig. 4, a and b, are similar in size, the magnifications are different, and 1–408-inhibited nuclei were much smaller than control nuclei (Fig. 2). TEM further revealed that 1–408-inhibited nuclei had numerous NPC-containing invaginations of the inner nuclear membrane (Fig. 4 b; arrows). These invaginations were morphologically distinct from the nuclear tunnels described by Fricker et al. (1997) in which the entire envelope invaginates to form tubules extending into the nuclear interior. Nuclei inhibited by 1.5–3 μM fragment 1–408 also appeared to have a higher density of NPCs than control nuclei, consistent with ongoing NPC assembly into growth-arrested nuclei.
Figure 4.
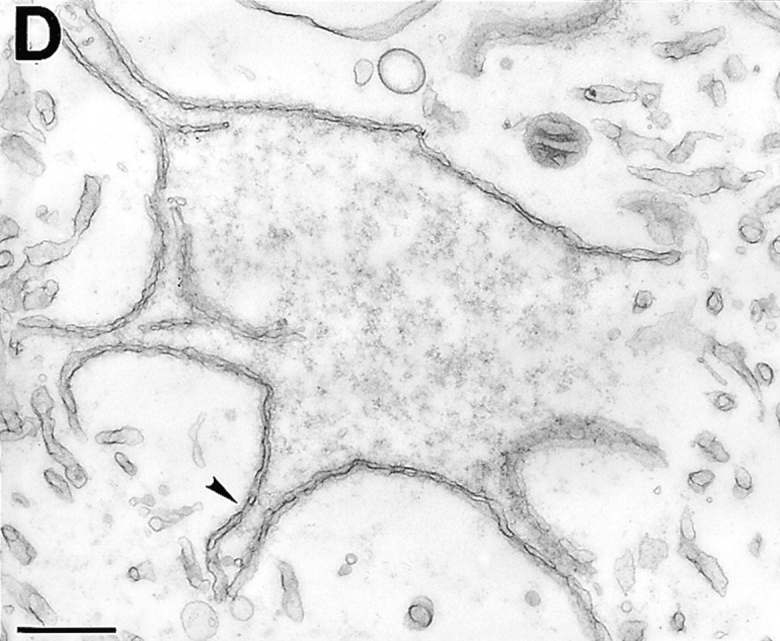
TEM of nuclei inhibited by human LAP2 fragments. (a) Positive (buffer) control: nucleus assembled for 3 h. (b) Nucleus assembled for 3 h in a reaction containing 3 μM fragment 1–408. Insets in a and b are higher magnifications to show NPCs. Arrows in b indicate regions where the inner membrane appears to invaginate. (c and d) Nuclei assembled for 3 h in a reaction containing 10 μM LAP2 fragment 1–187. Arrowheads indicate NPCs. Bars, 500 nm.
TEM of nuclei assembled in the presence of 5 or 10 μM fragment 1–187 revealed that the chromatin was covered, but not enclosed, by nuclear membranes that contained NPCs (Fig. 4, c and d; two nuclei assembled in 10 μM 1–187 are shown). The envelope patches were concave relative to the chromatin, in contrast to the rounded convex shapes of control nuclei (Fig. 4 a) and nuclei inhibited by fragment 1–408 (Fig. 4 b). We did not detect huge vesicles by TEM that might have corresponded to those seen by phase-contrast microscopy; we speculate that either these structures break during sample preparation, or that deeply concave sections of envelope might give the illusion of vesicles by light microscopy. The 1–187-arrested nuclei were similar to 1–408-arrested nuclei in two ways. First, they both had more NPCs than control nuclei, demonstrating that these LAP2 fragments do not interfere with NPC assembly. Second, they both had invaginations of the inner membrane into the nucleus, which might represent a problem in remodeling (or disconnecting) membrane-chromatin attachments (see Discussion).
Our morphological results and import assays suggested that LAP2 proteins help shape the structural interface between the nuclear membranes and chromatin. We were particularly intrigued by the structural effects of fragment 1–187, which generated concave scallop-shaped envelopes. To our knowledge this phenotype is novel; for example, lamin-depleted nuclei are enclosed and round (Newport et al., 1990).
Lamins Accumulate in 1–408-arrested Nuclei, but Not 1–187-arrested Nuclei
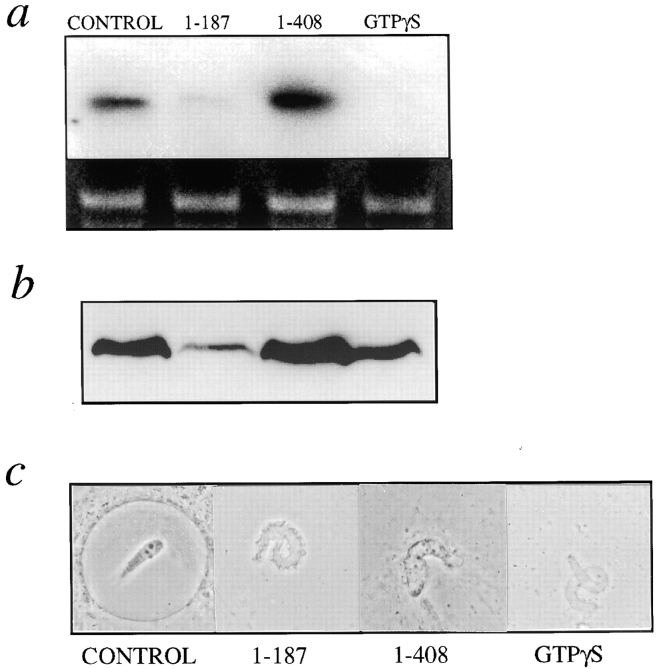
To ask if LAP2 fragments affected lamina assembly, we assayed the inhibited nuclei for lamin accumulation (Fig. 5 a). Nuclei were assembled for 3 h in reactions supplemented with either buffer, 3 μM fragment 1–408, or 5 μM fragment 1–187. Nuclei were then pelleted, subjected to SDS-PAGE, and immunoblotted with mAb 46F7, which is specific for lamin B3, the major lamin found in Xenopus eggs (Lourim and Khrone, 1993; Lourim et al., 1996) (Fig. 5 a). Identical results were obtained using an mAb raised against human lamin B (data not shown). Nuclei inhibited by fragment 1–408, but not those inhibited by fragment 1–187, accumulated lamins over time as shown by immunoblotting of pelleted nuclei (3-h time points shown; Fig. 5 a). The lamin signal in 1–408-inhibited nuclei was sometimes slightly stronger than in control nuclei (two of five experiments), perhaps reflecting the presence of additional lamin-binding proteins (i.e., fragment 1–408) in these nuclei. These lamin results were consistent with the structural results, since 1–408-inhibited nuclei were enclosed and active for nuclear protein import, and 1–187-inhibited nuclei were not enclosed and did not accumulate imported proteins such as lamins.
Figure 5.
Lamin accumulation in nuclei inhibited by LAP2 fragments. (a) Western blot of nuclei assembled in the presence of the indicated LAP2 fragment. Nuclei were assembled for 3 h and pelleted; the pelleted proteins were separated by 12% SDS-PAGE and immunoblotted with a monoclonal anti-lamin antibody (see Materials and Methods). (b) Lamins detected by indirect immunofluorescence in nuclei assembled for 3 h in (left to right): a control reaction (buffer added), a reaction containing 3 μM LAP2 fragment 1–408, and a reaction containing 5 μM LAP2 fragment 1–187. Nuclei were processed for immunofluorescence as described in Materials and Methods. Upper panels, contrast images; middle panels, DNA visualized with Hoechst; bottom panels, lamins visualized by a Texas red–conjugated secondary antibody.
We also used indirect immunofluorescence to detect lamins in inhibited nuclei (Fig. 5 b). Consistent with the lamin blots, lamins were not detected by indirect immunofluorescence in 1–187-inhibited nuclei. Lamins were detected both in control nuclei and nuclei inhibited by fragment 1–408, in close association with the nuclear envelope (Fig. 5 b). However, because the 1–408-inhibited nuclei were small, and the immunofluorescent signal quite bright (see Fig. 5 b), we could not determine unambiguously whether lamins were associated exclusively with the envelope, or if they might have also accumulated at inappropriate sites inside the nucleus.
Fragment 1–187 Blocks Lamina Assembly
As a negative control for the DNA replication experiments described below, we used GTPγS to inhibit vesicle fusion (Boman et al., 1992; Newport and Dunphy, 1992). No replication was observed in GTPγS-treated reactions, as expected, since these nuclei consist of small vesicles bound to the chromatin surface. However, the GTPγS-arrested nuclei accumulated a low level of lamins (Fig. 6 b; see also Wiese et al., 1997), suggesting that some interactions involving lamins may have proceeded to a limited extent even though vesicle fusion was inhibited by GTPγS. The modest accumulation of lamins on GTPγS-arrested nuclei contrasted significantly with 1–187-inhibited nuclei, which had only background amounts of lamins (compare to membranes alone; Fig. 5 a). We concluded that fragment 1–187 blocks lamin recruitment or attachment to the membrane–chromatin interface. Because there is strong evidence that the NH2-terminal region of LAP2 binds to chromatin (Furukawa et al., 1998), we further concluded that fragment 1–187 binds competitively to, and blocks, the chromatin partners for endogenous LAP2 proteins. These findings therefore suggest that endogenous LAP2 isoforms must engage their chromatin partner as a prerequisite for lamina assembly.
Figure 6.
DNA replication and lamin blots of nuclei inhibited by LAP2 fragments. Nuclei were assembled for 3 h in the presence of α[32P]dCTP plus either buffer, 3 μM LAP2β fragment 1–408, 5 μM LAP2 fragment 1–187, or 1 mM GTPγS. (a) Total DNA was prepared from aliquots of each sample, electrophoresed on a 0.8% agarose gel, and the gel dried and exposed to film (see Materials and Methods). Upper panel shows the autoradiograph of dried agarose gel; the lower panel shows the ethidium stain of the same gel as a control for DNA loading. (b) Protein from each sample was blotted and probed using a monoclonal anti-lamin antibody (see Materials and Methods). (c) Corresponding phase-contrast images of a typical nucleus from each sample in a.
DNA Replication Assays
DNA replication normally occurs in in vitro assembled nuclei if the lamina is properly assembled (Blow and Watson, 1987; Newport et al., 1990; Leno and Laskey, 1991; Cox, 1992). Therefore, replication is used as a marker for the assembly of a structurally intact nucleus. Since nuclei inhibited by fragment 1–187 did not accumulate lamins, we predicted that these nuclei would be unable to replicate. We did not know what to expect with 1–408-inhibited nuclei, which accumulated lamins. Fragment 1–408 is predicted to bind lamins (Foisner and Gerace, 1993; Furukawa et al., 1998), and might somehow disrupt lamina assembly, and hence, secondarily disrupt replication.
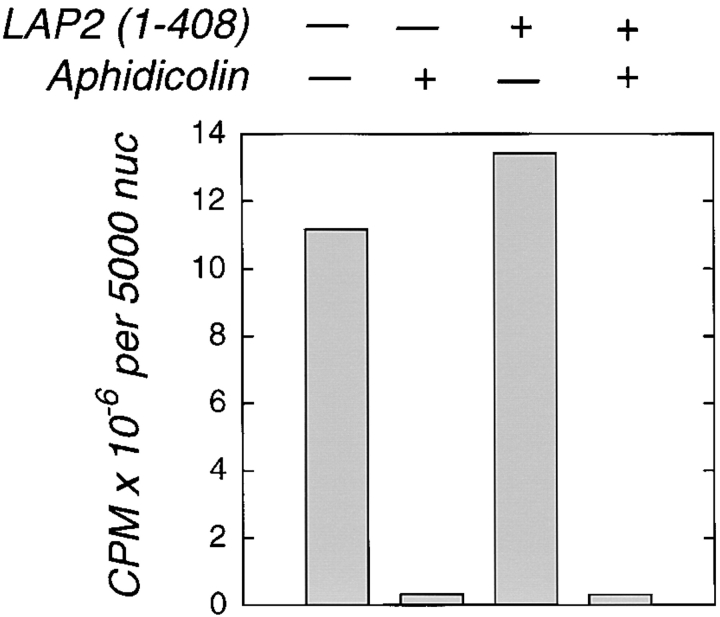
To test DNA replication, nuclei were assembled in the presence of [32P]dCTP for 3 h, with or without added LAP2 fragments, and the DNA was analyzed by agarose gel electrophoresis and autoradiography (Fig. 6 a). Aliquots were processed in parallel for Western blotting with an anti-lamin antibody (Fig. 6 b), and a representative nucleus from each sample is shown by phase-contrast (Fig. 6 c). No replication activity was detected in 1–187-arrested nuclei (Fig. 6 a), as predicted from their lack of enclosure. However, nuclei assembled in the presence of 3 μM fragment 1–408 consistently replicated at levels as high (three experiments) or higher (eight experiments) than control nuclei (Fig. 6 a). In the experiment shown, 1–408-inhibited nuclei also accumulated lamins at higher levels than controls (Fig. 6 b). The increased replication signal in 1–408-inhibited nuclei was not due to unequal loading (ethidium stain of DNA; Fig. 6 a, lower panels). No replication was detected in negative controls treated with GTPγS to prevent envelope formation (Boman et al., 1992; Newport and Dunphy, 1992), or in the presence of 50 μg/ml aphidicolin (Fig. 7), a specific inhibitor of DNA polymerase α (Ikegami et al., 1978). Note the modest degree of enhancement by fragment 1–408 in Fig. 7, an example of the low end of experimental variation in the extent of enhancement. We concluded that although the LAP2 fragment 1–408 blocked the expansion of nascent nuclei in vitro, it unexpectedly enhanced DNA replication activity. Based on densitometry quantitation of eight experiments, nuclei arrested by fragment 1–408 replicated an average of 2.5-fold better than controls.
Figure 7.
Replication of LAP2-arrested nuclei is aphidicolin-sensitive. Nuclei were assembled for 3 h with or without either LAP2 fragment 1–408 (3.3 μM) or aphidicolin (50 μM), in the presence of α[32P]dCTP. Reactions were digested with proteinase K, run on agarose gels, and label incorporated into high molecular weight DNA was quantitated by PhosphorImager (see Materials and Methods).
Enhanced [32P]dCTP Incorporation Is Not Due to Rereplication
We considered two different mechanisms for the increase in [32P]dCTP incorporation: fragment 1–408 might cause rereplication, which would represent a loss of cell cycle control, or it might enhance semiconservative DNA replication, the efficiency of which can vary from 30 to 100% in Xenopus egg extracts (Leno and Laskey, 1991).
The first possibility, rereplication, was tested by equilibrium density substitution in the presence of a dense nucleotide derivative, BrdU (Fig. 8 a). These reactions also contained [32P]dCTP, which allowed us to detect replicated strands, and quantitate the extent of replication (see Materials and Methods). The positive control (no LAP2 fragment) was expected to undergo a single round of semiconservative replication to yield DNA with one light strand and one heavy strand, which would migrate as a single peak in a CsCl gradient. If the enhanced replication were due to rereplication, we would detect a second peak of heavy–heavy DNA at a higher density, and we would not expect the heavy–light DNA from LAP2-treated nuclei to have significantly more radioactivity than the positive control. The experiment was done using both fractionated/reconstituted extract (data not shown) and fresh 10,000 g crude cytoplasm (Fig. 8 a). We found that LAP2-arrested nuclei yielded a single peak of radiolabeled DNA that comigrated at exactly the same refractive index, and hence, density, as the positive control, indicating a single round of semiconservative DNA replication. Furthermore, the peak of incorporated nucleotide was at least fourfold higher than the positive control, consistent with fragment 1–408 enhancing the efficiency of semiconservative replication. These results effectively ruled out the possibility that LAP2 fragment 1–408 causes rereplication.
Figure 8.
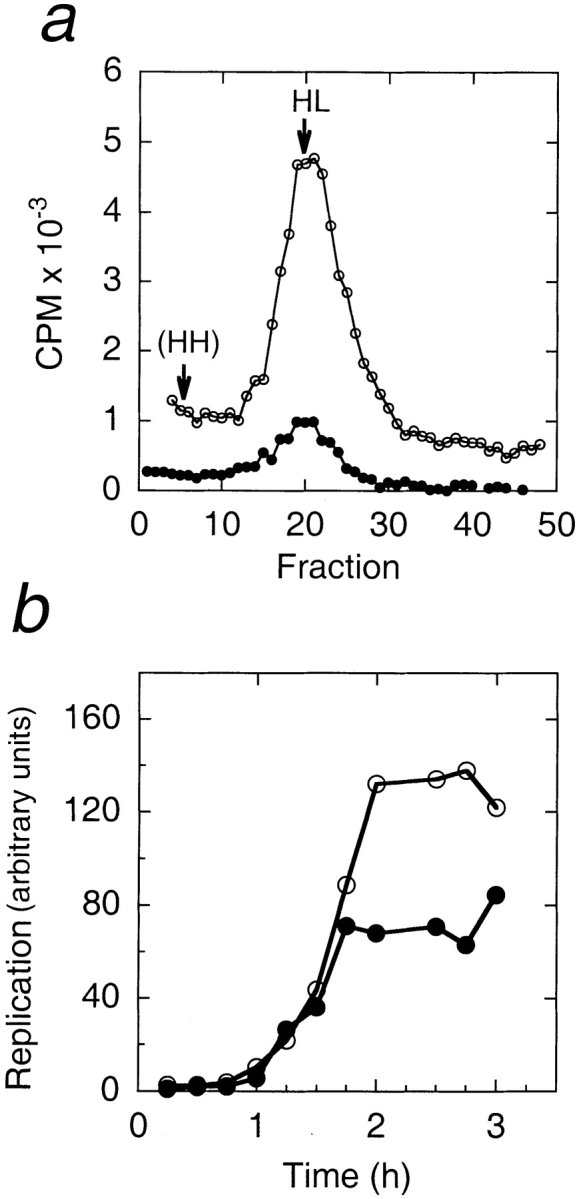
BrdU density substitution experiment, and replication time course. (a) Density substitution experiment. Nuclei were assembled for 2 h in reactions containing BrdU and α[32P]dCTP, in the presence or absence of 3.3 μM fragment 1–408. The substituted DNA was purified and separated by centrifugation on equilibrium CsCl gradients (see Materials and Methods). Replicated DNA (cpm of incorporated α[32P]dCTP) in each gradient fraction is plotted against gradient fractions, which had equal densities as determined by refractive index measurements. HL indicates the density of heavy–light DNA (1.746 g/ml), and HH the expected density of heavy–heavy DNA (1.80 g/ml; Walter et al., 1998). Filled circles indicate reactions lacking LAP2; open circles indicate reactions containing 3.3 μM LAP2 fragment 1–408. (b) Replication time course. Nuclei were assembled in reactions containing α[32P]dCTP, in the presence or absence of fragment 1–408. Aliquots were removed every 15 min for 3 h, and processed by running purified DNA on agarose gel, and quantitating the incorporated [32P]dCTP by PhosphorImager (see Materials and Methods). Filled circles indicate reactions lacking LAP2; open circles indicate reactions containing 3.3 μM LAP2 fragment 1–408.
To examine the time course of replication in LAP2-arrested nuclei, nuclei were assembled in reactions containing α[32P]dCTP in the presence or absence of fragment 1–408; aliquots were removed from each reaction every 15 min, and the incorporated radiolabel was quantitated (Fig. 8 b; see Materials and Methods). The time course of replication was initially indistinguishable between LAP2-arrested nuclei and positive controls: there was a lag phase of 45 min, followed by DNA synthesis. The only difference was that α[32P]dCTP incorporation into LAP2-arrested nuclei continued at the same rate for ≥15 min longer than positive controls, reaching a plateau that was, in this case, ∼1.8-fold higher than the positive control (Fig. 8 b).
The replication results collectively led us to two conclusions. First, the majority of our extracts, which were made from unactivated eggs, were not 100% efficient for replication. Based on the amount of replication enhancement seen in these experiments, which ranged from 0 to ∼4-fold (average, 2.5-fold), we estimated that the efficiency of replication in our extracts varied from 20% to 100%. Second, in extracts that were <100% efficient for replication, LAP2 fragment 1–408 increased the efficiency of semiconservative DNA replication, when present at low (2–4 μM) concentrations. Possible mechanisms for the enhancement of replication efficiency by LAP2 are considered in the Discussion.
Xenopus Oocytes Express LAP2 cDNAs Closely Related to Mammalian LAP2β
Given the effects of human LAP2 fragments in Xenopus nuclear assembly extracts, it was essential to determine if Xenopus LAP2 had the same structural effect on nuclear assembly. To do this, and compare the Xenopus and human LAP2 proteins, we screened a stage VI Xenopus oocyte cDNA library using radiolabeled full-length human LAP2β as a probe (see Materials and Methods). We rescreened 20 of nearly 300 positives, and identified three cDNAs coding for Xenopus LAP2β homologues, which were designated clones 2, 3, and 4 (see Fig. 9 a, and Materials and Methods).
Figure 9.
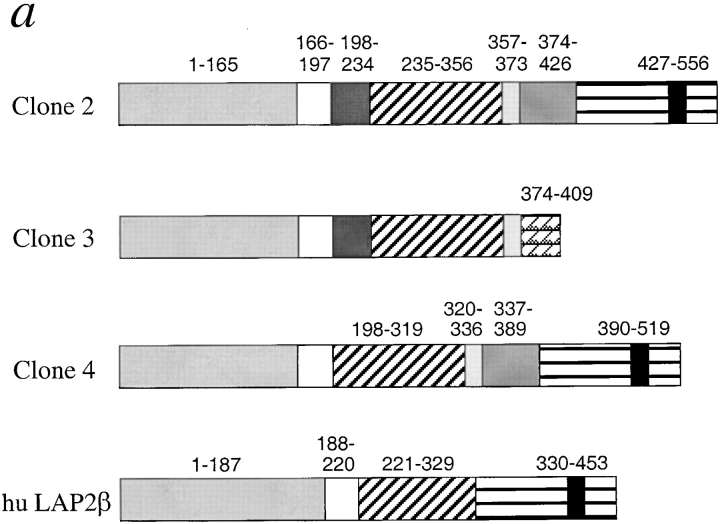
Comparison of predicted Xenopus LAP2 proteins to human LAP2β. (a) Schematic diagrams of translated Xenopus cDNAs and human LAP2β protein. Homologous regions between Xenopus and human LAP2 proteins are shaded similarly. Numbers denote residues that begin and end each block of homology. The black box in the striped COOH-terminal region denotes the predicted transmembrane span. The predicted masses of the proteins encoded by the Xenopus LAP2 cDNAs are 62.841 kD (clone 2), 46.363 (clone 3), and 58.674 kD (clone 4). DNA sequence data are available from GenBank under accession numbers AF048815 (clone 2), AF048816 (clone 3), and AF048817 (clone 4). (b) Alignment of human LAP2β and Xenopus LAP2-clone 2 proteins using BOXSHADE (see Materials and Methods). Black shading indicates identity, gray shading indicates similarity.
The three Xenopus LAP2 proteins are compared schematically to human LAP2β in Fig. 9 a. Clone 2 was the longest Xenopus LAP2 cDNA, encoding a protein of predicted mass 62.841 kD. Except for a single-base deletion at nucleotide 1119 in clone 3, which is either a mutation or the result of an alternative splicing event (see below), the three Xenopus cDNAs were identical at the nucleotide level except for two regions: nucleotides 595–705 and 1068–1278 in clone 2. These two regions encoded polypeptide inserts that we named insert A (37 residues, 198–234), insert B (17 residues, 357–373), and insert C (53 residues, 374–426), as diagrammed in Fig. 9 a. Clone 2 had all three inserts, whereas clone 3 lacked insert C, and clone 4 lacked insert A, suggesting that inserts A and C were alternatively spliced exons. Although insert B was present in all three Xenopus cDNAs, it was absent from human LAP2 and is therefore either a new exon, or a nonhomologous extension of the neighboring exon. All three putative new exons were located at exon boundaries in the mouse genomic sequence (Berger et al., 1996): insert A between mouse exons 5 and 6, and inserts B and C between mouse exons 8 and 9. We concluded that inserts A and C (and probably B) represent bona fide LAP2 exons in Xenopus. Exons homologous to inserts A, B, and C have not yet been reported in mammals. Overall, these results suggested that Xenopus clones 2, 3, and 4 were splicing variants related to mammalian LAP2β. Notably, clone 3 encoded a putative β-related isoform that lacks a transmembrane domain, similar to the ζ (zeta) isoform in mice (Berger et al., 1996).
Three regions were broadly similar between Xenopus and human LAP2β proteins: an NH2-terminal region (gray and white boxes in Fig. 9 a), a middle region (diagonal bars), and a COOH-terminal region (horizontal stripes). The protein encoded by clone 2 is compared with human LAP2β in detail in Fig. 9 b. The NH2-terminal region of clone 2 (residues 1–197) was 70% identical and 80% similar to the same region in human LAP2 (residues 1–220). The middle region was less conserved: residues 234–352 of clone 2 were 39% identical and 53% similar to human LAP2β residues 220–325. At the COOH-terminal region, Xenopus residues 427–556 were 63% identical and 74% similar to human LAP2β residues 330–453. We concluded that the middle region of LAP2β is the least conserved between species, in contrast to the more-conserved NH2-terminal and COOH-terminal domains.
Xenopus LAP2 Fragment 1–164 Inhibits Nuclear Growth
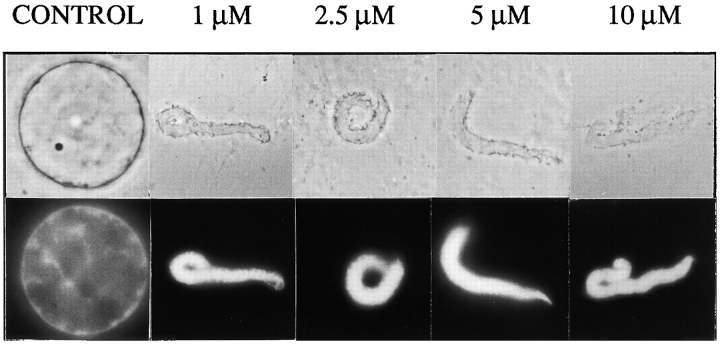
To assess the effects of Xenopus LAP2 on nuclear assembly, we focussed on the NH2-terminal domain. We expressed and purified the Xenopus LAP2 fragment consisting of residues 1–164, which are homologous to human fragment 1–187, and added it to Xenopus nuclear assembly reactions at concentrations ranging from 1 to 10 μM (Fig. 10). The Xenopus LAP2 fragment had structural effects identical to those of human LAP2 fragment 1–187, producing scalloped, nonenclosed envelopes at concentrations of 1, 2.5, and 5 μM (Fig. 10). At 10 μM, fragment 1–164 interfered with membrane targeting to the chromatin surface (Fig. 10), paralleling the effects of human fragment 1–187 at 30 μM (Fig. 3 b). We noted that Xenopus fragment 1–164 was inhibitory at concentrations two to five times lower than the comparable human fragment. We concluded that the human and Xenopus NH2-terminal fragments of LAP2 had the same effects on nuclear assembly in vitro. These results strongly suggested that the human polypeptides interacted with bona fide LAP2 binding partners in Xenopus extracts, with slightly lower efficiency, presumably due to species-specific differences in amino acid sequence. We concluded that the human LAP2 fragments used in our experiments affected nuclear assembly and DNA replication by competing for the binding partners of endogenous LAP2 proteins.
Figure 10.
Inhibition of nuclear assembly by Xenopus LAP2 fragment 1–164. Nuclei were assembled for 3 h in reactions containing the indicated concentration of fragment 1–164. Upper panels show phase-contrast images; lower panels show DNA as visualized by Hoechst. Xenopus fragment 1–164 gives the same arrest phenotype as human fragment 1–187, but at lower concentrations.
Discussion
Consistent with previous results from microinjected HeLa cells (Yang et al., 1997), our in vitro results show that a LAP2β fragment capable of binding to lamins has the effect of blocking nuclear expansion after enclosure. This finding confirms the importance of LAP2β and lamins in mediating nuclear growth. We further show that nuclei growth-arrested by the chromatin-and-lamin-binding nucleoplasmic domain of LAP2β (residues 1–408) were enhanced in their efficiency of semi-conservative DNA replication; this result has important new implications for LAP2 function, as discussed below. In contrast to Yang et al. (1997), who found that residues 1–85 of the conserved NH2-terminal domain had no effect in vivo, the complete NH2-terminal chromatin-binding domain of both human and Xenopus LAP2 strongly inhibited nuclear assembly, producing a scalloped envelope morphology and blocking lamina assembly. The implications of this phenotype are discussed below.
Is Chromatin Binding by LAP2 Proteins a Prerequisite for Lamin Assembly?
Several studies suggest that the LAP2 isoforms are collectively responsible for dynamically organizing the lamina: the biochemical demonstration that rat LAP2β binds lamin B (Foisner and Gerace, 1993), the sequential colocalization of LAP2α with lamin B and lamin A during nuclear assembly in vivo (Dechat et al., 1998), the in vivo and in vitro nuclear growth arrest by lamin-binding fragments of LAP2β (Yang et al., 1997; this study), and the block to lamin accumulation caused by the NH2-terminal chromatin-binding domain of LAP2 (human residues 1–187; this study). The finding, that nuclei arrested by fragment 1–187 did not accumulate lamins, was unexpected since this shared domain of LAP2 binds to chromatin, not lamins. We suggest that when the recombinant chromatin-binding domain of LAP2 is added to assembly reactions, it occupies chromatin sites and prevents endogenous LAP2 proteins from attaching to chromatin. To explain how fragment 1–187 then prevents lamin assembly, we propose that the endogenous LAP2 proteins need to bind to chromatin as a prerequisite for binding to lamins and promoting lamin assembly.
We characterized three cDNAs from Xenopus, which appear to be new β-related isoforms of LAP2. These cDNAs include three putative novel exons, referred to as inserts A, B, and C. Insert A is positioned immediately after the conserved NH2-terminal domain. Inserts B and C interrupt the minimal lamin-binding region of LAP2β (identified as residues 298–373 in rat LAP2β; see Yang et al., 1997). Interestingly, the minimal lamin-binding region is not encoded by a single exon, but spans exons 8 and 9 (mice; Berger et al., 1996). Xenopus inserts B and C are located precisely between mice exons 8 and 9, inserting 17 and 53 residues, respectively, into the middle of the lamin-binding region. Lamin-binding activity is influenced by the COOH-terminal region of LAP2, since residues 298–452 of rat LAP2β bind to lamins fivefold better than minimal residues 298–373 in a yeast two-hybrid assay (Furukawa et al., 1998). In view of our hypothesis that LAP2 chromatin-binding activity may regulate or promote lamin recruitment, it will be interesting to determine the lamin-binding and other activities of Xenopus LAP2 isoforms that have, or lack, each new exon.
LAP2 Proteins May Influence Chromatin Structure
One explanation for why fragment 1–408 (the chromatin-and-lamin-binding fragment) arrests the expansion of nascent nuclei is that it may contribute to the formation of excess or unregulated connections between lamins, membranes, and chromatin. This possibility is supported by the extensive invagination of the inner membrane seen in nuclei arrested by fragment 1–408. However an alternative possibility, that this chromatin-and-lamin-binding LAP2 fragment inhibits chromatin decondensation, is supported by our finding that high concentrations (54 μM) of fragment 1–408 caused the sperm chromatin to remain smaller than the size expected of chromatin swelled by exposure to egg cytosol (Fig. 2 a; see Newport and Dunphy, 1992). The association of LAP2 with chromatin is relatively strong, since immunoaffinity-purified LAP2β binds saturably to mitotic chromosomes with an affinity of 40–80 nM (Foisner and Gerace, 1993). We hypothesize that at high concentrations, recombinant LAP2β may either block chromatin decondensation, or promote condensation.
Interestingly, a chromatin binding partner for LAP2 has been provisionally identified by two-hybrid analysis in yeast as BAF (barrier to autointegration factor; Furukawa, 1999). BAF localizes to the nucleus during interphase, and to chromosomes during mitosis (Furukawa, 1999). BAF is a small novel cellular protein (89 residues) that was identified because it facilitates the efficient integration of HIV DNA into the cell's genome (Chen and Engelman, 1998; Lee and Craigie, 1998); in the absence of BAF, the viral DNA molecule tends to integrate intramolecularly into itself. Lee and Craigie (1998) propose that BAF acts by crossbridging and thereby compacting the viral DNA molecule. The normal cellular role of BAF is not known. If further experiments confirm that BAF and LAP2 are indeed binding partners, it will be interesting to test the idea that LAP2 influences chromatin structure by affecting BAF activity.
Concentration-dependent Effects of LAP2 Fragment 1–408 on DNA Replication
The minimal lamin-binding region of LAP2β (rat residues 298–373) inhibits the initiation, but not the progression, of DNA replication in vivo (Yang et al., 1997). Consistent with this, the full nucleoplasmic region of human LAP2β (fragment 1–408) also inhibited DNA replication at concentrations above 6 μM (data not shown). However at lower concentrations (1–3 μM), which reproducibly inhibited nuclear expansion, fragment 1–408 enhanced DNA replication activity by an average of 2.5-fold in ∼80% of our experiments. Thus, by varying the concentration of recombinant protein in the reaction, we uncovered a positive role for LAP2β in replication.
Based on density-substitution experiments, we eliminated the possibility that increased nucleotide incorporation was due to rereplication. A second possibility, that fragment 1–408 triggers extreme levels of repair synthesis, seems unlikely, but cannot be ruled out by our present data. Our results favor a third possibility, that LAP2β fragment 1–408 increases the efficiency of semiconservative DNA replication. This increased efficiency could be an indirect effect of LAP2 on nuclear size, since the small arrested nuclei may achieve higher concentrations of imported replication factors. For example, chromatin can replicate efficiently in the absence of nuclei in vitro, by sequential exposure to cytosolic extracts and 25-fold concentrated nucleoplasmic extracts (Walter et al., 1998). We can provisionally rule out such a size based model for one simple reason: 1–408-arrested nuclei were always small, but replication was not enhanced in ∼20% of our experiments (see below). We think this extract-to-extract variation is an important clue about the mechanism of enhancement.
Logically, replication enhancement is only possible in extracts that are less than 100% efficient. Xenopus egg extracts can vary in replication efficiency from 30% to 100% (Cox and Leno, 1990; Leno and Laskey, 1991; Walter et al., 1998). The cause of this variation is not yet known, but we hypothesize that it may reflect differences between extracts in the efficiency with which prereplication complexes assemble onto chromatin. The prereplication complex consists of MCM proteins, the cdc6 protein, and six ORC proteins; this complex can only be assembled when there are no cyclin-dependent kinases (CDKs) active in the cell (see Dillin and Rine, 1998, and references therein). In somatic cells, such a situation only exists for a narrow window of time between anaphase (when the mitotic cyclin-dependent kinases are inactivated) and early G1 (when G1-phase kinases are activated). According to the two-step model for replication control, which is widely supported by evidence from yeast and Xenopus, the prereplication complex is the obligatory precursor of the replication complex, and is removed during replication, thereby providing a mechanism to limit replication to a single round per cell cycle (see Stillman, 1996; Hua et al., 1997; Jalepalli and Kelly, 1997).
Eggs have high levels of mitotic CDK activity (also known as maturation promoting factor activity), which maintains them in a metaphase-arrested state until fertilization. Fertilization triggers a wave of intracellular Ca2+ release, which destroys the Ca2+-sensitive cytostatic factor stabilizing maturation promoting factor (Lorca et al., 1993; Masui, 1996). Many investigators mimic fertilization by either exposing eggs to Ca2+ or electric shock, before making extracts. In contrast, our extracts are from unactivated eggs; activation is not essential to obtain extracts competent for interphase nuclear assembly (Wilson and Newport, 1988), probably because unactivated eggs are exposed to contaminating Ca2+ ion during extract preparation. To explain our extract-to-extract variation in both replication enhancement per se (no enhancement in ∼20% of experiments) and the degree of enhancement (up to fourfold; averaging 2.5-fold), we hypothesize that in many of our extracts the mitotic CDK activity is not fully inactivated. Trace CDK activity might decrease the number of prereplication complexes that can assemble, and thus reduce the efficiency of replication. Further experiments are needed to test this hypothesis.
Hypothesis: LAP2 Proteins Serve as Downstream Effectors of Lamina Assembly in Promoting DNA Replication, Perhaps by Influencing Chromatin Structure
Nuclei assembled in lamin-depleted extracts cannot undergo DNA replication (Newport et al., 1990; reviewed by Gant and Wilson, 1997), initially suggesting that lamina assembly is required for DNA replication initiation or progression. However as noted above, replication can occur in the absence of nuclei if the chromatin is exposed to concentrated nucleosolic extracts (Walter et al., 1998). The replication-promoting components of these nucleosolic extracts have not yet been identified, and are likely to include multiple factors potentially including LAP2 isoform(s). Based on our evidence that LAP2 may affect chromatin structure, and that the full nucleoplasmic portion of LAP2β (fragment 1–408) enhances the efficiency of DNA replication, we hypothesize that LAP2 isoform(s) may act as downstream effectors of lamina assembly by promoting chromatin conformations favorable either to the assembly of prereplication complexes, or the progression of replication complexes. Further experiments are needed to determine if the putative chromatin-influencing activities of LAP2 proteins depend on lamin assembly, if they regulate lamin assembly, or both.
The idea that LAP2β might promote replication by affecting chromatin structure has a precedent when one considers LBR, the lamin B receptor. LBR interacts with a chromatin partner named Hp1 (Ye and Worman, 1996; Ye et al., 1997), which mediates repressive higher-order chromatin structure in Drosophila (reviewed by Elgin, 1996). In turn, Drosophila Hp1 is known to bind ORC1 (Pak et al., 1997). Thus, LBR (an inner nuclear membrane protein) binds to Hp1, which can bind a core component of the prereplication complex. The meanings and mechanisms of these interactions are not yet clear. However, it appears that LBR, and perhaps LAP2, may influence chromatin structure in ways that could modulate the competence of chromatin for DNA replication, and also conceivably its competence for transcription. Our identification of new exons in the lamin-binding region of Xenopus LAP2 isoforms increases the potential for subtle differences in functions of the various LAP2 isoforms. Further analysis of the effects of LAP2 isoforms on nuclear dynamics, chromatin structure, DNA replication, and potentially the transcriptional competence of chromosomes will be of interest.
Acknowledgments
We thank Dale Shumaker for his computer graphics expertise, Karen Chan for preparation of human LAP2 expression vectors, and Pamela Tuma for help with densitometry quantification. C.A. Harris particularly thanks John Siekierka for valuable advice and discussions. We thank Kenny Lee, Dale Shumaker, Jutta Beneken, Dan Leahy, and especially Carolyn Machamer for discussions and critical comments on the manuscript. K.L. Wilson is grateful to Carl Smythe, Julian Blow, and Chris Hutchison for enlightening discussions about replication.
This work was supported by a grant from the National Institutes of Health to K.L. Wilson.
Abbreviations used in this paper
BAF
barrier to autointegration factor
BrdU
bromodeoxyuridine
CDK
cyclin-dependent kinase
LAP
lamin-associated polypeptide
LBR
lamin B receptor
MWB
membrane wash buffer
NPC
nuclear pore complex
Footnotes
T.M. Gant's current address is California Pacific Medical Center, Geraldine Brush Cancer Research Institute, 2330 Clay Street, Stern Building, San Francisco, CA 94115-1932.
References
- Alsheimer M, Fecher E, Benavente R. Nuclear envelope remodelling during rat spermiogenesis: distribution and expression pattern of LAP2/ thymopoietins. J Cell Sci. 1998;111:2227–2234. doi: 10.1242/jcs.111.15.2227. [DOI] [PubMed] [Google Scholar]
- Bauer DW, Murphy C, Wu Z, Wu CH, Gall JG. In vitro assembly of coiled bodies in Xenopusegg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P, Dabauvalle MC, Scheer U. In vivo assembly of prenucleolar bodies in Xenopusegg extract. J Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger R, Theodor L, Shoham J, Gokkel E, Brok-Simoni F, Avraham KB, Copeland NG, Jenkins NA, Rechavi G, Simon AJ. The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res. 1996;6:361–370. doi: 10.1101/gr.6.5.361. [DOI] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nature Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Watson JV. Nuclei act as independent and integrated units of replication in a Xenopuscell-free DNA replication system. EMBO (Eur Mol Biol Organ) J. 1987;6:1997–2002. doi: 10.1002/j.1460-2075.1987.tb02463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Delannoy MR, Wilson KL. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J Cell Biol. 1992;116:281–294. doi: 10.1083/jcb.116.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp Cell Res. 1990;186:169–176. doi: 10.1016/0014-4827(90)90223-w. [DOI] [PubMed] [Google Scholar]
- Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS. DNA replication in cell-free extracts from Xenopuseggs is prevented by disrupting nuclear envelope function. J Cell Sci. 1992;101:43–53. doi: 10.1242/jcs.101.1.43. [DOI] [PubMed] [Google Scholar]
- Cox LS, Leno GH. Extracts from eggs and oocytes of Xenopus laevisdiffer in their capacities for nuclear assembly and DNA replication. J Cell Sci. 1990;97:177–184. doi: 10.1242/jcs.97.1.177. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R. Detergent-salt resistance of LAP2α in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 1998;17:4887–4902. doi: 10.1093/emboj/17.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Rine J. Roles for ORC in M phase and S phase. Science. 1998;279:1733–1737. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- Elgin SC. Heterochromatin and gene regulation in Drosophila. . Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Ellis DJ, Whitfield WGF, Hutchison CJ. GST-lamin fusion proteins act as dominant negative mutants in Xenopusegg extracts and reveal the function of the lamina in DNA replication. J Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. In press. [DOI] [PubMed]
- Furukawa K, Panté N, Aebi U, Gerace L. Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO (Eur Mol Biol Organ) J. 1995;14:1626–1636. doi: 10.1002/j.1460-2075.1995.tb07151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fritze CE, Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- Gant TM, Wilson KL. Nuclear assembly. Ann Rev Cell Dev Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- Gerace L, Foisner R. Integral membrane proteins and dynamic organization of the nuclear envelope. Trends Cell Biol. 1994;4:127–131. doi: 10.1016/0962-8924(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L. The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO (Eur Mol Biol Organ) J. 1993;12:4413–4424. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline S, Chan HK, Natarajan A, Siekierka JJ, Goldstein G. Three distinct human thymopoietins are derived from alternatively spliced mRNAs. Proc Natl Acad Sci USA. 1994;91:6283–6287. doi: 10.1073/pnas.91.14.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline SW, Mathew S, Siekierka JJ, Goldstein G. Structure and mapping of the human thymopoietin (TMPO) gene and relationship of human TMPO beta to rat lamin-associated polypeptide 2. Genomics. 1995;28:198–205. doi: 10.1006/geno.1995.1131. [DOI] [PubMed] [Google Scholar]
- Hozak P, Jackson DA, Cook PR. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J Cell Sci. 1994;107:2191–2202. doi: 10.1242/jcs.107.8.2191. [DOI] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Yan H, Newport J. A role for cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CJ, Bridger JM, Cox LS, Kill IR. Weaving a pattern from disparate threads: lamin function in nuclear assembly and DNA replication. J Cell Sci. 1994;107:3259–3269. doi: 10.1242/jcs.107.12.3259. [DOI] [PubMed] [Google Scholar]
- Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-α. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- Ishijima Y, Toda T, Matsushita H, Yoshida M, Kimura N. Expression of thymopoietin beta/lamina-associated polypeptide 2 (TP beta/LAP2) and its family proteins as revealed by specific antibody induced against recombinant human thymopoietin. Biochem Biophys Res Commun. 1996;226:431–438. doi: 10.1006/bbrc.1996.1373. [DOI] [PubMed] [Google Scholar]
- Jallepali PV, Kelly TJ. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? . Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lawlis SJ, Keezer SM, Wu J-R, Gilbert DM. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopusegg extracts. J Cell Biol. 1996;135:1207–1218. doi: 10.1083/jcb.135.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leno GH, Laskey RA. DNA replication in cell-free extracts from Xenopus laevis. . Methods Cell Biol. 1991;36:561–579. doi: 10.1016/s0091-679x(08)60297-6. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. The germinal vesicle material required for sperm pronuclear formation is located in the soluble fraction of egg cytoplasm. Exp Cell Res. 1983;148:481–491. doi: 10.1016/0014-4827(83)90169-6. [DOI] [PubMed] [Google Scholar]
- Lorka T, Cruzalegui FH, Fesquet D, Cavadore J-C, Méry J, Means A, Dorée M. Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopuseggs. Nature. 1993;366:270–273. doi: 10.1038/366270a0. [DOI] [PubMed] [Google Scholar]
- Lourim D, Krohne G. Membrane-associated lamins in Xenopusegg extracts: identification of two vesicle populations. J Cell Biol. 1993;123:501–512. doi: 10.1083/jcb.123.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D, Kempf A, Krohne G. Characterization and quantitation of three B-type lamins in Xenopusoocytes and eggs: increase of lamin LI protein synthesis during meiotic maturation. J Cell Sci. 1996;109:1775–1785. doi: 10.1242/jcs.109.7.1775. [DOI] [PubMed] [Google Scholar]
- Maison C, Pyrpasopoulou A, Theodoropoulos PA, Georgatos SD. The inner nuclear membrane protein LAP1 forms a native complex with B-type lamins and partitions with spindle-associated mitotic vesicles. EMBO (Eur Mol Biol Org) J. 1997;16:4839–4850. doi: 10.1093/emboj/16.16.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal S, Man NT, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- Martin L, Crimaudo C, Gerace L. cDNA cloning and characterization of lamina-associated polypeptide 1C (LAP1C), an integral protein of the inner nuclear membrane. J Biol Chem. 1995;270:8822–8828. doi: 10.1074/jbc.270.15.8822. [DOI] [PubMed] [Google Scholar]
- Masui Y. A quest for cytoplasmic factors that control the cell cycle. Prog Cell Cycle Res. 1996;2:1–13. doi: 10.1007/978-1-4615-5873-6_1. [DOI] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopuseggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Mills AD, Blow JJ, White JG, Amos WB, Wilcock D, Laskey RA. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J Cell Sci. 1989;94:471–477. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano A, Koga R, Ogawa M, Kurano Y, Kawada J, Okada R, Hayashi YK, Tsukahara T, Arahata K. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Wilson KL. Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol. 1991;36:607–634. doi: 10.1016/s0091-679x(08)60299-x. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Finlay DR, Forbes DJ. In vitrotransport of a fluorescent nuclear protein and exclusion of non-nuclear proteins. J Cell Biol. 1986;103:2091–2102. doi: 10.1083/jcb.103.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Dunphy W. Characterization of the membrane binding and fusion events during nuclear envelope assembly using purified components. J Cell Biol. 1992;116:295–306. doi: 10.1083/jcb.116.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E, Simos G, Georgatos SD, Giannakouros T. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J Biol Chem. 1996;271:8365–8372. doi: 10.1074/jbc.271.14.8365. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Pak DTS, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin replication complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Paulin-Levasseur M, Blake DL, Julien M, Rouleau L. The MAN antigens are non-lamin constituents of the nuclear lamina in vertebrate cells. Chromosoma. 1996;104:367–379. doi: 10.1007/BF00337226. [DOI] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO (Eur Mol Biol Org) J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Tschodrich-Rotter M, Peters R, Krohne G. Properties of fluorescently labeled Xenopus lamin A in vivo. . Eur J Cell Biol. 1994;65:70–81. [PubMed] [Google Scholar]
- Simos G, Georgatos SD. The inner nuclear membrane protein p58 associates in vivowith a p58 kinase and the nuclear lamins. EMBO (Eur Mol Biol Org) J. 1992;11:4027–4036. doi: 10.1002/j.1460-2075.1992.tb05496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Molec Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Weast, R.C. 1967. Handbook of Chemistry and Physics, 48th edition. Chemical Rubber Publishing Co. Cleveland, OH. D-144, D-149.
- Wiese C, Goldberg MW, Allen TD, Wilson KL. Nuclear envelope assembly in Xenopusextracts visualized by scanning EM reveals a transport-dependent “envelope smoothing” event. J Cell Sci. 1997;110:1489–1502. doi: 10.1242/jcs.110.13.1489. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Newport J. A trypsin-sensitive receptor on membrane vesicles is required for nuclear envelope formation in vitro. J Cell Biol. 1988;107:57–68. doi: 10.1083/jcb.107.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Wiese C. Reconstituting the nuclear envelope and endoplasmic reticulum in vitro. . Sem Cell Dev Biol. 1996;7:487–496. [Google Scholar]
- Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Evans CD, Blobel G. The lamin B receptor of the nuclear envelope inner membrane: a polytopic protein with eight potential transmembrane domains. J Cell Biol. 1990;111:1535–1542. doi: 10.1083/jcb.111.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J Cell Biol. 1997;139:1077–1087. doi: 10.1083/jcb.139.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to DrosophilaHP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]