Identification of Novel Listeria monocytogenes Secreted Virulence Factors following Mutational Activation of the Central Virulence Regulator, PrfA (original) (raw)
Abstract
Upon bacterial entry into the cytosol of infected mammalian host cells, the central virulence regulator PrfA of Listeria monocytogenes becomes activated and induces the expression of numerous factors which contribute to bacterial pathogenesis. The mechanism or signal by which PrfA becomes activated during the course of infection has not yet been determined; however, several amino acid substitutions within PrfA (known as PrfA* mutations) that appear to lock the protein into a constitutively activated state have been identified. In this study, the PrfA activation statuses of several L. monocytogenes mutant strains were subjected to direct isogenic comparison and the mutant with the highest activity, the prfA(L140F) mutant, was identified. The prfA(L140F) strain was subsequently used as a tool to identify gene products secreted as a result of PrfA activation. By use of two-dimensional gel electrophoresis followed by liquid chromatography-electrospray ionization-tandem mass spectroscopy analyses, 15 proteins were identified as up-regulated in the prfA(L140F) secretome, while the secretion of two proteins was found to be reduced. Although some of the proteins identified were known to be subject to direct regulation by PrfA, the majority have not previously been associated with PrfA regulation and their expression or secretion may be influenced indirectly by a PrfA-dependent regulatory pathway. Plasmid insertion inactivation of the genes encoding four novel secreted products indicated that three of the four have significant roles in L. monocytogenes virulence. The use of mutationally activated prfA alleles therefore provides a useful approach towards identifying gene products that contribute to L. monocytogenes pathogenesis.
Listeria monocytogenes is a gram-positive facultative intracellular bacterial pathogen capable of causing severe disease in humans following ingestion of contaminated food products (66). As an environmental pathogen, L. monocytogenes has several disparate lifestyles, which include life as a saprophyte in soil, water, and contaminated silage and life inside mammalian hosts (28). While little about patterns of bacterial gene expression in natural environments outside of host cells is known, L. monocytogenes gene expression profiles have been shown to shift dramatically when the bacterium transitions from planktonic growth in broth culture to the cytosol of infected mammalian host cells (10, 37). Many of the bacterial factors required for L. monocytogenes pathogenesis are regulated by a transcriptional regulator known as PrfA (47).
PrfA-dependent gene products play critical roles in every major step of bacterial pathogenesis, from host cell internalization of the bacteria and phagosomal escape to spread of the bacteria to adjacent cells (64, 66). L. monocytogenes induces its uptake into cells by using a variety of bacterial cell surface-associated molecules, including a family of PrfA-dependent internalin gene products (11, 17, 23). Once internalized, L. monocytogenes escapes from the intracellular vacuole through expression of two PrfA-dependent genes: hly, encoding the pore-forming toxin listeriolysin O (LLO) (12, 25, 39, 50), and plcA, encoding a phosphatidylinositol-specific phospholipase C (8). Upon entry into the cytosol, L. monocytogenes produces ActA, a PrfA-dependent gene product that directs host cell actin polymerization and enables bacterial spread into nearby host cells (40, 60). L. monocytogenes then escapes from the double membrane vacuole of the newly infected cell through the expression of plcA, hly, and an additional PrfA-dependent phospholipase encoded by plcB (59, 65). PrfA thus plays a critical role in L. monocytogenes pathogenesis by regulating the expression of genes required for multiple aspects of host infection (47).
The regulation of prfA expression and protein activity is quite complex and includes transcriptional (22), posttranscriptional (33), and posttranslational (52) mechanisms. While each regulatory mechanism is important for proper modulation of prfA expression and activity, the posttranslational regulation of PrfA is perhaps the least well understood. PrfA activation appears to require some form of protein modification or cofactor binding, and although the nature of the signal that mediates PrfA activation is not yet known, insight into the functional consequences of PrfA activation has been obtained through the isolation and characterization of PrfA mutants that lock the protein into a constitutively active state (referred to as prfA* mutants) (52, 57, 67, 72). When grown in vitro, prfA* mutants appear to mimic the natural activation of PrfA that occurs in L. monocytogenes within the cytosol of host cells where PrfA-dependent gene expression is highly up-regulated. Posttranslation activation of PrfA is required for L. monocytogenes virulence, as mutations in prfA that prevent activation result in severe attenuation in murine infection models (M. D. Miner, G. C. Port, J. C. Chang, H. G. A. Bouwer, and N. E. Freitag, submitted for publication).
As PrfA is a key regulator of L. monocytogenes virulence, we sought to identify the spectrum of gene products both directly and indirectly regulated by PrfA in order to uncover additional bacterial factors that contribute to L. monocytogenes pathogenesis. As part of this approach, levels of PrfA-dependent gene expression were directly compared between several prfA* mutant isogenic strains, thereby revealing the existence of two classes of prfA* mutants: moderately activated prfA* mutants [prfA(E77K) and prfA(G155S) mutants] and highly activated prfA* mutants [prfA(G145S) and prfA(L140F) mutants]. Profiles of secreted proteins derived from the most active prfA* mutant, the prfA(L140F) mutant, were then compared to profiles of secreted proteins isolated from wild-type as well as Δ_prfA_ mutant strains. Fifteen proteins were found to be up-regulated as a result of high-level PrfA activation, whereas two proteins were absent from the PrfA L140F secretome in comparison to wild-type and Δ_prfA_ mutant strains. As anticipated, novel PrfA activation-dependent gene products which were found to contribute to L. monocytogenes pathogenesis were identified.
MATERIALS AND METHODS
Bacterial strains and plasmids.
L. monocytogenes and Escherichia coli strains used in this study are listed in Table 1. All L. monocytogenes and E. coli strains were grown at 37°C in brain heart infusion (BHI) media (Difco Laboratories, Detroit, MI) and Luria broth (LB) (Invitrogen Corp., Grand Island, NY), respectively. For selection of pPL2 or its recombinant plasmids, chloramphenicol was used at 25 μg/ml for E. coli and at 5 to 10 μg/ml for L. monocytogenes. Streptomycin at 200 μg/ml was used to select for L. monocytogenes following bacterial conjugation and for the elucidation of bacterial CFU from the organs of infected mice (10403S is resistant to streptomycin).
TABLE 1.
L. monocytogenes and E. coli strains used in this study
| Strain | Description | Associated designation | Reference(s) or source |
|---|---|---|---|
| DH5α | E. coli strain for constructing recombinant plasmids | ||
| SM10 | E. coli strain as conjugation donor for pPL2 plasmid | ||
| NF-L100 | 10403S wild type | 3,18 | |
| DP-L2161 | 10403S Δ_hly_ | hly | 36 |
| DP-L3903 | 10403S with Tn_917_ insertion | 1 | |
| NF-L1003 | NF-L890 (Δ_prfA_) with actA-gus-plcB transcriptional fusion | 72 | |
| NF-E1019 | E. coli DH5α with pPL2-P_plcA_-prfA | 72 | |
| prfA mutants | |||
| NF-L1009 | NF-L1003 tRNAArg::pPL2 (Δ_prfA_ plus pPL2_i_)a | Δ_prfA_ | 72 |
| NF-L1011 | NF-L1003 tRNAArg::pPL2-prfA(L140F) (Δ_prfA_ plus L140F_i_) | prfA(L140F) | 72 |
| NF-L1041 | NF-L1003 tRNAArg::pPL2-prfA 476 (Δ_prfA_ plus WT_i_) | Wild-type prfA | 72 |
| NF-L1185 | NF-L1003 tRNAArg::pPL2-prfA(E77K) (Δ_prfA_ plus E77K_i_) | prfA(E77K) | This work |
| NF-L1186 | NF-L1003 tRNAArg::pPL2-prfA(G155S) (Δ_prfA_ plus G155S_i_) | prfA(G155S) | This work |
| NF-L1226 | NF-L1003 tRNAArg::pPL2-prfA(G145S) (Δ_prfA_ plus G145S_i_) | prfA(G145S) | This work |
| Insertion mutants | |||
| NF-L1239 | NF-L100 with pKSV7 single-crossover insertion within actA | _actA_ib | This work |
| NF-L1240 | NF-L100 with pKSV7 single-crossover insertion within lmo0135 | lmo0135i | This work |
| NF-L1241 | NF-L100 with pKSV7 single-crossover insertion within tcsA | _tcsA_i | This work |
| NF-L1242 | NF-L100 with pKSV7 single-crossover insertion within lmo2219 | lmo2219i | This work |
| NF-L1243 | NF-L100 with pKSV7 single-crossover insertion within lmo2417 | lmo2417i | This work |
| NF-L1245 | NF-L100 with pKSV7 single-crossover insertion near but not in lmo2219 | Silent pKSV7 insertion | This work |
| Δ_flaA_ mutant | |||
| NF-L1246 | NF-L965 tRNAArg::pPL2 (Δ_flaA_ plus pPL2_i_) | Δ_flaA_ | This work |
Construction of L. monocytogenes recombinant strains.
L. monocytogenes strains NF-L1009, NF-L1011, and NF-L1041 (72) are previously described derivatives of the 10403S parent strain. Briefly, all strains contain an in-frame prfA deletion as well as a transcriptional fusion of the reporter gene gus to actA within the L. monocytogenes chromosome. In addition, each strain contains the integrated plasmid vector pPL2 as empty vector (NF-L1009), with a wild-type copy of prfA (pPL2 _prfA_-WT [NF-L1041]), or with a prfA* allele [pPL2-prfA(L140F) (NF-L1011)]. Isogenic strains containing additional prfA* mutations were constructed as follows. Point mutations were introduced into pPL2 _prfA_-WT from pNF-E1019 by use of a QuikChange site-directed mutagenesis kit (Stratagene) with the oligonucleotides listed in Table S1 in the supplemental material according to the manufacturer's protocol to generate the prfA(E77K), prfA(G155S), and prfA(G145S) alleles. Plasmids containing the desired mutations within prfA in pPL2 were verified by sequencing. The resulting strains NF-L1185 [prfA(E77K)], NF-L1186 [prfA(G155S)], and NF-L1226 [prfA(G145S)] each contain a single copy of the integration vector pPL2 with the designated prfA allele inserted into the L. monocytogenes chromosome.
NF-L1246 (wild-type chromosomal prfA) was derived from nonmotile NF-L965 (Δ_flaA_) (57) conjugated with pPL2 vector in order to confer chloramphenicol resistance and to enable direct comparison of swimming motility with that of pPL2-complemented prfA* strains.
Measurement of GUS activity.
β-Glucuronidase (GUS) activity was measured as previously described (56, 57, 72), with some minor changes. Briefly, overnight cultures grown in BHI containing 5 μg/ml chloramphenicol (BHI cam5) were diluted 1:50 and grown with shaking at 37°C for 8 h. The optical density at 600 nm (OD600) was measured, and two 500-μl culture aliquots were collected hourly for all strains except for the prfA(L140F) (NF-L1011) and prfA(G145S) (NF-L1226) mutant strains, for which two 50-μl aliquots were collected (reflective of the increased GUS activity present in these two highly PrfA activated strains). Bacterial cells were recovered by centrifugation at 16,000 × g for 5 min, the supernatants were removed, and the bacterial pellets were frozen at −80°C for further analysis. For enzymatic assays, bacterial pellets were quickly thawed and resuspended in 100 μl or 1 ml ABT buffer (0.1 M potassium phosphate, pH 7.0, 0.1 M NaCl, 0.1% Triton). GUS activity was measured as described by Youngman (73), with the substitution of 4-methylumbelliferyl-β-d-glucuronide in place of 4-methylumbilliferyl-β-d-galactoside (Sigma Chemical Co., St. Louis, MO). Data were derived from duplicate samples taken from three independent experiments.
Isolation of bacterial secreted proteins.
For the isolation of secreted proteins, overnight cultures were diluted 1:20 into a final volume of 200 ml of prewarmed BHI cam5 and grown at 37°C for approximately 3 h to mid-log phase (OD600 of ∼0.6) with shaking. Culture OD600 was determined by using a spectrophotometer (UV-1201 UV-VIS spectrophotometer; Shimadzu Scientific Instruments, Inc., Columbia, MD).
Secreted proteins were isolated as previously described by Lenz and Portnoy (43), with minor modifications. Briefly, bacteria in 200 ml of mid-log-phase cell culture were pelleted by centrifugation at 20,000 × g for 5 min at 4°C and 160 ml of the resulting supernatants was collected. Trichloroacetic acid (TCA) was added to the supernatants to reach a final concentration of 5% TCA. Proteins were TCA precipitated on ice for 30 min. The supernatants were then centrifuged at 10,500 × g for 10 min at 4°C, the supernatants were discarded, and the resulting protein pellets were air dried. The TCA-precipitated protein pellets were washed with 4 ml of ice-cold acetone and centrifuged at 10,500 × g for 10 min at 4°C, decanted, and air dried again. The final pellets were resuspended in 400 μl of sodium dodecyl sulfate (SDS) boiling buffer containing 5% SDS, 10% glycerol, and 60 mM Tris, pH 6.8. Samples were boiled for 5 min before being frozen at −20°C until further analysis.
Protein concentrations were determined by bicinchoninic acid (BCA) analysis according to the manufacturer's protocol (Pierce, Rockford, IL). The absence of contaminating cytosolic proteins in the secreted protein samples was verified by Western analysis for the absence of PrfA protein in the secreted protein samples (data not shown).
Western analyses.
Recombinant LLO and rabbit polyclonal LLO antiserum were obtained from Diatheva, Italy. IRDye 680 goat anti-rabbit immunoglobulin G fluorescently labeled secondary antibody was obtained from Li-Cor Biosciences, Lincoln, NE. For Western analysis, 45.5 μg of secreted proteins from each strain was separated on NuPage Novex 10% bis-Tris gels and run according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). The proteins were then transferred onto nitrocellulose membranes at 30 V for 60 min. Membranes were processed and proteins were visualized using a Li-Cor Odyssey imager according to the manufacturer's protocol, with Odyssey blocking buffer (Li-Cor, Lincoln, NE). Primary antiserum was diluted 1:1,000, while secondary antibody was diluted 1:20,000. Proteins were detected on the Odyssey imager by scanning in the 680-nm wavelength and quantified by comparison with 8 ng of loaded recombinant LLO protein using Odyssey infrared imaging system application software, version 2.1 (Li-Cor, Lincoln, NE). Secreted proteins were isolated and analyzed from three independent experiments.
2-DE of protein samples.
Two-dimensional gel electrophoresis (2-DE) was performed by Kendrick Labs, Inc. (Madison, WI), according to the method of O'Farrell (48). Briefly, 5% β-mercaptoethanol was added to the protein samples and shipped overnight on dry ice to Kendrick Labs. Protein samples from each strain were loaded in equal concentrations based upon BCA analysis results, up to a maximum of 50 μl/gel (resulting in 250 to 500 μg/gel depending on the sample set). Isoelectric focusing (IEF) was carried out in glass tubes (inner diameter, 2.0 mm) by using 2.0% pH 4 to 8 ampholines (Gallard-Schlesinger Industries, Inc., Garden City, NY) for 9,600 V·h (IEF tube gel separation using carrier ampholines allows SDS-solubilized samples to be analyzed by 2-DE). One microgram of an IEF internal standard, tropomyosin (_M_r 33,000, pI 5.2), was added to the samples. After IEF, the tube gels were equilibrated for 10 min in buffer O (10% glycerol, 2.3% SDS, 0.0625 M Tris, pH 6.8) and sealed to the top of a stacking gel that overlays 10% acrylamide slab gels (0.75 mm thick). The SDS slab gel electrophoresis was carried out for 4.5 h at 12.5 mA/gel. The following proteins were added as molecular weight standards to the agarose that sealed the tube gel to the slab gel: myosin (_M_r 220,000), phosphorylase A (_M_r 94,000), catalase (_M_r 60,000), actin (_M_r 43,000), carbonic anhydrase (_M_r 29,000), and lysozyme (_M_r 14,000) (Sigma Chemical Co., St. Louis, MO). Following electrophoresis, the gels were stained with Coomassie brilliant blue R-250 and dried between two sheets of cellophane. Samples were prepared from three independent experiments, and duplicate gels were run for each sample. 2-DE gels were compared manually to identify protein spots unique to particular strains.
MS protein sample preparation and LC-ESI-MS-MS.
Coomassie-stained protein spots which were unique to the 2-DE gels of the prfA(L140F) mutant (NF-L1011), wild-type prfA (NF-L1041), or the Δ_prfA_ mutant (NF-L1009) were cut out and subjected to liquid chromatography-electrospray ionization-tandem mass spectroscopy (LC-ESI-MS-MS) analysis at the Fred Hutchinson Cancer Research Center Proteomics Facility, Seattle, WA.
Coomassie-stained gel slice trypsin digestions were performed as described by Shevchenko et al. (58), omitting the alkylation and reduction steps. Following digestion, samples were desalted using a micro-C18 ZipTip (Millipore) and dried. Samples were resuspended in 7 μl of 0.1% trifluoroacetic acid and analyzed by LC-ESI-MS-MS using a Microtech Scientific UltraPlus2 high-pressure liquid chromatograph coupled in-line with a ThermoElectron LCQ mass spectrometer. The instrumental configuration is described by Gatlin et al. (24) and used a pulled 100-μm capillary as both the electrospray tip and the chromatographic column. The capillary was packed with 5-μm-particle-size, 100-Å-pore-size Magic C18 (Michrom Bioresources) packing material. High-pressure liquid chromatography solvents were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B), and the separation gradient was 5% B to 40% B over 40 min. MS data were collected in a datum-dependent mode in which an MS scan was followed by MS-MS scans of the three most abundant ions from the preceding MS scan.
MS data were searched against four combined L. monocytogenes protein databases from TIGR (F2365, H7858, and F6854; http://www.tigr.org/tdb/listeria/) and the Pasteur Institute (EGD-e; http://genolist.pasteur.fr/ListiList/) by use of a software search algorithm (Institute for Systems Biology, Seattle, WA). Results were considered valid if at least two peptides identified to a protein and if the peptide matches had raw scores greater than 200 for +1 ions, 300 for +2 ions, and 300 for +3 ions, Z-scores greater than 4, and percent ions of greater than 15%. For further stringency, each protein was labeled as presented in Tables 4 and 5 if it was identified by MS in at least two out of three independent experiments. Proteins were given gene names based upon the sequenced EGD-e strain from the ListiList server (http://genolist.pasteur.fr/ListiList/).
TABLE 4.
Polypeptides absent or down-regulated in the PrfA L140F secretome
| Functional class | Gene designation | Protein name | Protein description | Spot no. | Peptide matcha | Coverage (%)b | Predicted pI | Predicted mass (kDa) | Observed pI | Observed mass (kDa) | References for virulence contribution |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Motility | lmo0690 | FlaA | Flagellin | 40 | 30 | 63.1 | 4.7 | 30.4 | 5.9 | 32.5 | 16,49 |
| Energetics | lmo0013 | QoxA | AA3-600 quinol | 41 | 28 | 45.1 | 6.4 | 41.6 | 7.0 | 34.5 | |
| oxidase subunit II | 42 | 7 | 20.7 | 6.9 | 34.6 |
TABLE 5.
Comparison of plaque formations by the L. monocytogenes gene disruption mutants
| Strain | Associated gene designation or description | Plaque size (% of wild type)a |
|---|---|---|
| NF-L100 | Wild type | 100 ± 9.9 |
| NF-L1239 | _actA_i | 0 |
| NF-L1239 | lmo0135i | 100.5 ± 7.6 |
| NF-L1239 | _tcsA_i | 95.4 ± 10.6 |
| NF-L1239 | lmo2219i | 86.3 ± 5.6*** |
| NF-L1239 | lmo2417i | 99.6 ± 6.0 |
Swimming motility assays.
Bacterial swimming motility was measured as previously described (57, 72), with slight modifications. Briefly, swimming motility was evaluated on semisolid (0.3% [wt/vol] agar) BHI cam5 plates stab inoculated from isolated colonies and incubated for 48 h at 37°C or at room temperature as indicated. Motility was quantified as the diameter of the swimming colony, measured at 24 and 48 h, minus the diameter of a nonmotile Δ_flaA L. monocytogenes_ deletion mutant (NF-L1246), and wild-type (NF-L1041) motility was set to 100%. Results were obtained from triplicate samples of two independent experiments.
Plasmid insertional disruption mutagenesis of target genes in the L. monocytogenes chromosome.
Five candidate genes and three control genes were selected for mutational analyses in L. monocytogenes by plasmid-based insertional disruption. Briefly, a 500-bp fragment of internal coding sequences of each candidate or control gene was chosen such that upon a single-crossover event a gene disruption was generated through targeted plasmid integration. Oligonucleotides (see Table S1 in the supplemental material) were designed for PCR amplification of internal coding sequences within the following genes: actA, gap, rpoB, lmo0135, tcsA, lmo1438, lmo2219, and lmo2417. The resulting PCR products were digested with XbaI and PstI and ligated into the temperature-sensitive plasmid vector pKSV7 (61) digested with the appropriate restriction enzymes.
As an integration control to determine the effect on L. monocytogenes fitness following plasmid integration, a pKSV7 construct originally intended for allelic exchange of lmo2219 was utilized (this construct results in a silent plasmid insertion and does not cause the insertional inactivation of any L. monocytogenes gene product). Briefly, oligonucleotides (see Table S1 in the supplemental material) were designed for PCR amplification of approximately 600 bp both upstream and downstream of the start and stop codons of lmo2219, respectively, and fused together via splicing by overlap extension PCR as previously described (31). The resulting PCR product was subcloned into pKSV7 following digestion with XbaI.
Plasmid constructs were subsequently electroporated into electrocompetent L. monocytogenes as previously described (30), incubated in BHI at 30°C for 2 h, plated onto BHI cam10 plates, and incubated at 30°C for 2 days. Single colonies were picked and patched onto fresh BHI cam10 plates and incubated overnight at 30°C. Colonies were then picked and inoculated into 3 ml BHI cam10 media and passaged for 3 days with 1:1,000 dilutions at 40°C with vigorous shaking. The 3 days of passage at high temperature (40°C) prohibits the temperature-sensitive pKSV7 plasmid from replicating and selects for plasmid integration in the presence of antibiotic. For each mutant, plasmid integration was confirmed by PCR using one primer outside of the region of integration and another for the plasmid (see Table S1 in the supplemental material).
L2 plaque assays.
Plaque assays were performed with murine L2 fibroblasts as described previously (63), with a multiplicity of infection of ∼1:3. Plaque size was measured using a comparator (Finescale, Orange, CA). The average diameters of at least 20 plaques from two independent experiments were determined for each strain, with the average wild-type (NF-L100) diameter being set to 100%.
Mouse intravenous infections.
All animal procedures were IACUC approved. L. monocytogenes was grown in BHI or BHI cam10 for strains with plasmid insertions, with shaking at 37°C, to an OD600 of ∼0.6. Bacterial cells were pelleted, washed in phosphate-buffered saline (PBS), and resuspended in PBS to reach a final concentration of approximately 4 × 104 bacteria/ml. Six to eight Swiss Webster mice (Charles River Laboratories, New York, NY) at 12 to 15 weeks old were infected via tail vein injection with 0.5 ml of bacterial culture (2 × 104 CFU). Three days postinfection, mice were sacrificed and livers and spleens were harvested. Organs were placed in 10 ml of 0.2% NP-40 (Nonidet P-40), tissues were homogenized using a Tissue Master 125 (Omni International, Marietta, GA), and 10-fold serial dilutions were plated onto BHI plates containing 200 μg/ml streptomycin. To ensure that plasmid integration was maintained throughout the mouse infection, at least 100 colonies/organ/mouse were replica plated onto BHI cam7.5 plates.
An actA insertional disruption mutant was used as a negative control, and this mutant was largely cleared from infected mice: no bacteria were recovered from infected livers, and of those mice with bacteria in the spleen, 5 log fewer mutant than wild-type CFU were recovered (∼103 versus ∼108) (data not shown). Following replica plating onto BHI cam7.5 plates to ensure plasmid maintenance during infection, ∼0.1% of colonies tested lost the integrated plasmid (with the exception of the lmo0135i mutant (lmo0135 plasmid insertion mutant), for which 3.4% of recovered colonies lost the integrated plasmid), and 54.5% of the actA insertion mutants recovered lost the integrated plasmid (data not shown).
RESULTS
Direct comparison of levels of PrfA-dependent gene expression in L. monocytogenes strains containing different prfA* alleles.
A number of mutations within prfA that have been reported to lead to the constitutive expression of virulence genes in bacterial strains grown in broth culture (prfA* mutations) have been identified over the past several years (52, 57, 67, 72). While some prfA* mutants have been analyzed using functional assays (57, 67), a direct comparison of levels of virulence gene expression between multiple isogenic prfA* mutants has not yet been reported. To identify the prfA mutation conferring the highest level of PrfA activation, four isogenic strains containing prfA* mutations were chosen for direct comparison: prfA(G145S) (52), prfA(E77K), prfA(G155S), and prfA(L140F) (57, 67, 72) strains.
Whereas the prfA(E77K) and prfA(G155S) mutations can easily be introduced into the 10403S parent strain via allelic exchange (57), the prfA(G145S) and prfA(L140F) mutations appeared to confer subtle effects on bacterial growth that interfere with allelic exchange, as several attempts to reconstruct each of the mutations in wild-type strains proved unsuccessful (72; unpublished data). Therefore, to enable isogenic comparison of the four prfA* alleles, each prfA mutation was introduced into a Δ_prfA L. monocytogenes_ background by use of a pPL2-based integration vector (41) that contained the selected mutant prfA coding sequence along with all of the promoters that regulate prfA expression (Table 1). These _prfA_-pPL2 vector constructs were then integrated into the bacterial chromosome in single copy within the tRNAArg gene without gene disruption (41). A similar construct containing wild-type _prfA_-pPL2 has previously been shown to functionally complement a prfA deletion strain (72). Drug selection (chloramphenicol) was maintained throughout the course of in vitro experiments to ensure retention of the integrated plasmid vectors.
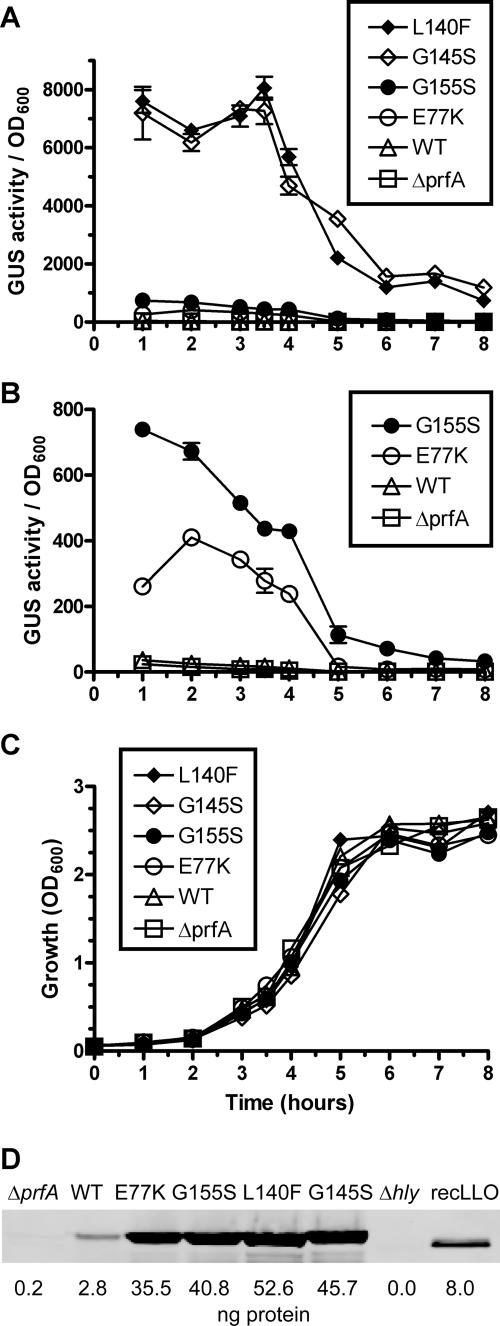
To quantitate and compare the levels of PrfA activation in strains containing the mutant prfA alleles, the reporter gene gus, encoding GUS, was introduced as a transcriptional fusion downstream of actA within the L. monocytogenes chromosome (56). actA is a PrfA-dependent virulence gene that is normally expressed at low levels in bacteria grown outside of host cells but is highly expressed following the activation of PrfA, a transition that normally occurs in bacteria located within the host cell cytosol (56; Miner et al., submitted). All pPL2-complemented prfA* strains exhibited similar growth characteristics in broth culture (Fig. 1C). As expected, strains that lacked functional PrfA (Δ_prfA_) (NF-L1009) or that contained a complementing wild-type copy of prfA on pPL2 (NF-L1041) exhibited very low levels of actA expression during growth in broth culture (Fig. 1A and B). However, the levels of actA expression for the various prfA* mutant strains were found to be greatly increased in comparison. The prfA(E77K) mutant (NF-L1185) and the prfA(G155S) mutant (NF-L1186) exhibited significantly increased levels of actA expression (17-fold and 26-fold, respectively, at peak levels of expression) (Fig. 1B), while the prfA(L140F) mutant (NF-L1011) and the prfA(G145S) mutant (NF-L1226) were the most active PrfA* mutants tested (Fig. 1A), with peak expression levels that were 480-fold and 434-fold greater, respectively, than those of strains containing wild-type prfA (Table 2).
FIG. 1.
actA expression levels and LLO secretion in isogenic L. monocytogenes prfA* mutants grown in BHI broth culture at 37°C. Units of GUS activity were determined as described in Materials and Methods, following normalization for bacterial culture OD600. Data shown are from duplicate samples and are representative of three separate experiments, expressed as averages ± standard deviations. (A) actA expression of wild-type prfA (WT), prfA(L140F), prfA(G145S), prfA(G155S), prfA(E77K), and Δ_prfA_ isogenic strains. (B) actA expression as described for panel A, excluding the highly active prfA(L140F) and prfA(G145S) strains and expanded for clarity. (C) Growth curves of all strains in BHI broth culture at 37°C, as determined by OD600. (D) Secreted proteins were TCA precipitated from bacterial supernatants during the mid-log phase of bacterial growth in BHI (OD600 of ∼0.6). Proteins were separated using SDS-polyacrylamide gel electrophoresis, and LLO was detected by Western analysis using rabbit polyclonal antiserum directed against LLO followed by a fluorescently labeled goat anti-rabbit immunoglobulin G (IRDye 680) secondary antibody. Proteins were visualized using an Odyssey imager, and quantification was determined using Odyssey infrared imaging system application software, version 2.1. Strain genotypes are noted above each lane, and values below indicate nanograms of protein detected compared to 8 ng of recombinant LLO (recLLO). Data shown are representative of three separate experiments.
TABLE 2.
actA expression as measured by GUS activity/OD600 for prfA mutant strains and LLO secretion as measured by Western analysis
| Strain | Genotype or description | _actA_-GUS activity | LLO quantification | ||
|---|---|---|---|---|---|
| Avg valuea | Fold Δb | Secreted protein (ng)c | Fold Δb | ||
| NF-L1041 | Wild-type prfA | 17 ± 1 | 1 | 2.8 | 1 |
| NF-L1009 | Δ_prfA_ | 10 ± 0 | 0.6 | 0.2 | 0.1 |
| NF-L1185 | prfA(E77K) | 279 ± 26 | 17 | 35.5 | 12.7 |
| NF-L1186 | prfA(G155S) | 437 ± 3 | 26 | 40.8 | 14.6 |
| NF-L1226 | prfA(G145S) | 7,273 ± 456 | 434 | 45.7 | 16.3 |
| NF-L1011 | prfA(L140F) | 8,055 ± 390 | 481 | 52.8 | 18.9 |
The prfA* mutant strains were also compared by monitoring secreted LLO protein levels via Western analysis. Comparison of secreted protein fractions derived from the various prfA* strains tested yielded LLO profiles that were consistent with mid-level activation of PrfA as conferred by the prfA(E77K) and prfA(G155S) mutant alleles, with higher levels of activation conferred by the prfA(L140F) and prfA(G145S) mutations (Fig. 1D). Strains containing the prfA(L140F) allele secreted the greatest amounts of LLO (an 18.9-fold increase in comparison to the amount for the wild type) (Table 2). The differences in magnitude of the relative severalfold increases for actA and hly expression as measured between the prfA* and wild-type strains likely reflect the differential requirements of these promoters for PrfA activation. The hly promoter exhibits significant expression in broth culture in the absence of PrfA activation, whereas the actA promoter is expressed at low to undetectable levels under identical conditions (56). As the prfA(L140F) mutation appeared to confer the most substantial degree of PrfA activation as measured by both hly and actA expression, strains containing this mutation were chosen for the analyses of the effects of PrfA activation on L. monocytogenes secreted protein profiles.
Mutational activation of PrfA leads to altered patterns of protein secretion.
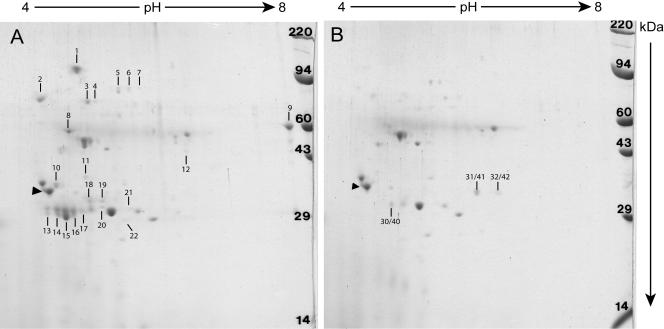
Bacterial virulence factors are often either secreted or surface localized to promote interaction and engagement with host cell components. In order to identify proteins whose production and/or secretion was directly or indirectly dependent upon PrfA activation, supernatants were collected during the mid-log phase of bacterial culture growth, corresponding to the time at which PrfA* activity was determined to be maximal (Fig. 1), and proteins were collected via TCA precipitation and analyzed by 2-DE.
Strains lacking functional PrfA (Δ_prfA_) or containing wild-type prfA produced secreted protein profiles that appeared essentially identical (data not shown), consistent with the lack of PrfA activation during bacterial growth in broth culture (Fig. 1 and Table 2). The profile of secreted proteins derived from the mutationally activated PrfA* prfA(L140F) strain was strikingly different from wild-type or Δ_prfA_-derived profiles and showed enhanced expression of 22 polypeptide species as detected via Coomassie staining (Fig. 2). Three species were also notably absent in the PrfA*-derived supernatants but present in both the wild-type and Δ_prfA_ supernatant fractions (Fig. 2). By comparison, prfA(G145S)-derived secreted protein profiles looked indistinguishable from those of prfA(L140F) strains, while the moderately activated PrfA* prfA(E77K) and prfA(G155S) mutants yielded similar yet less intense polypeptide patterns (data not shown), a result consistent with the high PrfA activation state of the prfA(G145S) mutant and the lower-level PrfA activation state of the prfA(E77K) and prfA(G155S) mutants (Fig. 1 and Table 2).
FIG. 2.
2-DE Coomassie-stained gels of secreted proteins isolated from isogenic L. monocytogenes prfA mutants. Secreted proteins were TCA precipitated from bacterial supernatants following approximately 3 h of bacterial growth in BHI (OD600 of ∼0.6). Protein concentration was determined by BCA analysis, and equal amounts of protein were separated by 2-DE. Following electrophoresis, the gels were Coomassie stained and dried. Tropomyosin was included as a pI control (indicated by arrowhead), and a molecular size ladder (far right) was included. Polypeptide spots that were strain unique (indicated by vertical lines and numbers) were cut out and subjected to MS analysis. (A) L. monocytogenes prfA(L140F) mutant (NF-L1011). (B) L. monocytogenes wild-type prfA (NF-L1041).
The differentially regulated polypeptide spots were excised from 2-DE gels and identified via LC-ESI-MS-MS. MS data were searched against all available L. monocytogenes protein databases, including two serotype 1/2a strains (EGD-e and F6854) and two serotype 4b strains (F2365 and H7858). From the 22 polypeptide spots up-regulated in the activated PrfA* strain, 15 proteins were identified (Table 3). Four of these, ActA, LLO, PlcB, and InlC, are encoded by genes that are known to be directly PrfA regulated via PrfA binding sites located within their promoter regions (47). Of the remaining 11 up-regulated proteins, only lmo2219 has been associated with PrfA regulation, as lmo2219 contains a potential PrfA binding site within its putative promoter region and was determined by microarray analyses to be up-regulated in a prfA(G145S) mutant (46). The 10 remaining up-regulated proteins, however, have not previously been associated with PrfA-dependent regulation, and the genes encoding these proteins all lack discernible PrfA binding sites within their predicted promoters or immediate upstream regions. The majority of these proteins have predicted functions that encompass four functional classes: cell wall modification enzymes, components of protein secretion, ABC transporters, and unknown lipoproteins. The absence of apparent PrfA binding sites within the promoters and/or immediate upstream regions suggests that the PrfA*-dependent secretion of these gene products may occur via an indirect mechanism, such as the PrfA*-dependent expression of another cytosolic regulator, or alternatively via a secretion pathway that is dependent upon PrfA activation.
TABLE 3.
Polypeptides unique or up-regulated in the PrfA L140F secretome
| Functional class | Gene designation | Protein name | Protein description | Spot no. | Peptide matcha | Coverage (%)b | Predicted pI | Predicted mass (kDa) | Observed pI | Observed mass (kDa) | Reference(s) or source for virulence contribution |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Virulence | lmo0202 | LLO | Listeriolysin O | 9 | 124 | 72.0 | 8.2 | 58.7 | 7.7 | 60.0 | 38,50 |
| lmo0204 | ActA | Actin assembly- | 1 | 60 | 46.3 | 4.7 | 70.3 | 5.9 | 125.0 | 14,40 | |
| inducing protein | 2 | 48 | 26.6 | 5.6 | 80.0 | ||||||
| 11 | 19 | 26.0 | 5.5 | 37.0 | |||||||
| lmo0205 | PlcB | Phospholipase C | 13-17 | 7 | 23.5 | 8.4 | 33.3 | 5.8 | 30.0 | 65 | |
| lmo1786 | InlC | Internalin C | 21 | 9 | 27.7 | 6.5 | 33.1 | 5.8 | 30.0 | 15,21 | |
| ABC transporter | lmo0135 | Similar to oligopeptide ABC transport system substrate-binding proteins | 8 | 40 | 53.6 | 4.8 | 58.3 | 5.9 | 58.0 | This study | |
| lmo2196 | OppA | Similar to pheromone ABC transporter (binding protein) | 3 | 30 | 49.6 | 5.1 | 62.6 | 6.0 | 80.0 | 4 | |
| lmo2417 | Conserved lipoprotein (putative ABC transporter binding protein) | 13-17 | 4 | 15.6 | 5.1 | 30.7 | 5.8 | 30.0 | |||
| Cell wall | lmo1438 | Similar to penicillin-binding protein | 5 | 10 | 17.5 | 8.3 | 79.9 | 6.2 | 85.5 | 29 | |
| modifying | 6 | 31 | 43.0 | 6.3 | 85.5 | ||||||
| 7 | 36 | 51.5 | 5.8 | 83.0 | |||||||
| lmo2522 | Similar to hypothetical cell wall binding protein from B. subtilis | 13-17 | 6 | 15.9 | 5.8 | 29.1 | 5.8 | 30.0 | |||
| 17 | 5 | 23.1 | 6.5 | 14.0 | |||||||
| 20 | 33 | 63.9 | 5.7 | 30.5 | |||||||
| lmo2691 | MurA or NamA | Similar to autolysin, _N_-acetylmuramidase | 3 | 2 | 7.1 | 10.3 | 63.6 | 6.0 | 80.0 | 9,42 | |
| 3 and 4 | 7 | 14.7 | 6.0 | 78.0 | |||||||
| lmo2754 | PBP5 | Similar to d-alanyl-d-alanine carboxypeptidase (penicillin-binding protein 5) | 12 | 20 | 42.5 | 7.6 | 48.1 | 6.8 | 45.0 | 29 | |
| Antigenic lipoprotein | lmo1388 | TcsA | CD4+ T-cell-stimulating (surface) antigen, lipoprotein | 10 | 18 | 59.4 | 4.8 | 38.4 | 5.3 | 35.0 | This study |
| lmo2637 | Conserved lipoprotein (putative) | 18 | 22 | 36.1 | 6.6 | 32.7 | 6.0 | 33.0 | |||
| 19 | 2 | 10.7 | 5.7 | 32.0 | |||||||
| Secretion | lmo0292 | HtrA | Similar to heat shock protein HtrA serine protease | 2 | 8 | 21.6 | 4.3 | 52.8 | 5.6 | 80.0 | 62,70 |
| lmo2219 | PrsA2 | Similar to posttranslocation molecular chaperone (protein export protein) (PrsA from B. subtilis) | 19 | 13 | 39.2 | 5.6 | 32.7 | 5.7 | 32.0 | 10, this study | |
| 20 | 17 | 43.7 | 5.6 | 31.0 |
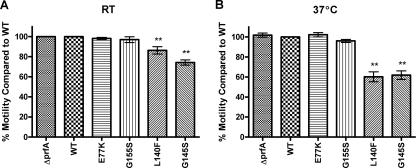
The secretion of QoxA and FlaA is reduced in prfA* strains, and the observed decrease in flagellar motility of prfA* strains correlates with the lack of FlaA secretion.
Two proteins, QoxA and FlaA, were identified as present in the wild-type and Δ_prfA_-derived supernatants but absent in the supernatants derived from the prfA(L140F) strain (Table 4). The observed reduction in FlaA secretion was intriguing given that it has previously been shown that selected prfA* mutants exhibited defects in swimming motility whereas strains lacking prfA exhibited increased motility in comparison to wild-type strains (57, 72). Consistent with these results, a direct comparison of each prfA* mutant strain on soft agar indicated that PrfA* activity is inversely proportional to swimming motility (Fig. 3). Bacterial strains containing the most active prfA* alleles [prfA(L140F) and prfA(G145S) strains] exhibited the greatest defects in swimming motility (60% ± 5.0% and 62% ± 4.3%, respectively, of the level of motility observed for strains containing wild-type prfA) (Fig. 3B). The reduced secretion of FlaA protein by prfA* mutant strains establishes a functional link between PrfA activation and motility.
FIG. 3.
Swimming motility of isogenic L. monocytogenes prfA mutants in semisolid media. Single colonies were stab inoculated into BHI 0.3% agar plates and incubated at room temperature or at 37°C. Swimming motility was calculated as the diameter of the halo as measured at 24 and 48 h minus the diameter of the nonmotile Δ_flaA L. monocytogenes_ deletion mutant (NF-L1246). Motility data are representative of triplicate samples obtained from two separate experiments and are expressed as averages ± standard errors of the means. For statistical analysis, all strains were compared against pPL2 _prfA_-WT (NF-L1041) by use of the Dunnett test and were calculated using GraphPad InStat, version 3.06 (San Diego, CA). Swimming motility at (A) room temperature (RT) or (B) 37°C, with wild-type (WT) motility set at 100%. **, P < 0.01.
Identification of novel proteins secreted as a result of PrfA activation that contribute to L. monocytogenes virulence.
Four of the 15 up-regulated proteins identified from the PrfA L140F secretome have well-established roles in L. monocytogenes virulence: ActA, LLO, PlcB, and InlC (66). The potential contributions of the remaining 11 identified proteins have not been well defined, although some of these proteins (Lmo0292 [HtrA], Lmo1438, Lmo2196 [OppA], Lmo2219, Lmo2691 [MurA, or NamA], and Lmo2754 [PBP5]) have been implicated in pathogenesis (4, 9, 10, 29, 42, 62, 70). To explore the pathogenic potential of additional proteins secreted from L. monocytogenes as a result of PrfA activation, the genes encoding five up-regulated proteins were selected for targeted disruption using the following criteria: (i) the existence of previously published data suggesting PrfA-dependent expression, (ii) similarity of the identified gene to virulence genes described for other bacterial pathogens, and/or (iii) lack of an established role in virulence in L. monocytogenes. At least one gene was chosen from each functional class, with the exception of the class of known virulence genes. Of the secreted proteins identified, the only novel gene product encoded by a gene with both a potential PrfA binding site in its promoter and existing evidence for PrfA-dependent gene expression was the putative protein secretion chaperone lmo2219 (46). Two of the three ABC transporters, encoded by lmo0135 and lmo2417, were targeted for insertional disruption, as the third (Lmo2196, also known as OppA) had previously been associated with virulence (4). One of the cell wall-modifying enzymes, Lmo1438, was selected, as well as the antigenic lipoprotein encoded by lmo1388 (also known as tcsA), based on its previous demonstration of expression within host cells and its capacity to stimulate T cells (54).
The targeted genes were disrupted through plasmid integration using the temperature-sensitive pKSV7 plasmid and selection for single-crossover events. Plasmid vector insertion controls including pKSV7 with internal fragments of the essential housekeeping genes gap or rpoB or empty pKSV7 vector failed to produce any chloramphenicol-resistant colonies at 40°C, confirming the site specificity of plasmid integration and the essential roles of gap and rpoB in L. monocytogenes physiology (data not shown). Of the five genes selected for plasmid disruption, the strain containing the plasmid insertion targeted for lmo1438 failed to grow under selective conditions (data not shown); this result strongly suggests that lmo1438 is an essential gene in L. monocytogenes strain 10403S. The remaining four plasmid insertional disruption mutants all exhibited similar growth rates in broth culture (data not shown).
The ability of L. monocytogenes to enter host cells, escape from vacuoles, replicate within the cytosol, and spread to adjacent cells can be assessed based on the ability of bacteria to form zones of clearing or plaques in tissue culture monolayers (63). The number of plaques formed and the size of the plaques can be used as indications of bacterial invasion and cell-to-cell spread, respectively. To compare levels of intracellular growth of the targeted gene disruption mutants in tissue culture cells, murine L2 monolayers were infected with each mutant strain and plaques were measured 3 days postinfection. All of the gene disruption mutants formed plaques of approximately the same size and frequency as wild-type L. monocytogenes, with the exception of the lmo2219 mutant, which was observed to yield slightly smaller plaques (86% that of the wild type) (Table 5), suggestive of a modest intracellular growth defect. In comparison, a mutant containing an insertional disruption of actA failed to produce any plaques, resulting from the inability of this strain to spread to adjacent cells (Table 5) (6).
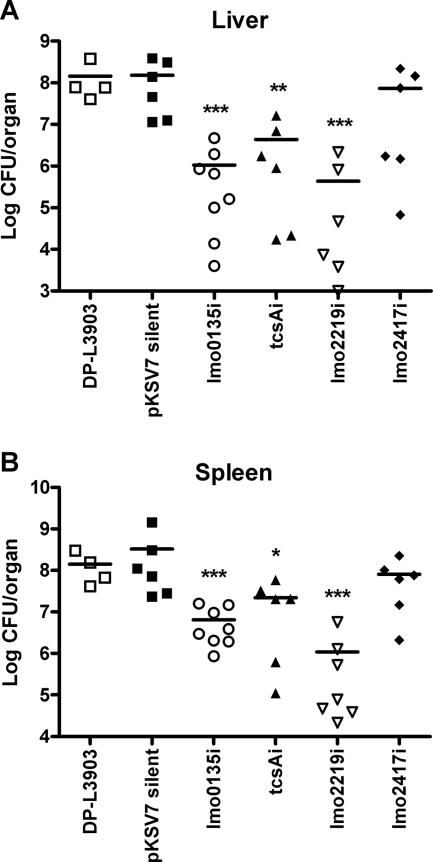
To address the potential contributions of the selected genes to animal infection, the gene disruption mutants were examined for the ability to replicate within the livers and spleens of infected mice. Twelve- to 15-week-old Swiss Webster mice were injected intravenously, and the livers and spleens were harvested at 72 h postinfection to assess bacterial load. Mutants were compared against two different reference strains: DP-L3903 (a 10403S derivative containing a silent Tn_917_ insertion that is fully virulent in mouse intravenous infections [1]) and NF-L1245, which contains a silent pKSV7 insertion within the L. monocytogenes genome. Both DP-L3903 and NF-L1245 were fully virulent, based on their ability to replicate within the livers and spleens of infected animals (Fig. 4). In contrast, three of the four gene disruption mutants showed significant defects in virulence, based on the reduced bacterial numbers recovered from both liver and spleen (Fig. 4). The tcsA insertion mutant (lmo1388) resulted in more than a 10-fold decrease in bacterial CFU recovered from the liver and spleen in comparison to wild-type strains, whereas the lmo0135 and lmo2219 gene disruption mutants were reduced in bacterial numbers by 2 log or more. The lmo2417 gene disruption mutant showed no significant defect. The lmo1388, lmo0135, and lmo2219 gene products therefore are not required for L. monocytogenes growth in broth culture but are required for full bacterial virulence in infected mice.
FIG. 4.
Bacterial growth in mouse livers and spleens following intravenous infection with L. monocytogenes gene disruption mutants. L. monocytogenes mutant strains were grown to mid-log phase in BHI broth, the bacteria were washed with PBS, and 2 × 104 CFU were injected via tail vein into six to eight 12- to 15-week-old Swiss Webster mice. Mice were euthanized at 3 days postinfection, organs were harvested and homogenized, and aliquots of tissue suspensions were plated on selective media for determination of bacterial CFU. Data are presented as scatter plots of CFU, with mean values indicated by horizontal bars. For statistical analysis, all strains were compared against the silent pKSV7 integration mutant NF-L1245 by use of unpaired t tests with two tails of transformed data, which were calculated using GraphPad InStat, version 3.06 (San Diego, CA). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Results were nearly identical when candidates were compared against DP-L3903, with the exception that the _tcsA_i spleen data were no longer significant (P = 0.0656). i, the pKSV7 plasmid insertion mutant.
DISCUSSION
PrfA activation is believed to be critical for the ability of L. monocytogenes to transition from life as an environmental organism to life within a mammalian host (28). Activation occurs following entry of L. monocytogenes into the cytosol of infected host cells (Miner et al., submitted) and leads to the expression of gene products that promote bacterial replication and spread to adjacent cells. Although significant progress towards identifying gene products directly regulated by PrfA has been made through the use of genome analysis and microarrays (26, 46), the full impact of PrfA activation on L. monocytogenes protein expression and bacterial cell physiology has not yet been examined. By using the mutational activation of PrfA as a tool for elucidating changes in protein production and secretion, this study has identified 17 proteins whose secretion is influenced by PrfA activation and has further implicated three novel proteins in L. monocytogenes virulence.
Current PrfA* mutants can be classified into at least two categories based on the expression of PrfA-dependent virulence genes in L. monocytogenes broth cultures: moderately active PrfA* mutants (PrfA E77K and PrfA G155S mutants) and highly active PrfA* mutants (PrfA G145S and PrfA L140F mutants). The existence of two distinguishable PrfA* mutant classes could indicate that PrfA is capable of undergoing a progressive conformational shift from an inactive species into a fully active state, with the moderately active class of mutants representing an intermediate step within that progression. Alternatively, the mid-level-activation mutants may be reflective of an artificial conformation imposed on PrfA structure by each of the two mutations. Structural comparisons between wild-type PrfA and PrfA G145S mutant proteins by Eiting et al. (19) have revealed a conformational difference that exists between the two proteins such that the helix-turn-helix region responsible for DNA binding appears more accessible in the PrfA G145S mutant. The ability of the prfA(E77K) and prfA(G155S) mutant strains to remain fully virulent (or even hypervirulent) in animal models of infection strongly suggests that the moderately active PrfA* mutants retain the ability to become fully activated within host cells (53, 57).
The proteins identified in this study as differentially secreted in the presence of one of the most active prfA* alleles, prfA(L140F), fall into several functional categories, including ABC transport components, cell wall modification enzymes, antigenic lipoproteins, secretion machinery components, metabolic enzymes, and components necessary for swimming motility. Recent studies have implicated some of these proteins in bacterial virulence, whereas the roles of others remain to be defined. For example, ABC transporters couple ATP hydrolysis with transport of small molecules across bacterial membranes, and oligopeptide ABC transporters have been shown to be involved not only in nutrient acquisition but also in protein secretion and quorum sensing in gram-positive bacteria (13, 27). Three ABC transport components were identified in the prfA(L140F) mutant secretome: Lmo0135, Lmo2196 (OppA), and Lmo2417. OppA has previously been shown to mediate transport of oligopeptides and to contribute to intracellular growth and survival of bacteria in bone marrow-derived macrophages and in the livers and spleens of infected mice (4). Based on the reduced numbers of the lmo0135 insertion mutant CFU recovered from the livers and spleens of infected mice (Fig. 4), the lmo0135 gene product contributes to L. monocytogenes pathogenesis in animals, perhaps by providing transport of a nutritional component required for growth within the host but not required for bacterial growth in vitro.
Interestingly, the largest class of secreted proteins identified from the prfA(L140F) strain consisted of gene products predicted to modify the structure and/or integrity of the bacterial cell wall (Lmo1438, Lmo2522, Lmo2691, and Lmo2754). The two penicillin-binding proteins identified, Lmo1438 and Lmo2754 (PBP5), have recently been shown to contribute to L. monocytogenes virulence, as gene disruption mutants injected intraperitoneally into mice yielded 2-log and 1-log reductions, respectively, in bacterial CFU recovered from the spleen (29). Strain differences may account for the inability to generate an insertional disruption mutant in lmo1438 in strain 10403S, whereas Guinane et al. (29) were successful in disrupting lmo1438 in the EGD-e background by using a similar method.
Additional proteins found to be up-regulated in prfA(L140F) supernatants included Lmo1388 (TcsA), Lmo2637, Lmo2219, and Lmo0292 (HtrA). TcsA was first identified in a screen for L. monocytogenes CD4+ T-cell-stimulating antigens (54), and it shares sequence similarity with the Bmp (_b_asic _m_embrane _p_rotein) family of proteins found in a variety of bacteria, including various Bacillus and Streptococcus species, as well as pathogenic spirochetes, including Treponema pallidum and Borrelia burgdorferi, in which the Bmp proteins are highly antigenic outer membrane proteins of unknown function (7). In L. monocytogenes, TcsA is located at the 5′ end of a series of genes which appear to comprise a sugar ABC transport system; thus, TcsA could potentially be involved in the bacterial transport of carbohydrates within the host. HtrA (also known as DegP) is a member of a highly conserved serine protease family and is thought to degrade misfolded proteins and potentially function as a chaperone. HtrA is known to contribute to virulence in several gram-negative and gram-positive bacterial pathogens, such as Salmonella enterica serovar Typhimurium (34), Pseudomonas aeruginosa (5), Streptococcus pneumoniae (32), Staphylococcus aureus (51), and Streptococcus pyogenes (35), presumably by assisting in the secretion and maturation of virulence factors (44). In L. monocytogenes, HtrA has been shown to be essential not only for bacterial resistance to high salt (71) and oxidative stress (70) but also for bacterial virulence in mice (62, 70).
A second potential secretion chaperone identified as a result of PrfA activation was Lmo2219. lmo2219 contains a putative PrfA binding site in its promoter region and has previously been associated with PrfA-dependent regulation by transcriptome analysis (46). Lmo2219 has significant homology to PrsA from Bacillus subtilis (45% identity, 63% similarity) and other peptidyl-prolyl isomerases and is thought to act as a chaperone for secreted proteins. A recent study identified the expression of lmo2219 as up-regulated inside of host cells, and a Δlmo2219 strain exhibited a defect in bacterial growth and spread within fibroblast monolayers as well as decreased growth in bone marrow-derived macrophages (10). In this study, the lmo2219 gene disruption mutant was found to exhibit the largest virulence defect of all mutants tested (Fig. 4). Two _prsA_-like genes with peptidyl-prolyl isomerase domains are present in the L. monocytogenes genome, lmo1444 and lmo2219, and we therefore propose to name them prsA1 and prsA2, respectively, following the annotation given by TIGR in their L. monocytogenes sequence databases (http://www.tigr.org/tdb/listeria/). In B. subtilis, prsA has been shown to be an essential gene (68). The presence of two _prsA_-like genes in L. monocytogenes and the ability to inactivate lmo2219 suggest the existence of some functional redundancy or perhaps a division of substrate specificity between the two chaperones whereby the lmo2219 gene product may contribute to the folding of extracellular virulence factors.
Finally, two proteins were identified as down-regulated or absent in the prfA(L140F) secretome: Lmo0013 (QoxA, quinol oxidase subunit II) and Lmo0690 (FlaA). In B. subtilis, QoxA, together with QoxB, QoxC, and QoxD, forms an oligomeric enzymatic complex on the cell membrane which catalyzes the transfer of electrons from quinol onto molecular oxygen to form water in the terminal step of the respiratory chain (55). However, terminal oxidases may take on alternate roles as electron scavengers in order to detoxify the intracellular environment triggered by the antimicrobial response during infection; such has been proposed with cytochrome bd in both Shigella flexneri (69) and Brucella abortus (20). FlaA, the major subunit of flagella, was also found to be down-regulated or absent in the prfA(L140F) secretome, a finding which correlates with the decrease in swimming motility observed for prfA* mutants (Fig. 3). The decrease in secreted FlaA suggests that activated PrfA may either down-regulate flaA gene expression or interfere with FlaA assembly. Evidence for PrfA-dependent regulation of L. monocytogenes motility gene expression exists: motA transcripts (encoding a component of the flagellar motor in the plasma membrane) appear to be repressed by PrfA (motA expression is twofold greater in a Δ_prfA_ strain at 37°C) (45), and flaA transcripts were reduced in the presence of the prfA(G145S) mutation at 20°C (52).
Very recently, Baumgartner et al. (2) undertook a proteomic analysis of lipoproteins in L. monocytogenes by deleting the prolipoprotein diacylglyceryl transferase (Lgt), which led to a release of lipoproteins into the culture supernatant. Twenty-six of the 68 predicted lipoproteins were identified in the extracellular proteome of the Δ_lgt_ strain, including most of the lipoproteins identified in this study. This suggests that PrfA activation results in the overexpression and/or hypersecretion of lipoproteins such that a significant number are shed into the extracellular environment. Alternatively, PrfA activation may result in an alteration of the bacterial cell membrane or cell surface that results in the release of lipoproteins; changes in L. monocytogenes prfA(L140F) mutant surface properties have been reported previously (72). Baumgartner et al. (2) further found that overexpression of wild-type prfA from a multicopy plasmid in the Δ_lgt_ strain resulted in the up-regulation of six proteins, four of which were identified in this study (ActA, InlC, Lmo2219 [PrsA2], and Lmo2196 [OppA]) and two which were not (InlA and Lmo0366). The difference in the numbers of proteins identified is likely reflective of both the higher activation state of the prfA(L140F) allele used here (versus the overexpression of nonactivated wild-type protein) and the presence of functional Lgt.
This study demonstrates that upon PrfA activation the L. monocytogenes secretome is highly enriched for proteins that contribute to virulence. Although targeted gene disruption via plasmid insertion does not exclude the possibility of polar effects of the insertions on downstream gene expression, the disruption mutants are still clearly useful for identifying regions of the L. monocytogenes chromosome that contribute to bacterial pathogenesis. Since several of the gene products identified were not described as up-regulated in a transcriptome analysis of the mutationally activated PrfA G145S strain (46) and lack obvious PrfA binding sites within their promoter regions, it appears that a PrfA-dependent product or pathway (rather than direct activation by PrfA) is responsible for their altered patterns of expression and/or secretion. These studies highlight the significant and broad-range influences of PrfA activation on L. monocytogenes protein expression profiles and on bacterial virulence.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Phil Gafken at the Fred Hutchinson Cancer Research Center Proteomics Facility for MS analysis and helpful discussions and Jon Johansen at Kendrick Labs for 2-DE gel analysis and advice. We also thank the members of the Freitag lab for helpful discussion, Ana Gervassi for critical reading of the manuscript, Maurine Miner for help with strain construction, and Lawrence Lee for help with mouse infections.
This work was supported by Public Health Service grants AI41816 and AI055651 (N.E.F.) and a National Science Foundation graduate research fellowship (G.C.P.) as well as by funding from the M. J. Murdock Trust.
Footnotes
▿
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Auerbuch, V., L. L. Lenz, and D. A. Portnoy. 2001. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69**:**5953-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgartner, M., U. Karst, B. Gerstel, M. Loessner, J. Wehland, and L. Jansch. 2007. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189**:**313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139**:**2005-2009. [PubMed] [Google Scholar]
- 4.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68**:**7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178**:**511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90**:**11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryksin, A. V., H. P. Godfrey, C. A. Carbonaro, G. P. Wormser, M. E. Aguero-Rosenfeld, and F. C. Cabello. 2005. Borrelia burgdorferi BmpA, BmpB, and BmpD proteins are expressed in human infection and contribute to P39 immunoblot reactivity in patients with Lyme disease. Clin. Diagn. Lab. Immunol. 12**:**935-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mo. Microbiol. 8**:**143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll, S. A., T. Hain, U. Technow, A. Darji, P. Pashalidis, S. W. Joseph, and T. Chakraborty. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J. Bacteriol. 185**:**6801-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74**:**1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13**:**23-31. [DOI] [PubMed] [Google Scholar]
- 12.Cossart, P., M. F. Vicente, J. Mengaud, F. Baquero, J. C. Perez-Diaz, and P. Berche. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57**:**3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73**:**241-268. [DOI] [PubMed] [Google Scholar]
- 14.Domann, E., J. Wehland, M. Rohde, S. Pistor, M. Hartl, W. Goebel, M. Leimeister-Wachter, M. Wuenscher, and T. Chakraborty. 1992. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11**:**1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domann, E., S. Zechel, A. Lingnau, T. Hain, A. Darji, T. Nichterlein, J. Wehland, and T. Chakraborty. 1997. Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogenes whose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats. Infect. Immun. 65**:**101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dons, L., E. Eriksson, Y. Jin, M. E. Rottenberg, K. Kristensson, C. N. Larsen, J. Bresciani, and J. E. Olsen. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72**:**3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16**:**251-261. [DOI] [PubMed] [Google Scholar]
- 18.Edman, D. C., M. B. Pollock, and E. R. Hall. 1968. Listeria monocytogenes L forms. I. Induction maintenance, and biological characteristics. J. Bacteriol. 96**:**352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiting, M., G. Hageluken, W. D. Schubert, and D. W. Heinz. 2005. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol. Microbiol. 56**:**433-446. [DOI] [PubMed] [Google Scholar]
- 20.Endley, S., D. McMurray, and T. A. Ficht. 2001. Interruption of the cydB locus in Brucella abortus attenuates intracellular survival and virulence in the mouse model of infection. J. Bacteriol. 183**:**2454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21**:**823-837. [DOI] [PubMed] [Google Scholar]
- 22.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61**:**2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65**:**1127-1141. [DOI] [PubMed] [Google Scholar]
- 24.Gatlin, C. L., G. R. Kleemann, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263**:**93-101. [DOI] [PubMed] [Google Scholar]
- 25.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1987. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55**:**1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294**:**849-852. [DOI] [PubMed] [Google Scholar]
- 27.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40**:**963-975. [DOI] [PubMed] [Google Scholar]
- 28.Gray, M. J., N. E. Freitag, and K. J. Boor. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74**:**2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guinane, C. M., P. D. Cotter, R. P. Ross, and C. Hill. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob. Agents Chemother. 50**:**2824-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins, D. E., C. Buchrieser, and N. E. Freitag. 2006. Genetic tools for use with Listeria monocytogenes, p. 620-633. In J. J. Ferretti et al. (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington D.C.
- 31.Horton, R. 1993. In vitro recombination and mutagenesis of DNA, p. 251-260. In B. White et al. (ed.), PCR protocols: current methods and applications. Humana Press, Totowa, NJ.
- 32.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72**:**3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110**:**551-561. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5**:**401-407. [DOI] [PubMed] [Google Scholar]
- 35.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69**:**5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, S., and D. A. Portnoy. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 62**:**5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188**:**556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kathariou, S., P. Metz, H. Hof, and W. Goebel. 1987. Tn_916_-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169**:**1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayal, S., and A. Charbit. 2006. Listeriolysin O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 30**:**514-529. [DOI] [PubMed] [Google Scholar]
- 40.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. _L. monocytogenes_-induced actin assembly requires the actA gene product, a surface protein. Cell 68**:**521-531. [DOI] [PubMed] [Google Scholar]
- 41.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184**:**4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 100**:**12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45**:**1043-1056. [DOI] [PubMed] [Google Scholar]
- 44.Lyon, W. R., and M. G. Caparon. 2004. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect. Immun. 72**:**1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michel, E., J. Mengaud, S. Galsworthy, and P. Cossart. 1998. Characterization of a large motility gene cluster containing the cheR, motAB genes of Listeria monocytogenes and evidence that PrfA downregulates motility genes. FEMS Microbiol. Lett. 169**:**341-347. [DOI] [PubMed] [Google Scholar]
- 46.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47**:**1613-1625. [DOI] [PubMed] [Google Scholar]
- 47.Miner, M. D., G. C. Port, and N. E. Freitag. 2007. Regulation of Listeria monocytogenes virulence genes, p. 139-158. In H. Goldfine et al. (ed.), Listeria monocytogenes: pathogenesis and host response, 1st ed. Springer, New York, NY.
- 48.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250**:**4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neil, H. S., and H. Marquis. 2006. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 74**:**6675-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167**:**1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rigoulay, C., J. M. Entenza, D. Halpern, E. Widmer, P. Moreillon, I. Poquet, and A. Gruss. 2005. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect. Immun. 73**:**563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripio, M. T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179**:**1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ripio, M. T., G. Dominguez-Bernal, M. Suarez, K. Brehm, P. Berche, and J. A. Vazquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res. Microbiol. 147**:**371-384. [DOI] [PubMed] [Google Scholar]
- 54.Sanderson, S., D. J. Campbell, and N. Shastri. 1995. Identification of a CD4+ T cell-stimulating antigen of pathogenic bacteria by expression cloning. J. Exp. Med. 182**:**1751-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santana, M., F. Kunst, M. F. Hullo, G. Rapoport, A. Danchin, and P. Glaser. 1992. Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J. Biol. Chem. 267**:**10225-10231. [PubMed] [Google Scholar]
- 56.Shetron-Rama, L. M., H. Marquis, H. G. A. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70**:**1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48**:**1537-1551. [DOI] [PubMed] [Google Scholar]
- 58.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68**:**850-858. [DOI] [PubMed] [Google Scholar]
- 59.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63**:**4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, G. A., and D. A. Portnoy. 1997. How the Listeria monocytogenes ActA protein converts actin polymerization into a motile force. Trends Microbiol. 5**:**272-276. [DOI] [PubMed] [Google Scholar]
- 61.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spollM gene. Biochimie 74**:**705-711. [DOI] [PubMed] [Google Scholar]
- 62.Stack, H. M., R. D. Sleator, M. Bowers, C. Hill, and C. G. M. Gahan. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71**:**4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58**:**3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109**:**1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vazquez-Boland, J. A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60**:**219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14**:**584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vega, Y., M. Rauch, M. J. Banfield, S. Ermolaeva, M. Scortti, W. Goebel, and J. A. Vazquez-Boland. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 52**:**1553-1565. [DOI] [PubMed] [Google Scholar]
- 68.Vitikainen, M., T. Pummi, U. Airaksinen, E. Wahlstrom, H. Wu, M. Sarvas, and V. P. Kontinen. 2001. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of α-amylase in Bacillus subtilis. J. Bacteriol. 183**:**1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Way, S. S., S. Sallustio, R. S. Magliozzo, and M. B. Goldberg. 1999. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri Virulence. J. Bacteriol. 181**:**1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson, R. L., L. L. Brown, D. Kirkwood-Watts, T. K. Warren, S. A. Lund, D. S. King, K. F. Jones, and D. E. Hruby. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74**:**765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wonderling, L. D., B. J. Wilkinson, and D. O. Bayles. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70**:**1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong, K. K., and N. E. Freitag. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186**:**6265-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Youngman, P. 1987. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other gram-positive bacteria, p 79-103. In K. Hardy et al. (ed.), Plasmids: a practical approach. IRL Press, Oxford, England.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]