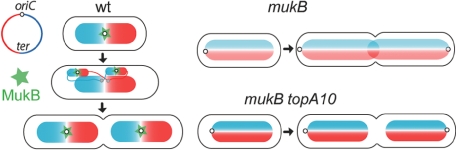
MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves (original) (raw)
Abstract
The circular Escherichia coli chromosome is organized by bidirectional replication into two equal left and right arms (replichores). Each arm occupies a separate cell half, with the origin of replication (oriC) at mid-cell. E. coli MukBEF belongs to the ubiquitous family of SMC protein complexes that play key roles in chromosome organization and processing. In mukBEF mutants, viability is restricted to low temperature with production of anucleate cells, reflecting chromosome segregation defects. We show that in mukB mutant cells, the two chromosome arms do not separate into distinct cell halves, but extend from pole to pole with the oriC region located at the old pole. Mutations in topA, encoding topoisomerase I, do not suppress the aberrant positioning of chromosomal loci in mukB cells, despite suppressing the temperature-sensitivity and production of anucleate cells. Furthermore, we show that MukB and the oriC region generally colocalize throughout the cell cycle, even when oriC localization is aberrant. We propose that MukBEF initiates the normal bidirectional organization of the chromosome from the oriC region.
Introduction
The Escherichia coli chromosome adopts a compact structure, the nucleoid, with each locus following a reproducible choreography throughout the cell cycle (reviewed in Espeli and Boccard, 2006). Bidirectional replication, initiating at oriC and terminating in the ter region opposite to oriC, defines two replication arms or replichores. For newborn cells with non-overlapping replication cycles, the two chromosome arms locate to different cell halves, later replicated markers being more pole proximal (Nielsen et al., 2006; Wang et al., 2006). This Left–Right () chromosome organization is regenerated quickly after replication. However, the mechanisms positioning the sister origins at the cell quarters and/or organizing the two chromosome arms are unknown.
_S_tructural _m_aintenance of _c_hromosomes (SMC) proteins are ubiquitous and required for many aspects of chromosomes segregation in eukaryotes and prokaryotes (Nasmyth and Haering, 2005). SMC dimers adopt a flexible V-shaped structure, whose open ends may come together in a reaction facilitated by partner proteins. Two of the best-studied SMC complexes are cohesins, which hold the sister chromatids together during mitosis, and condensins, which organize mitotic chromosomes. In E. coli, as in Bacillus subtilis, SMC impairment leads to thermosensitivity, and to the formation of ∼15% anucleate cells at permissive temperature, indicative of a defect in chromosome segregation (Niki et al., 1991; Britton et al., 1998). These phenotypes are suppressed by a decrease of topoisomerase I activity, while inhibition of gyrase is synthetically lethal with mutation in bacterial SMC (Sawitzke and Austin, 2000; Lindow et al., 2002), consistent with MukBEF acting in chromosome organization by organizing DNA supercoiling. The increase of negative supercoiling in topA mutants could directly compensate for the impairment of MukBEF organization activity, or the suppression could be an indirect consequence of the change in supercoiling arising as a consequence of TopoI depletion. A direct role of increased negative supercoiling in TopA– in suppressing the Muk– phenotype is supported by data showing that inhibition of gyrase activity reverses this suppression (Sawitzke and Austin, 2000); that negative supercoiling is increased in E. coli cells grown at low temperature (Stupina and Wang, 2005), when MukBEF is dispensable; and by the demonstration that MukB constrains DNA in a condensin-like fashion in vitro (Petrushenko et al., 2006).
Nevertheless, the primary role of bacterial SMC proteins remains elusive (reviewed in Strunnikov, 2006). Here, we have analysed the contribution of MukBEF to chromosome organization and have related this to MukB localization in living cells. We show that chromosome organization is changed in mukB mutant cells. The two chromosome arms do not separate into distinct cell halves and flank the ori region at mid-cell as in Muk+ cells, but rather extend from pole to pole with the oriC region located at the old pole. Furthermore, we show that MukB and the oriC region generally colocalize throughout the cell cycle, even when oriC localization is aberrant.
Results
Sister origins position aberrantly at opposite poles in mukB cells
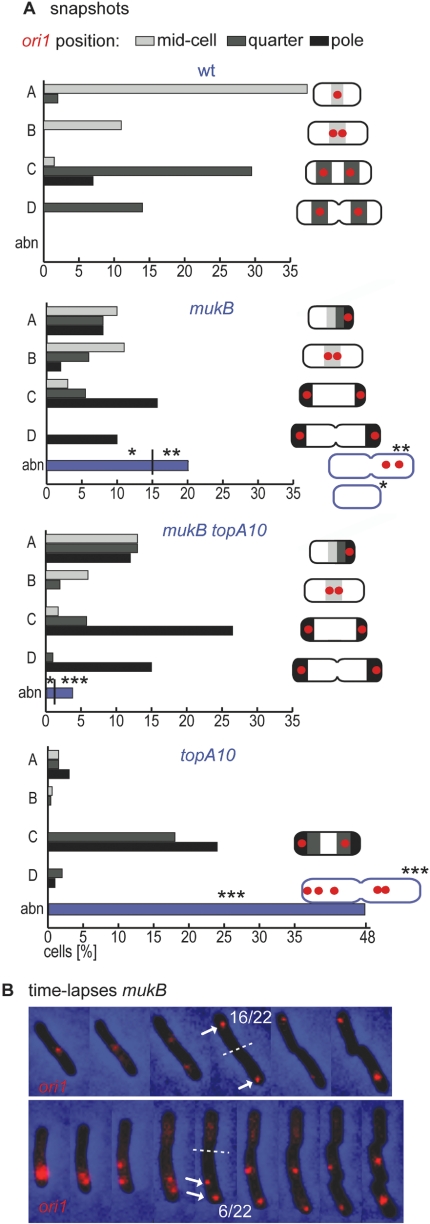
Tracking of the ori1 locus, located 15 kb counterclockwise of oriC, by fluorescent repressor-operator sites (FROS; Lau et al., 2003), revealed abnormal positioning and segregation of sister origins in mukB mutants. In wild-type cells growing at 22°C, without overlapping replication cycles, our snapshot data (Fig. 1A) were consistent with the segregation pathway previously described (Wang et al., 2006). ori1 is always close to mid-cell in newborn cells and after duplication at mid-cell, sister origins migrate in opposite directions towards the quarter positions, where they remain until cell division. In contrast, snapshots of mukB mutants showed that duplicated ori1 foci were most often positioned close to opposite old poles (types C and D; ∼70% of 2×ori1 cells), with focus duplication being inferred to occur either at an old pole or at mid-cell (type B).
Fig. 1.
Polar positioning of ori1 in mukB and mukB topA10 cells. A. Snapshot analysis of ori1 positioning in different strains as indicated (wt: IL02; mukB: OS27; mukB topA10: OS47; topA10: OS70). For about 500 cells of each strain, cells were first classified into the five types shown in the schematic. Types A–C correspond to non-septating cells with one ori1 focus (type A), two ori1 foci closely spaced (type B) and two ori1 foci well separated (type C); type D corresponds to septating cells with segregated two sister ori1 foci, and type abn corresponds to all the other cells judged abnormal. Cells were divided along the longitudinal axis in six equal parts: left pole, left quarter, mid-cell left, mid-cell right, right quarter and right pole. The ori1 position (histograms) was classed in either polar (black), quarter (dark grey) or mid-cell (light grey). The predominant position was represented by schematics on the right-hand side of the histograms. Three types of abnormal cells were distinguished: *lacking ori1; **containing two ori1 foci in the same cell half and ***containing more than two ori1 foci. B. Time-lapse analysis of ori1 in mukB cells. Examples of successful (top) and unsuccessful (bottom) segregation are illustrated. The arrows indicate the position of the sister ori1 foci at the time of division (dashed line). Images were taken every hour.
Time-lapse tracking confirmed the snapshot analysis of mukB mutants, with all the successful segregations (16/22 cases), showing sister ori1 segregations to opposite poles (Fig. 1B, top and Fig. S1A and B), rather than the quarter positions observed in wild-type cells. Consistent with this, 21/24 mukB newborn cells had ori1 close to an old pole. Duplication of ori1 foci occurred either at the pole (3/16; bottom panel) or after movement of a single focus to mid-cell (13/16; Fig. 1B, top). In the cases of unsuccessful ori1 segregation (6/22 cases), ori1 failed to duplicate (2 cases, Fig. S1C), and showed delayed ori1 duplication at a pole (2 cases; Fig. 1B, bottom) or at mid-cell (2 cases). Therefore, when sister ori1 loci are successfully segregated to mukB daughter cells, they position aberrantly close to the old poles. This result suggests that in the absence of MukBEF, chromosome organization is perturbed specifically, with a new type of organization that places newly replicated sister origins at the old poles.
The topA10 mutation does not suppress the aberrant ori positioning of mukB cells
Because impairment of topoisomerase I (topA10) suppresses the temperature-sensitivity and anucleate cell production of Muk– cells (Sawitzke and Austin, 2000), ori1 positioning was analysed in mukB topA10 mutant cells. As expected, the viability at 37°C in minimal or rich media was restored, and the population contained few anucleate cells (1% as compared with 15% in a mukB strain). At 22°C in minimal medium (and at 30°C), most segregated sister ori1 foci observed in the _mukB topA_10 snapshots showed polar positioning, with a similar pattern to the mukB strain (Fig. 1A). Consistent with this, in 15/17 newborn cells observed by time-lapse tracking, ori1 focus was close to the old pole (not shown). The _topA_10 single mutant cells exhibited a defect in cell division and aberrant ori1 positioning, apparently as a consequence of impaired chromosome segregation. This phenotype is quite distinct from that of mukB and mukB topA10 cells (Fig. 1A) and has, as its main features, a substantial fraction (48%) of filamentous cells with more than two ori1 foci (abn), and a corresponding deficiency in cells containing a single ori1 focus (type A). In cells with two separated sister ori1 foci, 57% of ori1 foci were at a pole (but very rarely both together at poles), while the others were at quarter cell, as in wild-type cells. Therefore, the phenotype of _mukB topA_10 cells, when judged by ori1 positioning, is similar to that of mukB cells, despite the fact that temperature-sensitivity and anucleate cell production is abolished in the double mutant. We conclude that the chromosome organization defect of mukB cells persists when the topA10 allele is introduced. Thus, this defect can be functionally separated from temperature-sensitivity and anucleate cell production.
Left–Right (<L-_ori_-R>) nucleoid organization is perturbed in mukB cells
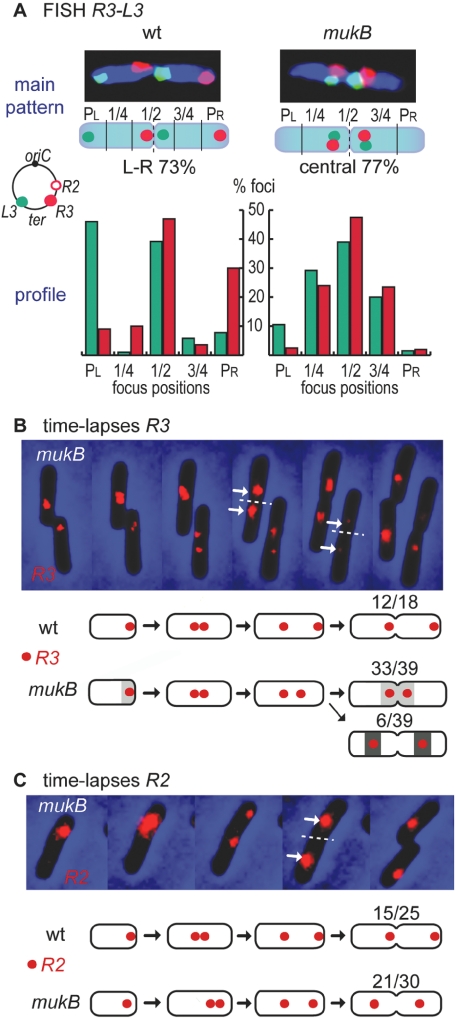
The above results show that the <L-_ori_-R> organization about the transverse axis of normal cells is altered when MukBEF is absent, with polar ori positioning being reminiscent of the type of organization in Caulobacter crescentus (Viollier et al., 2004) and in chromosome 1 of Vibrio cholera (Fogel and Waldor, 2005). To further probe nucleoid organization, mukB cells grown at 22°C were examined using two-colour fluorescent in situ hybridization (FISH) with probes to L3 and R3 chromosome loci located on opposite arms at positions 2269 and 872 kb respectively close to the ter region (Fig. 2A). The patterns of L3–R3 positioning (Fig. 2A, top), and the L3 and R3 population profiles (bottom), were analysed in cells containing duplicated foci for each locus. These four-foci cells were enriched for by the addition of the cell division inhibitor cephalexin for 2.5 h (one-third of a generation time) prior to analysis (Wang et al., 2006). A pattern in each sister nucleoid, arranged as <_L3–R3–L3–R3_> in pairs of sister nucleoids, was predominant (73%) for the wild-type strain, as reported earlier (Wang et al., 2006). This wild-type organization is also revealed by the L3–R3 population profile, in which L3 peaks at the left pole (PL) and at mid-cell, while R3 peaks at mid-cell and at the right pole (PR).
Fig. 2.
Perturbation of nucleoid organization in mukB cells. A. L3 and R3 positions by two-colour FISH. Analysis was restricted to four-foci cells having a normal nucleoid (as judged by DAPI staining). Foci from 123 MukB+ cells (AB1157) and 150 mukB cells (OS53) were binned into five positions from pole left (PL) to pole right (PR). The predominant four-focus patterns and the L3–R3 profiles are shown.
B. Time-lapse analysis of R3 by FROS. Images of mukB cells (OS55) were taken every hour, kept at room temperature (G ∼5.5 h, as judged by cell elongation). Below, schematics show the predominant segregation pathways of R3 loci in wild-type and mukB cells. C. Same as B for R2 locus (OS30).
The organization about a transverse axis was absent in mukB mutants. Instead, the two pairs of sister foci cluster in the central region of the cell close to the newly developing poles (77% of nucleoid pairs; also see the population profile). This result was confirmed by time-lapse tracking of the R3 locus using FROS in cells growing at room temperature (∼22°C). In the wild-type cells, segregation gives rise predominantly to asymmetric positioning of sister R3 (or R2) loci with respect to mid-cell, generating one cell with R3 close to the old pole and one cell with R3 close to the new pole (cartoons, Fig. 2B; Wang et al., 2006). In mukB mutants, the segregation led to a symmetrical pattern of sister R3 foci remaining in the central region, producing two daughter cells with a new pole-proximal R3 locus (time-lapse and cartoons, Fig. 2B and Fig. S2). Consistent with this, a symmetrical pattern about the cell quarter positions of sister R2 loci was observed in the mukB mutant (Fig. 2C). Therefore, the positioning patterns of R2, R3 and L3 loci suggest an absence of organization about a transverse cell axis in mukB cells. The results are consistent with chromosome organization having moved from being arranged about a transverse axis to apparently being organized about a longitudinal axis, with the two arms extending from the old pole to the new pole, with or without twisting about each other. However, proof of this would require a very extensive analysis with many pairs of loci analysed.
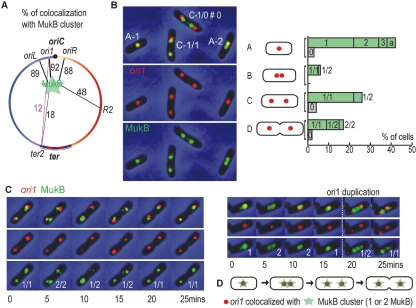
MukB clusters localize to the origin region
Knowing that MukBEF forms foci within cells, when assayed by immunocytochemistry (den Blaauwen et al., 2001), or with a fluorescent fusion protein (Ohsumi et al., 2001), we analysed their position and dynamics with respect to chromosomal loci. For simultaneous visualization, the chromosomal mukB gene was replaced by the functional mukB-gfp4 fusion gene, and FROS in cells grown at 30°C was used to track ori1 (3908 kb), oriL (3713 kb), oriR (4139 kb), R2 (366 kb) or ter2 (1801 kb) loci (Fig. 3A).
Fig. 3.
Colocalization of MukB with the origin region.
A. Colocalization is indicated as a number (%) on black lines linking MukB and each given loci positioned on the circular map of the E. coli chromosome (ori1, R2, ter2 loci from OS18 and OS29, OS69, OS19 cells respectively). Colocalization (%) of ter2 with ori1 in IL05 cells is indicated on the purple line.
B. Snapshot analysis of MukB-GFP and ori1 positioning. Images of representative wild-type cells (OS18; see histogram for legend). Histogram representing the proportion of the different MukB patterns for each cell type (A–D, Fig. 2). Green bars correspond to MukB/ori1 colocalization, and patterns 1, 2 and 3 correspond to one, two or three MukB foci colocalizing with one ori1 focus. When cells contains two ori1 foci, each ori1 focus can colocalize either with one or two MukB, generating pattern 1/1, 1/2 or 2/2. Grey bars and pattern 0 correspond to the absence of colocalization of one ori1 focus with MukB, including cells containing one ori1 focus or two ori1 foci (patterns 0/0, 0/1 and 0/2 are included in pattern 0). Pattern ‘a’ corresponds to cells with an extra MukB focus distant from the ori1/MukB colocalizing foci.
C. Time-lapse tracking simultaneously MukB and ori1 (without or with ori1 duplication, left and right respectively; OS18 cells). The number of MukB foci per cluster (bottom panel) and the time intervals between images (below panels) are indicated.
D. Representation of the cell-cycle dynamics of ori1 and MukB deduced from snapshot and time-lapse analysis.
MukB-GFP foci appeared either as a single focus or as two (and sometimes three) closely spaced foci (MukBEF clusters); interconversion between these states was reversible and dynamic (Fig. 3C). Thus, we have chosen to score the frequency of colocalization (%) of a given genetic locus with a MukB cluster, by examining at least 400 cells for each locus. A genetic locus is defined as colocalizing with a MukB cluster if there is an overlap between the genetic locus focus and a MukB focus (that may belong to a cluster) when images were overlaid.
For a region of at least 400 kb centred on oriC, the colocalization of ori loci with MukB clusters was greater than 88%, revealing a preferential colocalization with the origin region. Increasing the distance from oriC reduced gradually the colocalization with a MukB cluster (48% for R2 and 18% for ter2). Because the ter region crosses the ori region during each replication cycle (Wang et al., 2006), it is not surprising that we observe a low, but significant, colocalization of ter2 with both ori1 (purple line) and MukB. Within the cell population, the ratio of MukB foci to ori1 foci was 1.4. Different MukB/ori1 patterns were observed, but predominantly (83% of cells) each ori1 is colocalizated with one or two MukB foci (patterns 1 and 2 in cells with one origin; and 1/1, 1/2 and 2/2 in cells with two origins, Fig. 3B). Time-lapse tracking revealed that the MukB-GFP signal could reversibly interconvert between one and two MukB foci within a 5 min time frame, in situations where ori1 duplication did not occur (12/13 cases; Fig. 3C left and Fig. S3A). The duplication of MukB was observed either concomitantly (within 5 min, the time between two images) with ori1 (4/9 cases, Fig. 3C, right and Fig. S3B) or 5 min before (5/9 cases, not shown). The MukB cluster could interconvert within these intervals. Thus, dynamic MukB clusters tend to colocalized with ori1 throughout the cell cycle (Fig. 3D).
Mispositioned or non-duplicating origin regions remain colocalized with MukB
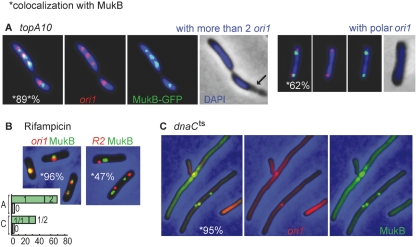
In order to test whether MukB positioning depends on ori1 location, the relative MukB and ori1 positioning were analysed in topA10 mutant cells grown at 30°C, where aberrant origin positioning occurs (Fig. 1A and 35% of origins are positioned at a nucleoid edge). Colocalization of a polar ori1 focus with a MukB cluster was substantial in normal-length cells (62%, Fig. 4A). Furthermore, the colocalization was maintained (89%) in the 48% of filamentous cells containing more than two ori1 foci (Fig. 1).
Fig. 4.
Mispositioned or non-duplicating ori1 foci colocalize with MukB. A. topA10 cells (OS70) containing more than two ori1 (left) or containing a polar ori1 (right).
B. Wild-type cells after 3 h of rifampicin treatment with tetO array inserted at either position ori1 (left, OS29 cells; see Fig. 3 for the histogram legend) or R2 (right, OS69 cells).
C. _dnaC_ts cells (OS82) grown at 37°C for 8 h with tetO at ori1. The percentages in panels A, B and C refer to the colocalization of ori1 or R2 loci with MukB clusters, as deduced from the examination of at least 400 cells.
Furthermore, the colocalization of ori1 and MukB was not affected when cells contained completely replicated chromosomes and were unable to re-initiate replication, by using rifampicin or a _dnaC_ts mutant (Fig. 4B and C). After 3 h of rifampicin treatment at 30°C, the MukB/ori1 ratio is close to 1, most MukB clusters being replaced by a single focus (compare Figs 3B and 4B left). The colocalization of R2, an _ori_-distal locus, with MukB was not increased after rifampicin treatment (Fig. 4B right). DnaC impairment for 8 h did not lead to loss of ori1 colocalization with MukBEF, although now there were additional MukBEF foci distant from ori1, but still colocalized with the nucleoid. The view that MukB focus formation requires DNA association is supported by the observation that out of ∼500 cells lacking an ori1 focus (> 90% of which do not exhibit any DAPI staining; data not shown), which were observed after DnaC impairment, none exhibited a MukB focus (two such cells without ori1 are shown in Fig. 4C). Therefore, the origin regions remained predominantly colocalized with MukBEF clusters during extensive periods of DNA replication arrest.
Discussion
We have revealed that in a mukB mutant, the two chromosome arms are not separated into distinct cell halves and the sister origins lose their normal positioning, migrating to the outside poles after duplication. A perturbation of the oriC region in a mukB mutant was reported by Weitao et al. (2000), but probably because of their use of faster growth conditions and the absence of time-lapse analysis, they concluded that oriC positioning was random rather than polar. The suppression of the thermosensitivity and of the formation of anucleated cells by topA10 mutation led to the model that MukB action, within a MukBEF complex, is limited to the organization of DNA supercoiling (Sawitzke and Austin, 2000). However, we observed that the aberrant ori1 positioning is not suppressed by a topA10 mutation. Thus, one interpretation that we favour is that the action of MukB involves more than the organization of DNA supercoils. Moreover, the absence of suppression in a strain where the indirect consequences of MukB loss should be reduced (as mukB topA10 double mutants have a normal viability) suggests that the aberrant positioning of the origin is a direct consequence of MukB loss.
In addition, we have shown the preferential colocalization of MukBEF with the ori region, with one, two or three MukB foci being apparently associated with a given ori region, interconversion of focus number being frequent. Previous work, using either immunocytochemistry (den Blaauwen et al., 2001) or fluorescent protein fusions (Ohsumi et al., 2001), has shown that MukBEF forms foci or clusters but did not track MuBEF position in relation to specific genetic markers, and did not use time-lapse analysis. Consequently, they did not report an apparent association with the ori region and the rapid interconversion of focus number associated with a given ori. Nevertheless, the results of Ohsumi et al. (2001) appear consistent with those reported here. We propose that the MukBEF complex acts in the origin region to initiate bidirectional organization after replication.
We consider three types of mechanism that could give rise to the observations of MukB-ori colocalization. The first involves a direct interaction of MukBEF with some feature of the ori region (Fig. 5). This could relate to some sequence motif(s) present in the ori region, or to some other aspect of ori biology. Alternatively, the ori region and MukBEF could be targeted by independent mechanisms to the same cellular compartment. This seems unlikely, given that MukB clusters still colocalize with ori1 when it is positioned aberrantly. A third possible mechanism is a rosette model in which MukBEF molecules bind discrete sites spread over the chromosome (for example, one per topological domain), and MukBEF–MukBEF interactions lead to a rosette-like structure, centred on the origin region.
Fig. 5.
Model of E. coli chromosome organization/segregation. Wild-type: Chromosomes segregation is facilitated by the organization initiated by MukB colocalizing with the ori regions. mukB (22°C) and mukB topA10: ‘MukB-free’ segregation is allowed only when the level of negative DNA supercoiling is increased (low temperature/topA10) but leads to an aberrant arrangement, with the two arms extending (twisted or not with each other) from the old poles to the new poles. For clarity, the two replichores are represented as untwisted. The chromosome organization in mukB topA10 cells is extrapolated from the aberrant origin positioning.
At present we favour the first mechanism, although we have failed to show a preferential association of MukBEF with ori sequences using chromatin immunoprecipitation assays that employed either real-time polymerase chain reaction (PCR) or microarrays assays. Indeed, in two independent microarray experiments, DNA fragments enriched by the co-immunoprecipition of MukB were not exclusively contained in the origin region, but scattered over the whole chromosome (M. Pinskaya et al., unpubl. data). There is no evidence that any SMC complex can recognize specific DNA sequences directly. Nevertheless, both cohesins (Nasmyth and Haering, 2005) and condensins (Wang et al., 2005a) can associate with specific chromosome regions. Such localization may reflect places of DNA loading and/or positions to which the SMC complex is directed [for example by transcription (Lengronne et al., 2004) or by other protein–DNA complexes]. In E. coli, such an association seems to occur within the ‘Ori macrodomain’, a ∼1 Mbp region that behaves as a single unit in some assays (Espeli and Boccard, 2006), but wherein loci can still behave independently (Fekete and Chattoraj, 2005).
The <_ori_-out _ter_-in> chromosome arrangement proposed for mukB cells (Fig. 5) is similar to that observed for C. crescentus and chromosome 1 of V. cholera, although both express functional SMCs, and the origins are actively transported to the poles, which could force structuring of the two chromosome arms about a polar origin (Viollier et al., 2004; Fogel and Waldor, 2005). Consistent with this, when the V. cholera chromosome I transport system is mutated, the origins adopt similar positions to E. coli origins (Fogel and Waldor, 2006), suggesting that the chromosome I of V. cholerae would also have a organization in absence of constraint on its origin positioning. Jun and Mulder (2006) have shown how entropy considerations alone may direct a given pattern of bacterial chromosome organization and segregation. Such a pattern may be modified by proteins that transport DNA, or which organize specific chromosomal regions. Our model of MukBEF action at sister ori regions is consistent with the positioning of ori at mid-cell as a consequence of space restriction: within a confined rod-shaped cell, an equal amount of chromosome DNA is placed around the left and the right ori sides. Thus, the organization of the two chromosome arms via MukBEF would facilitate the segregation of the sister chromosomes without the need to position any chromosome region along the cell.
Experimental procedures
The bacterial strains used are listed in Table S1. E. coli AB1157 strains containing tetO or lacO arrays were constructed as previously described (Wang et al., 2005b). We were unable to construct stable mukB strains containing both L3 and R3 operator arrays when the cognate repressors were present. TetR-CFP and LacI-CFP were expressed from pWX9 and pWX17 plasmids respectively, and both TetR-YFP and LacI-CFP from pWX6 plasmid (Wang et al., 2005b). To prevent replication blockage (Wang et al., 2006), AT (40 ng ml−1) and IPTG (0.5 mM) were added to the culture of strain containing the tetO/TetR-CFP system and the lacO/LacI-CFP system respectively. tetO/TetR-CFP was used for all FROS visualization in topA10 and mukB single and double mutants and the parental strain. tetO/TetR-CFP and lacO/LacI-CFP were both used for FROS visualization simultaneously with MukB-GFP. _mukB-gfp_4 (Ohsumi et al., 2001), mukB::km (Niki et al., 1991) and _dna_C2 (Zhou et al., 1997) alleles were introduced by transduction in AB1157 derivatives. topA10 allele (Biek and Cohen, 1989) was transduced to AB1157 mukB::km selecting transductants at 37°C, and then mukB-gfp allele was transduced. For snapshot experiments, except when indicated otherwise, cells were cultivated in M9 sodium-acetate supplemented with 100 μg ml−1 arginine, histidine, leucine, threonine and proline. For time-lapse experiments, cells were grown on a microscope slide coated with an agarose layer prepared with the medium used for snapshot experiment. The doubling time in liquid was ∼8 h at 22°C for MukB+ and mukB::km strains, and ∼4.8 h at 30°C for MukB+ cells, ensuring that newborn cells contain a non-replicating chromosome, as confirmed by flow cytometry (not shown). Microscopy using FROS and FISH was performed as described by Wang et al. (2006). Chromatin immunoprecipitation were realized with little modifications (see Supplementary material) to the protocol described previously (Kuras and Struhl, 1999) with E. coli AB1157 cells expressing MukB-GFP as the only MukB source, using anti-GFP antibodies (Roche). Microarrays and DNA hybridizations were realized by Oxford Gene Technology company (http://www.ogt.co.uk).
Acknowledgments
We thank Xindan Wang for the gift of strains, Xun Liu for motivating discussions and meticulous demonstration of the FISH technique, and Suckjoon Jun, Christian Lesterlin, Sean Kennedy and Sonia Trigueros for critical reading and comments on the manuscript. We also acknowledge Jim Sawitzke, S. Hiraga and H. Niki for strains. The work was funded by the Wellcome Trust. O.D. was supported by the Clarendon Postgraduate Award, R.R.-L. was supported by Conacyt and Clarendon Postgraduate Awards, and C.P. was supported by an EMBO Long-term Fellowship and the Wellcome Trust.
Supplementary material
The following supplementary material is available for this article:
Fig. S1
Time-lapse analysis of ori1 in mukB cells (OS27).
Fig. S2
Additional time-lapse analysis of R3 by FROS in mukB cells (OS55).
Fig. S3
Time-lapse tracking simultaneously MukB and ori1 [without (A) or with (B) ori1 duplication].
Table S1
Bacterial strains.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05881.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Biek DP, Cohen SN. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: mutations in the topA gene allow pSC101 replication in the absence of IHF. J Bacteriol. 1989;171:2066–2074. doi: 10.1128/jb.171.4.2066-2074.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, Lindqvist A, Lowe J, Nanninga N. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol Microbiol. 2001;42:1179–1188. doi: 10.1046/j.1365-2958.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Boccard F. Organization of the Escherichia coli chromosome into macrodomains and its possible functional implications. J Struct Biol. 2006;156:304–310. doi: 10.1016/j.jsb.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow JC, Britton RA, Grossman AD. Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J Bacteriol. 2002;184:5317–5322. doi: 10.1128/JB.184.19.5317-5322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol Microbiol. 2001;40:835–845. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai CH, Rai R, Rybenkov VV. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem. 2006;281:4606–4615. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV. SMC complexes in bacterial chromosome condensation and segregation. Plasmid. 2006;55:135–144. doi: 10.1016/j.plasmid.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Wang JC. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J Biol Chem. 2005;280:355–360. doi: 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005a;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005b;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T, Dasgupta S, Nordstrom K. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol Microbiol. 2000;38:392–400. doi: 10.1046/j.1365-2958.2000.02138.x. [DOI] [PubMed] [Google Scholar]
- Zhou P, Bogan JA, Welch K, Pickett SR, Wang HJ, Zaritsky A, Helmstetter CE. Gene transcription and chromosome replication in Escherichia coli. J Bacteriol. 1997;179:163–169. doi: 10.1128/jb.179.1.163-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1
Time-lapse analysis of ori1 in mukB cells (OS27).
Fig. S2
Additional time-lapse analysis of R3 by FROS in mukB cells (OS55).
Fig. S3
Time-lapse tracking simultaneously MukB and ori1 [without (A) or with (B) ori1 duplication].
Table S1
Bacterial strains.