Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response (original) (raw)
Abstract
Stress-induced eukaryotic translation initiation factor 2 (eIF2) α phosphorylation paradoxically increases translation of the metazoan activating transcription factor 4 (ATF4), activating the integrated stress response (ISR), a pro-survival gene expression program. Previous studies implicated the 5′ end of the ATF4 mRNA, with its two conserved upstream ORFs (uORFs), in this translational regulation. Here, we report on mutation analysis of the ATF4 mRNA which revealed that scanning ribosomes initiate translation efficiently at both uORFs and ribosomes that had translated uORF1 efficiently reinitiate translation at downstream AUGs. In unstressed cells, low levels of eIF2α phosphorylation favor early capacitation of such reinitiating ribosomes directing them to the inhibitory uORF2, which precludes subsequent translation of ATF4 and represses the ISR. In stressed cells high levels of eIF2α phosphorylation delays ribosome capacitation and favors reinitiation at ATF4 over the inhibitory uORF2. These features are common to regulated translation of GCN4 in yeast. The metazoan ISR thus resembles the yeast general control response both in its target genes and its mechanistic details.
Introduction
Phosphorylation of translation initiation factor 2 on serine 51 of its α subunit is a highly conserved point of convergence of signaling pathways adapting eukaryotic cells to diverse stressful conditions. Specific kinases have evolved to recognize the consequences of amino acid starvation (Dever et al., 1992), viral infection (Kostura and Mathews, 1989), iron deficiency (Chen et al., 1991), and accumulation of malfolded proteins in the ER (Harding et al., 1999) and convert these unrelated upstream stress signals to a single phosphorylation event. Phosphorylated eukaryotic translation initiation factor 2 (eIF2) α inhibits the guanine nucleotide exchange factor, eIF2B, attenuating eIF2-GTP-tRNAi met ternary complex formation and thereby reducing global translation initiation and polypeptide biosynthesis (Hinnebusch, 2000). Mutations in specific eIF2α kinases sensitize cells to the cognate stress (Harding et al., 2000b, 2001; Han et al., 2001; Zhang et al., 2002a; Anthony et al., 2004), and mutations that abolish eIF2α phosphorylation altogether, by converting serine 51 to alanine, broadly affect stress resistance (Scheuner et al., 2001). These observations attest to the functional importance of eIF2α phosphorylation as a convergence point in various stress-response pathways.
Part of the protective effect of eIF2α phosphorylation is exerted by attenuated global protein synthesis, which relieves chaperones of their load of unfolded client proteins (Harding et al., 2000b; Han et al., 2001; Scheuner et al., 2001), conserves amino acid pools for essential functions (Zhang et al., 2002b; Anthony et al., 2004) and attenuates metabolic consequences of protein synthesis (Tan et al., 2001; Harding et al., 2003; Lu et al., 2004). However eIF2α phosphorylation also promotes stress induced gene expression.
The mechanisms involved in activating this eIF2α(P)-dependent gene expression program have been studied in great detail in yeast. The crucial event is the paradoxically increased translation of the GCN4 mRNA, triggered by yeast's only eIF2α kinase, Gcn2p, which is activated by amino acid deprivation (Hinnebusch, 2000; Dever, 2002). Several short upstream ORFs (uORFs) mediate translational control of the GCN4 mRNA by repressing basal Gcn4p synthesis. However, under stress conditions that promote eIF2α phosphorylation and limit availability of eIF2-GTP-tRNAi met ternary complexes, these uORFs favor translation reinitiation at the Gcn4p coding region and synthesis of the effector transcription factor, which binds and activates numerous genes (Hinnebusch, 1997; Hinnebusch and Natarajan, 2002).
Vertebrates have four different eIF2α kinases and these activate numerous genes in an integrated stress response (ISR), promoting cell survival and stress resistance (Harding et al., 2000a, 2003; Scheuner et al., 2001; Okada et al., 2002; Lu et al., 2004). Activating transcription factor 4 (ATF4) is a transcription factor that activates many ISR target genes (Harding et al., 2003). It too is translationally activated in an eIF2α(P)-dependent manner (Harding et al., 2000a; Scheuner et al., 2001). We have analyzed the mechanisms involved in this key step of the ISR.
Results and discussion
A faithful reporter of ATF4 translational regulation
To determine if eIF2α phosphorylation is sufficient to up-regulate ATF4 translation we made use of a ligand activateable version of PERK. Addition of the AP20187 ligand leads to oligomerization of the chimeric Fv2E-PERK kinase and promotes eIF2α phosphorylation that is uncoupled from ER stress, which normally activates PERK. Incorporation of radiolabeled amino acids into ATF4 increased within minutes of Fv2E-PERK activation and highest levels were observed at relatively low doses of the activator associated with low levels of phosphorylated eIF2α. ATF4 translation was strictly dependent on eIF2α phosphorylation as it was altogether absent from EIF2A S51A mutant mouse fibroblasts lacking the regulatory phosphorylation site on eIF2α (Fig. 1 A).
Figure 1.
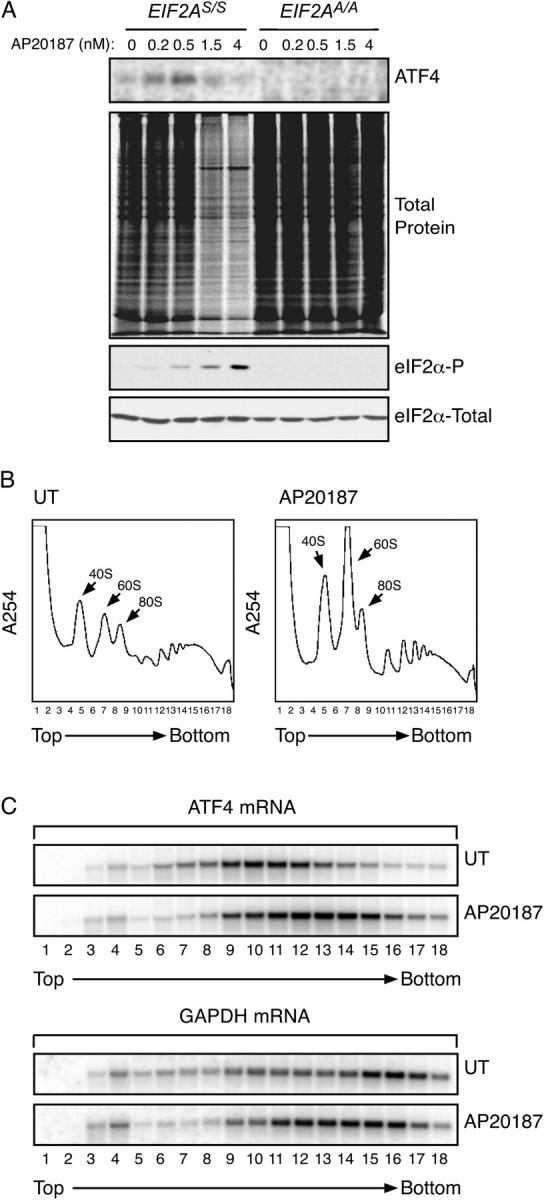
EIF2α phosphorylation is sufficient to up-regulate ATF4 translation. (A) Autoradiogram of SDS-PAGE of radiolabeled proteins after a brief labeling pulse of Fv2E-PERK expressing wild-type (EIF2A S/S) or mutant (EIF2A A/A) mouse fibroblasts pretreated for 30′ with the indicated concentration of the AP20187 activating ligand. The panels (from top to bottom) show immunoprecipitated radiolabeled endogenous ATF4, all radiolabeled newly synthesized proteins, and immunoblots of phosphorylated and total eIF2α. (B) Polysome profiles of mRNA isolated from untreated mouse fibroblasts and the same cells 60′ after induction of eIF2α phosphorylation by activation of the ligand-inducible Fv2E-PERK eIF2α kinase. Note the accumulation of ribosomal subunits and monosomes in the treated cells. (C) Northern blot analysis of ATF4 and GAPDH mRNA from fractions collected from the gradient shown in B. Note the shift to the right, (heavier) fractions in the peak of the ATF4 mRNA in the treated cells and the shift in opposite direction of the GAPDH peak.
Enhanced ATF4 translation correlated with a shift in the distribution of the mRNA to heavier fractions on a sucrose gradient. In untreated cells the ATF4 mRNA peak coincided with fractions engaged by approximately one ribosome. Fv2E-PERK activation caused the ATF4 mRNA to shift to a heavier fraction, reflecting engagement by two to three ribosomes. The normally well-translated GAPDH mRNA moved in the opposite direction (Fig. 1, B and C). These observations indicate low levels of translation initiation of the ATF4 mRNA in the basal state and its increase when eIF2α is phosphorylated, independently of stress.
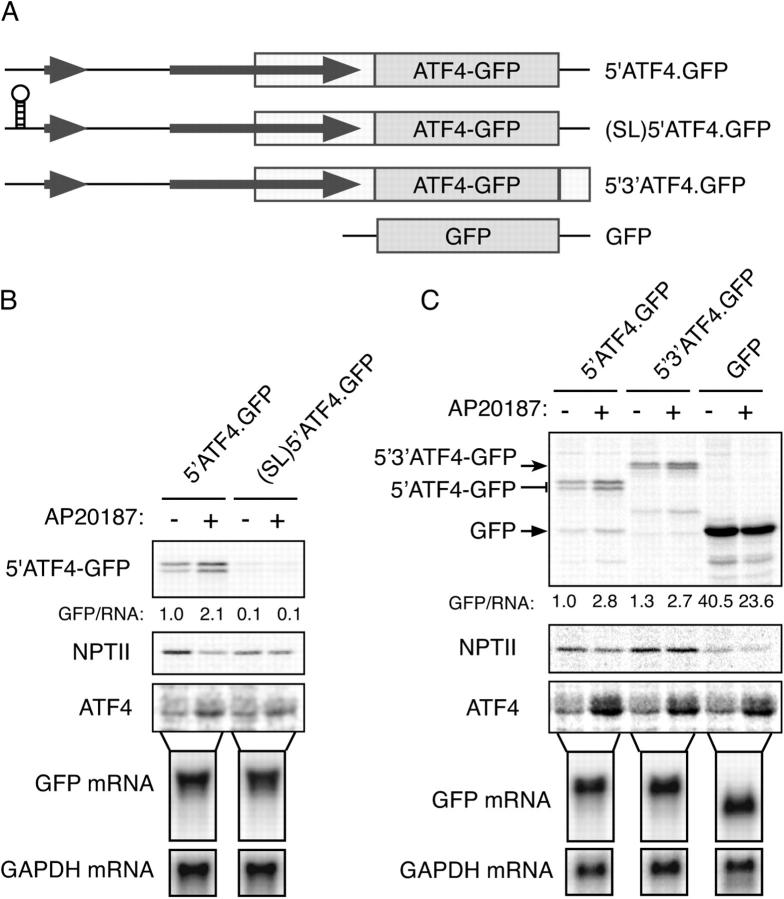
The ATF4 mRNA has two conserved uORFs implicated in translational regulation (Harding et al., 2000a). The 5′ uORF1 has three codons whereas the 3′ uORF2 is longer and extends into the ATF4 coding region. To establish a system for studying the impact of cis acting elements on ATF4 translation, we inserted the 5′ end of the ATF4 mRNA into a GFP reporter gene fusing the coding regions of ATF4 and GFP immediately 3′ of the termination codon of uORF2 (Fig. 2 A). A constitutive cytomegalovirus promoter drove transcription of this 5′ATF4-GFP reporter and SV40 viral signals specified termination and 3′ mRNA processing.
Figure 2.
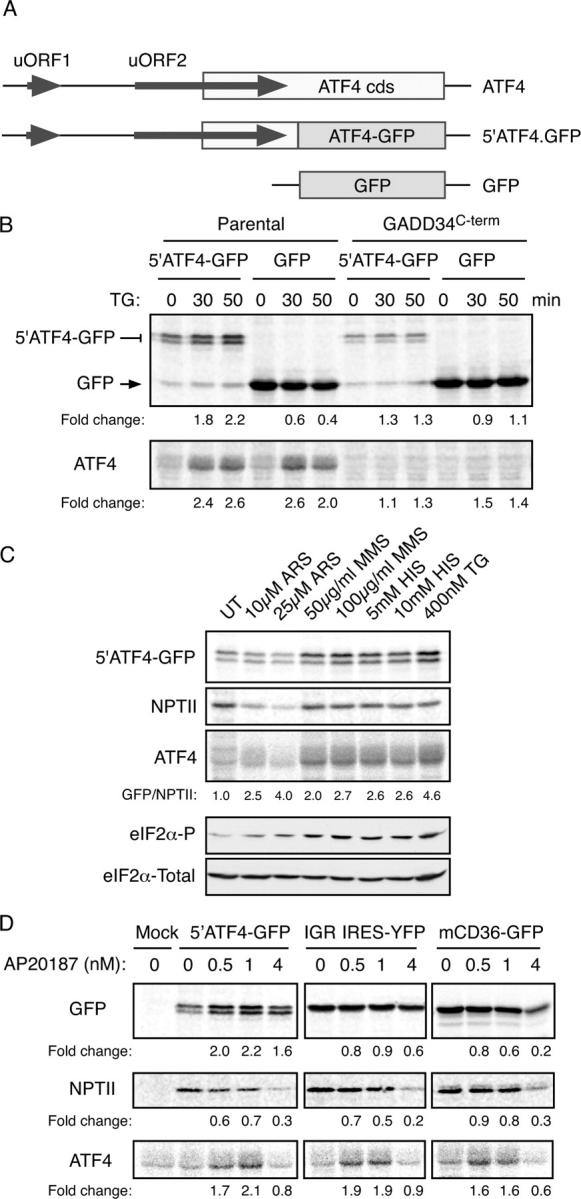
The 5′ end of the ATF4 mRNA mediates translational regulation of the gene. (A) Organization of the 5′ end of the mouse ATF4 mRNA, the derivative 5′ATF4-GFP reporter and the parental GFP reporter. (B) Autoradiogram of SDS-PAGE of radiolabeled proteins after a brief labeling pulse of wild-type CHO cells or cells stably overexpressing the COOH terminus of GADD34 (which blocks eIF2α phosphorylation) transfected with the indicated reporter plasmids. The cells were treated with the endoplasmic reticulum stress-inducing agent thapsigargin (TG) for the indicated time and the encoded GFP fusion proteins (top) and endogenous ATF4 (bottom) were immunoprecipitated with specific antibodies. (C) Autoradiogram of SDS-PAGE of radiolabeled proteins after a brief labeling pulse of CHO cells transfected with the ATF4-GFP reporter and pretreated for 30 min with the indicated concentration of arsenite (ARS), the electrophile methyl-methanesulfonate (MMS), histidinol (HIS, which activates GCN2), thapsigargin (TG), and the encoded GFP fusion proteins (top), the NPTII, encoded by a different gene on the same plasmid (second panel) and endogenous ATF4 (third panel) were immunoprecipitated with specific antisera. The GFP/NPTII ratio provides an estimate of the translational inducibility of the reporter in the treated cells. Immunoblots of phosphorylated (eIF2α-P) and total eIF2α from parallel lysates are shown in the bottom panels. (D) Autoradiogram of radiolabeled proteins from Fv2E-PERK expressing CHO cells treated with the indicated concentration of the activating AP20187 ligand for 30′ before and during the 20′ labeling pulse. The cells were transfected with the ATF4 reporter (5′ATF4-GFP), a cricket paralysis virus internal ribosome entry site reporter (IGR IRE5-YFP) or a GFP translational reporter derived from the 5′ end of mouse CD36 gene (mCD36-GFP). GFP, endogenous ATF4 and NPTII were immunoprecipitated as in C with specific antisera.
Treatment of cells with thapsigargin, an agent that causes ER stress and activates endogenous PERK (Harding et al., 1999), increased ATF4-GFP translation, detected by brief pulse labeling with 35S-radiolabeled met/cys followed by immunoprecipitation with an anti-GFP antiserum. This mirrored changes in endogenous ATF4, detected by immunoprecipitation from the same lysates (Fig. 2 B). Similar concordance between ATF4 and the reporter was observed in cells exposed to other ISR inducers (Fig. 2 C). In contrast, a reporter lacking the ATF4 5′ region was repressed by thapsigargin treatment. Translational induction was eIF2α(P) dependent as it was blocked by overexpressing GADD34C-term, an eIF2α-directed phosphatase (Novoa et al., 2001; Fig. 2 B; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200408003/DC1).
Reporter activation tapered off at highest doses of AP20187, which strongly repress global protein synthesis (reflected in reduced translation of the neomycin phosphotransferase, NPTII, marker carried on the same plasmid; Fig. 2 D). Translational inducibility by eIF2α(P) was specific to 5′ATF4 sequences as neither the internal ribosome entry site of the cricket paralysis virus mRNA, nor the uORF-containing 5′ fragment of the mouse CD36 gene, activated translation of the ATF4 reporter (Fig. 2 D).
Introduction of a stable stem loop at the 5′ end of ATF4-GFP mRNA markedly diminished reporter translation and blocked all stress inducibility without affecting mRNA levels (Fig. 3, A and B). Furthermore, the 5′ATF4 fragment directed low levels of translation initiation when incorporated as a second cistron downstream of the long luciferase coding region in a bi-cistronic reporter gene (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200408003/DC1). Together, these findings indicate that ribosomes scan the length of the mRNA to reach the ATF4 coding region and that the ATF4 5′ region is unable to direct efficient internal ribosome initiation. Replacement of the SV40 termination and mRNA processing signals of the reporter gene with the endogenous ATF4 mouse genomic fragment did not affect the reporter (Fig. 3 C), suggesting that communication between the 5′ and 3′ ends of the mRNA are unimportant to its translational regulation.
Figure 3.
ATF4 mRNA translation requires ribosomes scanning. (A) Predicted structure of the mRNA expressed by the reporter genes. Depicted is the stem loop introduced at the 5′ end of the mRNA and replacement of vector derived viral termination and poly-adenylation sequences (thin line) with ATF4 derived termination sequences (white rectangle). (B) Autoradiogram of SDS-PAGE of radiolabeled proteins from untreated and AP20187-treated CHO cells expressing Fv2E-PERK. The cells were transfected with ATF4-GFP reporters with or without the stable stem-loop structure at the 5′ end of the mRNA. Radiolabeled GFP, NPTII, and endogenous ATF4 were immunoprecipitated with a specific antisera. Cytoplasmic RNA from a parallel sample of untreated, transfected cells was resolved by Northern blot and hybridized to GFP and GAPDH probes, as indicated. The ratio of the labeled GFP to RNA signal in each lane is provided. (C) As in B, cells were transfected with ATF4.GFP reporters with a viral 3′ termination sequences (5′ATF4.GFP), or the genomic ATF4 3′ termination sequences (5′3′ATF4.GFP) or the parental GFP reporter.
Two 5′ uORFs mediate translational regulation of ATF4 by eIF2α phosphorylation
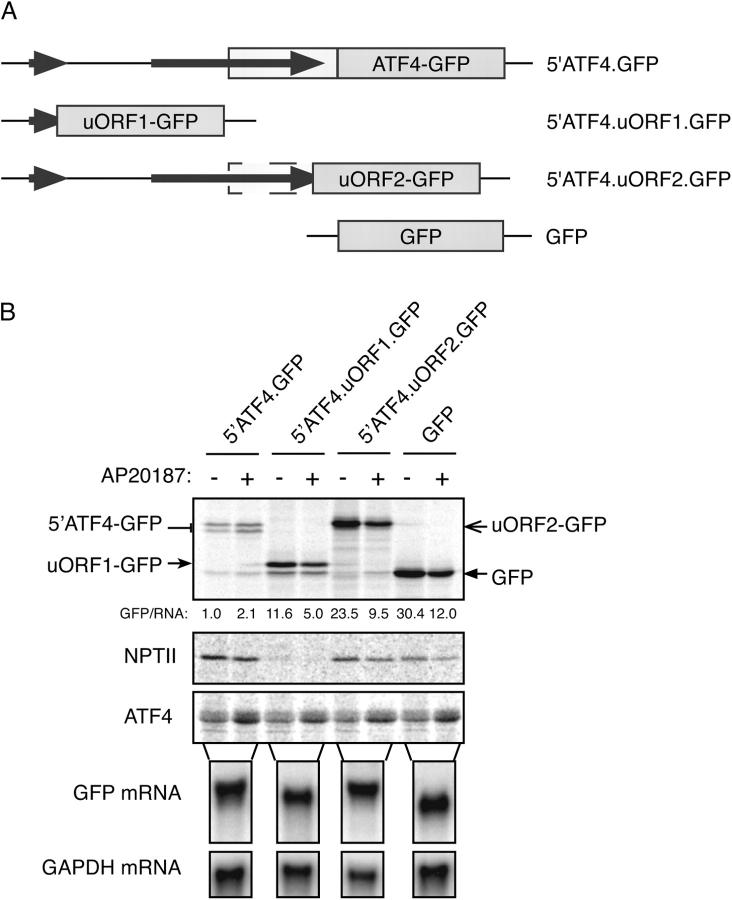
To determine if the scanning ribosomes initiate translation at the two uORFs we fused these to GFP and transfected the reporters into the Fv2E-PERK(+) CHO cells. Pulse labeling and immunoprecipitation showed high levels of translation initiation at both uORF1 and uORF2 under basal conditions (Fig. 4). Translation initiation at these uORFs was 10–20-fold higher than at ATF4. But unlike ATF4, which is translationally induced by eIF2α(P), both uORF1 and uORF2 were repressed by these conditions.
Figure 4.
Both ATF4 uORFs are translated under basal conditions. (A) Predicted structure of the mRNA expressed by the reporter genes used in these experiments. (B) Autoradiograms of SDS-PAGE of radiolabeled proteins from untreated and AP20187-treated Fv2E-PERK(+) CHO cells transfected with the ATF4-GFP reporter, or reporters fusing uORF1 or uORF2 to GFP. Cytoplasmic RNA from a parallel sample of untreated, transfected cells was resolved by Northern blot and hybridized to GFP and GAPDH probes. The low basal expression of NPTII in cells transfected with the uORF1-GFP is a reproducible if unexplained finding.
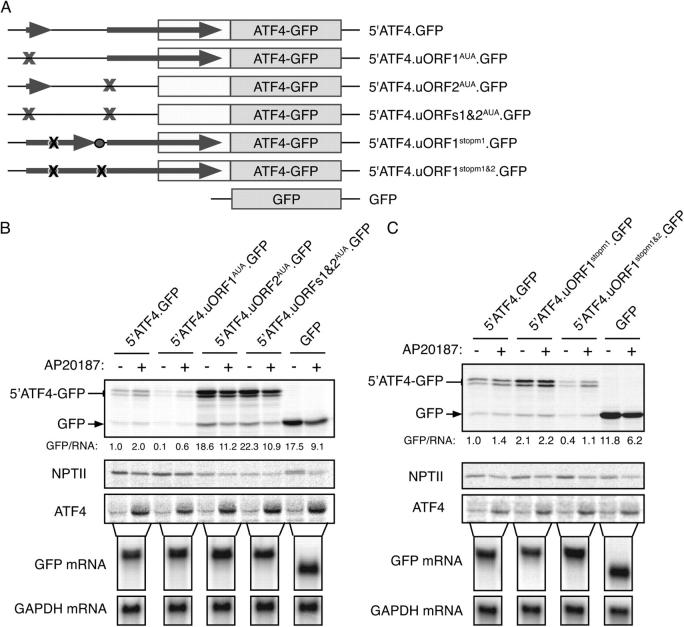
To measure the impact of the two uORFs on ATF4 translation and its regulation, we sequentially converted their AUG start codon to AUA in the context of the 5′ATF4-GFP reporter (Fig. 5 A). Loss of uORF1 markedly inhibited basal and induced ATF4-GFP translation, whereas loss of uORF2 markedly de-repressed basal ATF4-GFP translation and abolished inducibility (Fig. 5 B). The repressive effect of uORF2 on ATF4 translation is consistent with the highly processive nature of translation and with the lack of a mechanism for translating ribosomes to back track along the mRNA and reinitiate translation at AUGs located 5′ of the stop codon. Therefore, it is likely that all scanning ribosomes initiating translation at uORF2 are precluded from translating ATF4.
Figure 5.
Interplay of repressive and stimulatory uORFs regulates ATF4 translation. (A) mRNA expressed by the reporter genes used in these experiments. Where indicated, the initiating AUG codons of the uORFs were converted to AUA and the stop codons between the colinear uORF1 and uORF2 converted to UCG and UAC, respectively. (B) Autoradiograms of SDS-PAGE of radiolabeled proteins from untreated and AP20187-treated Fv2E-PERK(+) CHO cells transfected with the 5′ATF4-GFP reporter, or reporters with a mutation in the start codon of uORF1, uORF2 or both uORFs, or the parental GFP reporter. Cytoplasmic RNA from a parallel sample of untreated transfected cells was resolved by Northern blot and hybridized to GFP and GAPDH probes, as indicated. (C) As in B except cells were transfected with reporters in which the stop codons between the two uORFs were converted to Ser or Tyr codons.
Regulated reinitiation downstream of uORF1 accounts for most, but not all, eIF2α phosphorylation-mediated translation of ATF4
The stimulatory effect of uORF1 on ATF4 translation in the context of uORF2 is consistent with very efficient reinitiation at downstream AUGs. Such efficient translation reinitiation will direct a significant fraction of the ribosomes that have translated uORF1 to initiate at the repressive uORF2. By reducing formation of eIF2-GTP-tRNAi met ternary complexes, eIF2α phosphorylation reduces the probability that ribosomes terminating at uORF1 will acquire these complexes in time to translate the repressive uORF2, but may acquire them later, in time to initiate at the ATF4 AUG or subsequent downstream AUGs; note that initiation at both ATF4-GFP and the downstream GFP AUG (which is retained in the reporter) is coregulated (Fig. 3 C and Fig. 5 B). Reporters lacking uORF2 or both uORFs directed similar basal levels of ATF4-GFP translation (Fig. 5 B); as many ribosomes scanning the ATF4 mRNA initiate at uORF1 (Fig. 4 B), these observations suggest that uORF1 directs efficient reinitiation at downstream AUGs. Interestingly, loss of this hypothesized uORF1-dependent mechanism for “delivering” ribosomes to the ATF4 initiation codon does not altogether abolish up-regulation of ATF4-GFP by eIF2α phosphorylation (Fig. 5 B, compare lanes 3 and 4).
We had previously observed that deletion of the ATF4 mRNA region encompassing uORF1 de-repressed basal expression of an ATF4-luciferase reporter, which led us to conclude that uORF1 is repressive (Harding et al., 2000a). However, we now suspect that those observations were misleading as they were made on a deletion mutant that may have drastically distorted the mRNA. Furthermore, while this manuscript was under preparation, Vattem and Wek (2004) reported that a point mutation abolishing uORF1 does not significantly de-repress the ATF4-luciferase reporter used in our earlier work and has a markedly negative effect on ATF4-luciferase inducibility by pharmacological manipulations that promote eIF2α phosphorylation.
This model for translational reinitiation, orchestrated by availability of eIF2-GTP-tRNAi met ternary complexes predicts that shortening the distance between the stop codon of uORF1 and the start codon of uORF2 could favor reinitiation at ATF4, bypassing uORF2. Therefore, we converted the stop codon of uORF1 from a UAG to an UCG elongating uORF1 and reducing the inter-uORF distance from 87 to a mere 3 nts. As predicted (Abastado et al., 1991), this manipulation increased basal translation of ATF4-GFP and nearly eliminated its inducibility (Fig. 5 C). Eliminating both stop codons between uORF1 and uORF2 fused the two into one long repressive uORF that diminished both basal and induced translation of ATF4. Remarkably, eIF2α phosphorylation also elicited some residual induction of ATF4 in this reporter too (Fig. 5 C).
Similarities and differences between GCN4, ATF4, and other translationally regulated genes
Two uORFs account for most stress-induced translation of yeast GCN4 (Mueller and Hinnebusch, 1986; Mueller et al., 1987, 1988) and mouse ATF4. In both mRNAs, the 3′ most uORF represses basal translation of the protein coding region and the 5′ uORF1 is required to conditionally reverse this repression. This reversal is most easily explained by intrinsic high efficiency of reinitiation downstream of uORF1, which is further modulated by eIF2α phosphorylation and availability of eIF2-GTP-tRNAi met ternary complexes (Hinnebusch, 1997).
Mouse ATF4 has many features of a streamlined GCN4. The two partially redundant intervening yeast uORFs 2 and 3 are missing from the mouse mRNA. The 3′ uORF2 is lengthened considerably and extends into the coding region of ATF4, rendering it unconditionally repressive. By itself, uORF1 moderately represses basal Gcn4p translation (Mueller and Hinnebusch, 1986), whereas uORF1 has no measurable repressive effect on ATF4, suggesting higher efficiency of reinitiation downstream of the mammalian ORF1 than its yeast counterpart.
Although the aforementioned uORFs play a dominant role in regulating both GCN4 and ATF4, there is firm evidence for at least one alternative eIF2α phosphorylation-dependent mechanisms up-regulating translation that cannot be reconciled with regulated reinitiation. Two different ATF4 reporter constructs containing a single repressive uORF (Fig. 5 B, uORF1AUA; Fig. 5 C, uORF1stopm1&2; both of which extend past the start codon of ATF4-GFP) are translationally up-regulated by eIF2α phosphorylation. Similar observations have been made in GCN4 reporters with a single repressive uORF (Mueller and Hinnebusch, 1986; Tzamarias and Thireos, 1988). The nature of this alternative mechanism remains to be determined.
Loss of function mutations in subunits of the yeast GTP exchanger, eIF2B, or in eIF2 also de-repress GCN4 translation, supporting the hypothesis that eIF2α phosphorylation regulates GCN4 translation by reducing availability of eIF2-GTP-tRNAi met ternary complexes (Hinnebusch, 1997). Interestingly, some loss of function mutations in human genes encoding eIF2B subunits (a cause of childhood ataxia with cerebral hypomyelination; Fogli et al., 2002), also partially de-repress ATF4 (L. Kantor and O. Elroy-Stein, personal communication).
Other ISR target genes such as CHOP and GADD34 also have two or more uORFs and these strongly repress basal translation of the protein. However, no evidence for translational up-regulation of these genes in response to eIF2α phosphorylation has been uncovered (Jousse et al., 2001). Stress conditions associated with eIF2α phosphorylation have been reported to regulate translation initiation at internal AUGs in C/EBP family members (Calkhoven et al., 2000) and eIF2α phosphorylation plays a role in regulating the activity of the CAT1 mRNA's internal ribosome entry sequence (Yaman et al., 2003), however the relation of these processes to the regulation of ATF4 and GCN4 translation is unclear. ATF4 and GCN4 thus remain unique known examples of this unusual mode of translational regulation.
ATF4 accounts for <50% of the ISR (Harding et al., 2003; Lu et al., 2004). This suggests the existence of other effectors of eIF2α phosphorylation mediating gene expression. It will be interesting to determine if they too will prove to be translationally regulated by the ancient and conserved mechanism governing the expression of GCN4 and ATF4.
Materials and methods
Reporter gene construction
The 5′ end of mouse ATF4 cDNA was recovered by 5′RACE from mRNA of tunicamycin-treated mouse fibroblasts and was ligated between the HindIII-BamHI sites of pEGFP-N1 (CLONTECH Laboratories, Inc.), fusing the ATF4 and EGFP reading frames at Val40 of ATF4 to create the parental reporter 5′ATF4.GFP (plasmid sequence provided in Online supplement material). Point mutations described in the text were introduced into the ATF4 cDNA using over-lapping PCR mutagenesis. A stable stem-loop structure derived from the pRMF.pcDNA3 plasmid (Thoma et al., 2004) was inserted immediately downstream of the transcription start site of 5′ATF4.GFP to create the (SL) plasmid described in Fig. 3 B. A mouse genomic fragment extending 146 nts 5′ and 317 nts 3′ of the ATF4 poly-adenylation site was used to replace the GFP stop codon and transcriptional termination and poly-adenylation signals in the parental reporter, creating the 5′3′ATF4.GFP derivative, which fuses the last 21 residues of mouse ATF4 to GFP. Where indicated, the 5′ ATF4 fragment described above was fused in frame to GFP at the 3′ ends of uORF1 or uORF2. The mCD36.GFP plasmid was created by ligating a 316-nt fragment from the 5′ end of the mouse CD36 cDNA into pEGFP, fusing the coding region of CD36 to GFP at Gly12. The cricket paralysis virus intergenic region internal ribosome entry sequence, IGR IRES-YFP reporter (Wilson et al., 2000) was a gift of P. Sarnow (Stanford University, Stanford, CA).
Cell culture, cell transfection treatment, and metabolic labeling
Stable clones of mouse fibroblasts expressing Fv2E-PERK with defined EIF2A genotypes (Scheuner et al., 2001) have been described previously (Lu et al., 2004). Molecular dissection of the ATF4-GFP mRNA (Figs. 2–5) was performed in CHO cells stably expressing Fv2E-PERK established by the same procedure. CHO cells were chosen over mouse fibroblasts because of their enhanced transfectability.
Cells were transfected with the ATF4-GFP expressing plasmids using FuGENE 6 lipid-mediated gene transfer (1814443; Roche) and 24 h later were pretreated with thapsigargin (T9033; Sigma-Aldrich) at a final concentration of 400nM or AP20187 (ARIAD; www.ariad.com/regulation kits) 0.5 nM (unless otherwise indicated) for 30′ before and during a 20′ pulse label (200 μCi/ml, in DME with 10% of the normal methionine and cysteine content) with 35S TRANSlabel (MP Biomedical).
Immunoblot and immunoprecipitation
Total eIF2α was detected with a mAb to human eIF2α, a gift of the late E. Henshaw and phosphorylated eIF2α was detected with an epitope specific antiserum (RG0001; Research Genetics).
Radiolabeled endogenous ATF4 was immunoprecipitated from cell lysates as described previously (Harding et al., 2000a). Green fluorescent fusion proteins and NPTII were immunoprecipitated in identical conditions as ATF4 using 1 μL of commercial antiserum to GFP (A6455; Molecular Probes) or 2 μg of IgG to NPTII (N2008-05; U.S. Biologicals).
Northern blot and polysome profiles
Cytoplasmic RNA from transfected cells was recovered as described previously (Schreiber et al., 1989). Northern blots of MOPS-formaldehyde gels were hybridized to labeled cDNA probes from the GFP gene and GAPDH. Sucrose-density centrifugation of lysates of untreated and AP20187 treated Fv2E-PERK(+) mouse fibroblasts and analysis of mRNA content by Northern blot were performed as described previously (Harding et al., 2000a).
Online supplemental material
Fig. S1 is an immunoblot of GADD34C-term and eIF2α-P in the cells shown in Fig. 2 B. Fig. S2 is an autoradiogram of ATF4-GFP translation when inserted as a 3′ cistron. JCB.200408003.seq.txt is an EMBL formatted TEXT file with the sequences of the plasmids used in these studies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200408003/DC1.
Acknowledgments
We thank Donalyn Scheuner and Randy Kaufman (University of Michigan, Ann Arbor, MI) for the EIF2A S51A cells, the ARIAD Corporation for the inducible dimerization system, Peter Sarnow for the CPV IRES plasmid, and Mathias Hentze (EMBL, Heidelberg, Germany) for the stable stem loop sequence.
This work was supported by National Institutes of Health grants ES08681 and DK47119. D. Ron is a Scholar of the Ellison Medical Foundation.
Abbreviations used in this paper: ATF4, activating transcription factor 4; eIF2, eukaryotic translation initiation factor 2; ISR, integrated stress response; NPTII, neomycin phosphotransferase; uORF, upstream ORF.
References
- Abastado, J.P., P.F. Miller, B.M. Jackson, and A.G. Hinnebusch. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 11:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, T.G., B.J. McDaniel, R.L. Byerley, B.C. McGrath, D.R. Cavener, M.A. McNurlan, and R.C. Wek. 2004. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 279:36553–36561. [DOI] [PubMed] [Google Scholar]
- Calkhoven, C.F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Chen, J.J., M.S. Throop, L. Gehrke, I. Kuo, J.K. Pal, M. Brodsky, and I.M. London. 1991. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc. Natl. Acad. Sci. USA. 88:7729–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever, T.E. 2002. Gene-specific regulation by general translation factors. Cell. 108:545–556. [DOI] [PubMed] [Google Scholar]
- Dever, T.E., L. Feng, R.C. Wek, A.M. Cigan, T.F. Donahue, and A.G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 68:585–596. [DOI] [PubMed] [Google Scholar]
- Fogli, A., K. Wong, E. Eymard-Pierre, J. Wenger, J.P. Bouffard, E. Goldin, D.N. Black, O. Boespflug-Tanguy, and R. Schiffmann. 2002. Cree leukoencephalopathy and CACH/VWM disease are allelic at the EIF2B5 locus. Ann. Neurol. 52:506–510. [DOI] [PubMed] [Google Scholar]
- Han, A.P., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, M. Fleming, P. Leboulch, S.H. Orkin, and J.J. Chen. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, and D. Ron. 1999. Translation and protein folding are coupled by an endoplasmic reticulum resident kinase. Nature. 397:271–274. [DOI] [PubMed] [Google Scholar]
- Harding, H., I. Novoa, Y. Zhang, H. Zeng, R.C. Wek, M. Schapira, and D. Ron. 2000. a. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. b. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. [DOI] [PubMed] [Google Scholar]
- Harding, H., H. Zeng, Y. Zhang, R. Jungreis, P. Chung, H. Plesken, D. Sabatini, and D. Ron. 2001. Diabetes Mellitus and excocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in survival of secretory cells. Mol. Cell. 7:1153–1163. [DOI] [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, H. Zeng, I. Novoa, P. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272:21661–21664. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. Translational control of gene expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. CSHL Press, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 185–243.
- Hinnebusch, A.G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 1:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse, C., A. Bruhat, V. Carraro, F. Urano, M. Ferrara, D. Ron, and P. Fafournoux. 2001. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 29:4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostura, M., and M.B. Mathews. 1989. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol. Cell. Biol. 9:1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P.D., C. Jousse, S.J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R.J. Kaufman, D. Ron, and H.P. Harding. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, P.P., and A.G. Hinnebusch. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 45:201–207. [DOI] [PubMed] [Google Scholar]
- Mueller, P.P., S. Harashima, and A.G. Hinnebusch. 1987. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc. Natl. Acad. Sci. USA. 84:2863–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, P.P., B.M. Jackson, P.F. Miller, and A.G. Hinnebusch. 1988. The first and fourth upstream open reading frames in GCN4 mRNA have similar initiation efficiencies but respond differently in translational control to change in length and sequence. Mol. Cell. Biol. 8:5439–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa, I., H. Zeng, H. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of ATF6 and PERK in transcription during the mammalian unfolded protein response. Biochem. J. 366:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner, D., B. Song, E. McEwen, P. Gillespie, T. Saunders, S. Bonner-Weir, and R.J. Kaufman. 2001. Translational control is required for the unfolded protein response and in-vivo glucose homeostasis. Mol. Cell. 7:1165–1176. [DOI] [PubMed] [Google Scholar]
- Schreiber, E., P. Matthias, M.M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts”, prepared from small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, S., N. Somia, P. Maher, and D. Schubert. 2001. Regulation of antioxidant metabolism by translation initiation factor 2alpha. J. Cell Biol. 152:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, C., A. Ostareck-Lederer, and M.W. Hentze. 2004. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol. Biol. 257:171–180. [DOI] [PubMed] [Google Scholar]
- Tzamarias, D., and G. Thireos. 1988. Evidence that the GCN2 protein kinase regulates reinitiation by yeast ribosomes. EMBO J. 7:3547–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem, K.M., and R.C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., T.V. Pestova, C.U. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell. 102:511–520. [DOI] [PubMed] [Google Scholar]
- Yaman, I., J. Fernandez, H. Liu, M. Caprara, A.A. Komar, A.E. Koromilas, L. Zhou, M.D. Snider, D. Scheuner, R.J. Kaufman, and M. Hatzoglou. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 113:519–531. [DOI] [PubMed] [Google Scholar]
- Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D.R. Cavener. 2002. a. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., B.C. McGrath, J. Reinert, D.S. Olsen, L. Lei, S. Gill, S.A. Wek, K.M. Vattem, R.C. Wek, S.R. Kimball, et al. 2002. b. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22:6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]