Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages (original) (raw)
Abstract
Condensin is an evolutionarily conserved protein complex that helps mediate chromosome condensation and segregation in mitotic cells. Here, we show that condensin has two activities that contribute to meiotic chromosome condensation in Saccharomyces cerevisiae. One activity, common to mitosis, helps mediate axial length compaction. A second activity promotes chromosome individualization with the help of Red1 and Hop1, two meiotic specific components of axial elements. Like Red1 and Hop1, condensin is also required for efficient homologue pairing and proper processing of double strand breaks. Consistent with these functional links condensin is necessary for proper chromosomal localization of Red1 and Hop1 and the subsequent assembly of the synaptonemal complex. Finally, condensin has a Red1/Hop1-independent role in the resolution of recombination-dependent linkages between homologues in meiosis I. The existence of distinct meiotic activities of condensin (axial compaction, individualization, and resolution of recombination-dependent links) provides an important framework to understand condensin's role in both meiotic and mitotic chromosome structure and function.
Keywords: SMC-family; chromosome structure; double strand breaks; meiosis; S. cerevisiae
Introduction
In meiosis, two rounds of chromosome segregation follow one round of DNA replication. In the first division, meiosis I, homologous chromosomes pair and separate; whereas in the second division, meiosis II, sister chromatids segregate from each other. The segregation of sister chromatids during meiosis II is functionally equivalent to their segregation during mitosis and is thought to occur by a very similar mechanism. However, the pairing and segregation of homologues during meiosis I requires meiosis specific processes. Double strand breaks (DSBs) are induced by a special endonuclease (Keeney et al., 1997). These DSBs are repaired by recombination pathways that ensure the formation of crossovers between homologues, thereby promoting proper meiosis I segregation (Roeder, 1997; Zickler and Kleckner, 1999).
Meiotic chromosome segregation requires proper meiotic chromosome structure. The rod-shaped meiotic chromosome is similar to the mitotic chromosomes in appearance, suggesting an underlying mitotic-like organization. However, in most organisms, meiosis I cells assemble a specialized trilaminar structure between homologues called the synaptonemal complex (SC). The SC modulates recombination, which is essential for homologue segregation. The presence of both meiosis-specific and mitotic-like features of meiotic chromosomes raises several questions. Do the common features of meiotic and mitotic chromosomes reflect a shared underlying mechanism of organization? If so, is the machinery responsible for this shared mechanism important for the assembly of meiosis-specific structures like the SC? Finally, how do mitotic-like and meiosis-specific features of meiotic chromosomes contribute to chromosome recombination and segregation during meiosis I?
The meiotic chromosome structure has been proposed to be built upon mitotic components that are expressed in meiosis (Zickler and Kleckner, 1999; Stack and Anderson, 2001). One structural component of mitotic chromosomes is the evolutionarily conserved protein complex, condensin. The 13S condensin complex is composed of two subcomplexes, an 8S core containing the two Smc-family subunits and the 11S regulatory subcomplex containing the three non-Smc subunits (Strunnikov et al., 1995; Hirano et al., 1997). In budding yeast, the five subunits are named as Smc2, Smc4, Brn1, Ycg1, and Ycs4 (Freeman et al., 2000; Lavoie et al., 2002). Condensin has been shown to be important for the establishment and maintenance of mitotic chromosome condensation in vitro, using Xenopus laevis egg extracts, and in vivo, using mutational analyses in both budding and fission yeasts (Hirano and Mitchison, 1994; Saka et al., 1994; Strunnikov et al., 1995; Lavoie et al., 2000, 2002; Bhalla et al., 2002). However, in other organisms condensin inactivation has led to only small changes in chromosome condensation, suggesting the possibility of alternative pathways for chromosome compaction (Bhat et al., 1996; Steffensen et al., 2001; Hagstrom et al., 2002; Kaitna et al., 2002). Interestingly, condensin inactivation in mitotic cells of all organisms results in chromosome bridging at anaphase, presumably the failure to resolve links between sister chromatids (Saka et al., 1994; Bhat et al., 1996; Ouspenski et al., 2000; Steffensen et al., 2001; Hagstrom et al., 2002; Lavoie et al., 2000, 2002). This bridging has been assumed to be the consequence of incomplete or improper chromosome compaction.
Given its function in mitotic chromosome condensation, condensin was a likely candidate to participate in the organization of meiotic chromosomes. However, a recent analysis in Caenorhabditis elegans suggests that condensin subunits Mix-1 (homologue of Smc2) and Smc-4 are required for meiosis II, but not for meiosis I (Hagstrom et al., 2002). A similar conclusion was suggested from antisense depletion studies of condensin in activated Xenopus eggs (Watrin et al., 2003). Because meiosis II is very similar to mitotic chromosome segregation, these observations are consistent with the notion that condensin function is limited only to mitotic-like chromosome divisions. However, there are two sets of condensin like complex in C. elegans, raising the possibility that a role for condensin in meiosis I might be obscured by functional redundancy. Similarly condensin's role in meiosis I in the Xenopus study may have been obscured by the persistence of significant condensin activity after depletion, particularly given the huge stockpiles of condensin in the egg. Thus, it is important to assess the role of condensin in meiosis I in another organism.
The study of condensin in budding yeast meiosis has several strengths. Only a single copy of each condensin subunit has been found in the budding yeast genome (Freeman et al., 2000), eliminating the complexity of functional redundancy. Conditional temperature-sensitive mutants in several condensin subunits have already been identified and well characterized (Freeman et al., 2000; Lavoie et al., 2002). These conditional alleles permit the inactivation of condensin specifically in meiosis. The evaluation of condensin role in meiotic chromosome organization is facilitated by the fact that meiotic chromosomes in budding yeast are highly compacted and individualized like meiotic chromosomes in other eukaryotes. Finally, numerous studies have led to the identification and characterization of key components required for proper recombination and formation of the SC. These include the meiosis specific endonuclease, Spo11 (Keeney et al., 1997), the meiosis specific cohesin, Rec8 (Klein et al., 1999), components of the axial–lateral (Red1 and Hop1) and transverse elements (Zip1) of SC (Sym et al., 1993; Smith and Roeder, 1997; Woltering et al., 2000), and components that mediate repair of DSBs, Dmc1, and Rad51 (Bishop et al., 1992). Here we use these tools to address the role of condensin in meiotic chromosome organization. Our results suggest that condensin contributes to meiosis both through its ability to facilitate mitotic-like compaction of meiotic chromosomes and through additional meiosis-specific activities.
Results
Condensin is present in meiosis and is essential for sporulation
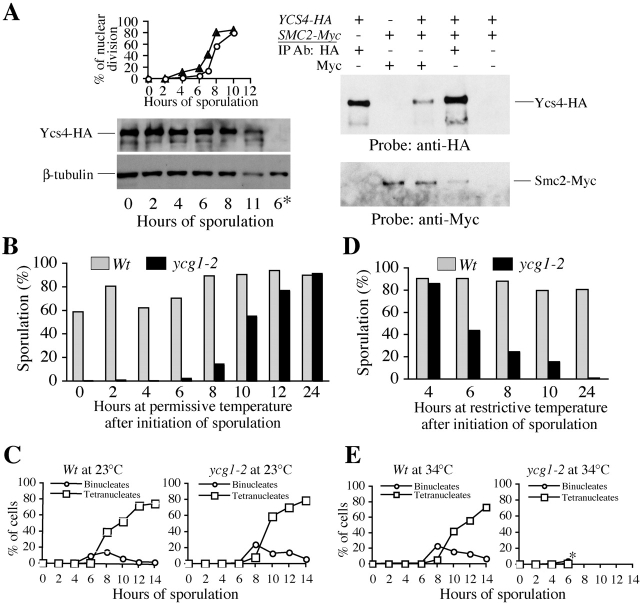
To investigate the potential requirement for condensin during meiosis in budding yeast, we examine its meiotic expression (Fig. 1 A). We used Smc2 as representative of the 8S core subcomplex, and Ycs4 of the 11S regulatory subcomplex. A COOH-terminal HA-tagged allele of YCS4 was incorporated into the endogenous YCS4 locus, providing the sole functional copy in the genome. This strain undergoes meiosis (∼80% sporulation efficiency), and generates viable spores (>95%) although its progression through meiosis I is slightly delayed (Fig. 1 A, top left). The Ycs4-HA protein is present at a constant level throughout meiosis (Fig. 1 A, bottom left). A similar result is observed for a functional version of Smc2 tagged with the Myc epitope (unpublished data). Therefore, condensin is present in meiosis. In addition, 5 h after induction of meiosis, the myc-tagged Smc2 is able to pull down HA-tagged Ycs4 by immunoprecipitation, and vice versa (Fig. 1 A, right). These results suggest that condensin subunits form a complex in yeast meiotic cells, similar to mitotic cells (Freeman et al., 2000).
Figure 1.
Presence of condensin in yeast meiotic cells and its role in sporulation. (A) At indicated times after the initiation of sporulation, meiotic cells were fixed and assayed for nuclear division. Protein extracts were made simultaneously to evaluate condensin protein level. The percentage of nuclear division (meiosis I + meiosis II) is shown for untagged strain (NH144, ▴) and the tagged YCS4-3HA strain (2892, ○; top left). The Ycs4-3HA protein levels were monitored by Western blot probing with an anti-HA antibody (bottom left). The same blot was reprobed with a β-tubulin antibody to confirm sample loading. The last lane (6*) was loaded with protein extract made from an untagged strain (NH144) after 6 h of sporulation. To assess condensin complex assembly in meiosis, proteins were extracted from cells after 5 h of sporulation, and subjected to immunoprecipitation and Western blotting (right). The first two lanes were loaded with extracts from single tagged strains, 2892 (YCS4-3HA) and 2933 (SMC2-6Myc), respectively. The last three lanes were from strain 2937 that has both condensin subunits tagged. (B–E) Inactivation of the condensin complex through the ycg1-2 allele leads to defective nuclear division and sporulation. (B) Aliquots from synchronous meiotic cultures of wild-type (strain 2864) and ycg1-2 (2863) were shifted up from 23°C to 34°C at indicated times after the initiation of sporulation and were assayed for sporulation efficiency (Materials and methods). (C) Nuclear division monitored by DAPI-staining of fixed cells from wild-type and ycg1-2 after initiation of sporulation at 23°C. (D) Sporulation efficiency was determined in cultures that were shifted to 34°C upon induction of sporulation (time 0) and then shifted down from 34°C to 23°C at indicated times. (E) Nuclear division monitored by DAPI-staining of fixed cells from wild-type and ycg1-2 after initiation of sporulation at 34°C. The asterisk indicates that after 8 h of sporulation, mutant cells undergo massive nuclear fragmentation, which prevents assaying nuclear division.
The presence of condensin in sporulating cells suggests that it is functionally important for this process. To test function of condensin in sporulation, we used condensin conditional mutants (smc2-8, ycg1-2, and ycs4-2). Mitotic cultures of wild-type and condensin mutants were grown at 23°C (permissive), placed in sporulation medium, and incubated at 23°C or 34°C. Wild-type cultures make spores at either temperature but there is a slight reduction at 34°C (Fig. 1, B and D). In contrast, only 5% of ycg1-2 cells make spores at 34°C compared with 90% at 23°C (permissive). The low sporulation frequency is also observed with the ycs4-2 (∼2%) and smc2-8 (∼9%) mutants at 34°C. The slightly higher sporulation in smc2-8 is consistent with the fact that mitotic cultures of smc2-8 grow slowly at 34°C, whereas the ycs4-2 and ycg1-2 strains fail to grow at all. Most spores produced by condensin mutants are nonviable at 34°C. For example, only 21% of spores from ycg1-2 are viable after sporulation at 34°C. In contrast, more than 95% of spores are viable when ycg1-2 sporulates at 23°C. These data demonstrate that condensin is required for sporulation.
To determine the latest time for condensin function, synchronous meiotic cultures of wild-type and ycg1-2 were shifted at different times from permissive (23°C) to nonpermissive (34°C) to initiate the inactivation of condensin in mutant cells (Fig. 1 B). Wild-type cells sporulate relatively efficiently independent of the time of shift to 34°C (Fig. 1 B, gray bars). In contrast, mutant cells are able to sporulate well only when inactivation of ycg1-2 protein is delayed until 10–12 h in sporulation medium (Fig. 1 B, black bars). By this time 55–67% of meiotic cells have already completed both nuclear divisions (Fig. 1 C). Thus, condensin function is required late in sporulation consistent with a role in meiotic chromosome segregation.
To determine the earliest requirement for condensin function, we performed a complementary temperature shift-down assay, activating ycg1-2 and presumably condensin at different times after the initiation of meiosis. Efficient sporulation of the mutant requires activation of ycg1-2 protein before 6 h into meiosis (Fig. 1 D). Because ycg1-2 mutant cells are compromised for nuclear division at 34°C (Fig. 1 E), we use wild-type cells to infer the minimal time when onset of nuclear divisions in ycg1-2 could have occurred. Meiotic S-phase occurs ∼4 h after initiation of sporulation at 34°C (not depicted), followed by the onset of nuclear divisions (MI and MII) between 6–8 h after the initiation of sporulation (Fig. 1 E). Together, our results are consistent with a requirement for condensin function in a window of meiosis sometime at or after meiotic S-phase until the completion of meiotic chromosome segregation.
Condensin is a component of the axial core of meiotic chromosome
As a first step to determine meiotic function of condensin, we examined its localization on meiotic chromosomes. Nuclei from meiotic cells expressing epitope-tagged condensin subunits were spread onto slides and processed for indirect immunofluorescence. In early meiosis, Ycs4-HA is heavily enriched on the rDNA similar to mitotic cells (unpublished data). As cells progress into meiosis, condensin staining appears over the amorphous nuclear DNA. By pachytene, when individual chromosomes can easily be resolved, Ycs4-HA is present as semi-continuous foci along the entire length of the synapsed chromosomes (Fig. 2 A) except at the ribosomal DNA locus, where Ycs4-HA is more concentrated (Fig. 2 B). By anaphase I, Ycs4-HA becomes more enriched in regions of the chromosomes that are proximal to the spindle poles (Fig. 2 A). Because Smc2-Myc shows a similar pattern (unpublished data), these localization experiments suggest that the chromosome association of condensin is dynamic.
Figure 2.
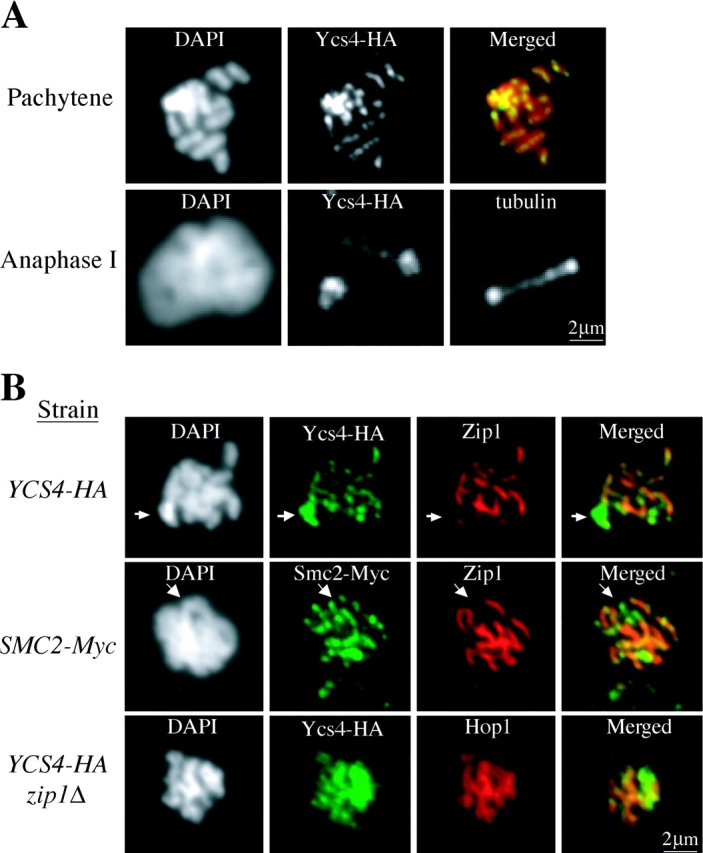
Distribution of condensin on meiotic chromosomes. (A) Condensin subunit Ycs4 localizes to meiotic chromosomes. Nuclei from meiotic cells of strain 2892 were spread onto slides and processed for indirect immunofluorescence using anti-HA and anti-tubulin antibodies. Note that Ycs4-3HA staining is restricted to the mid-line of synapsed chromosomes. At anaphase I, Ycs4-3HA concentrates in the polar regions of the separating chromosomes. (B) Condensin is associated with the core of synapsed chromosomes. Nuclei from meiotic cells of listed strains were spread onto slides and processed for indirect immunofluorescence using anti-HA, anti-Myc, anti-Zip1, and anti-Hop1 antibodies. Although both Smc2-6Myc and Ycs4-3HA staining are fragmented on synapsed chromosomes, Zip1 staining is continuous. The arrows indicate the ribosomal DNA region on chromosome XII, where Zip1 does not localize. Note that chromosomal association of Ycs4-3HA is independent of Zip1.
In pachytene cells, the chromosomal staining of condensin subunits lies in the middle of the bulk chromatin stained by DAPI (Fig. 2, A and B). This corresponds to the position of the SC. To better assess the localization of condensin and SC, spread nuclei were processed for immunofluorescence and stained with antibodies to detect condensin and components of the SC including Zip1, a marker for the central element, and Hop1 or Red1, markers for the axial–lateral elements (Smith and Roeder, 1997; Dong and Roeder, 2000). Indeed, condensin subunits localize to the same axes–core as SC markers (Fig. 2 B and see next paragraph). Next, we examined condensin localization in meiotic cells lacking the central element because of a deletion of the ZIP1 gene. Even in the absence of Zip1, condensin continues to localize with axial–lateral components, Hop1 and Red1 (Fig. 2 B and not depicted). Therefore, condensin associates with the core of meiotic chromosomes.
Condensin is important for meiotic chromosome compaction
Together, the axial localization of condensin and its established role in mitotic chromosome compaction (for review see Hirano, 2002) suggested that condensin might have a role in meiotic chromosome compaction. The compaction of yeast meiotic chromosomes has two observable properties. First is the end to end length, which reflects axial organization and longitudinal compaction. To assay this aspect of compaction, we constructed wild-type and condensin mutant strains that contained a GFP marker 35 kb away from centromere V. This allowed us to identify chromosome V in spread nuclei and to measure the length of the meiotic axis as defined by Rec8 staining (Fig. 3 A). These strains also included the ndt80 mutation, which inactivates a meiotic transcription factor and causes homogenous arrest at pachytene (Xu et al., 1995). By arresting the cells, differences between wild-type and condensin mutants could not be attributed to cell cycle differences. Association of Rec8 with the chromosomes is not significantly altered in condensin mutants based on indirect immunofluorescence and chromosome immunoprecipitation analysis (unpublished data). At 34°C the axial length of chromosome V at pachytene is 0.9 μm in ndt80 cells, compared with 1.3 μm in ycg1-2 ndt80 cells and 1.5 μm in ycs4-2 ndt80 cells, increases of 44 and 67%, respectively (Fig. 3 B). These data suggest that a chromosome axis can be formed independent of proper condensin function, but condensin is required to achieve the proper longitudinal compaction of this axis.
Figure 3.
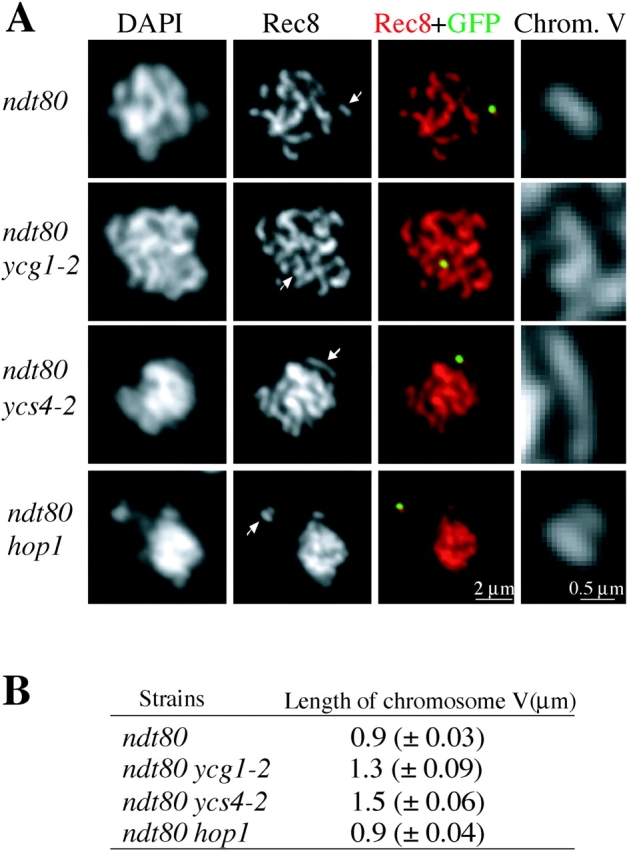
Condensin contributes to total chromosome compaction at pachytene. (A) Representative micrographs of spread nuclei from ndt80 cells arrested in pachytene. Strains harboring ndt80 (2880_), ycg1-2 ndt80 (2879)_, ycs4-2 ndt80 (2881), and hop1 ndt80 (2973) were harvested after 12 h of sporulation. Chromosomal DNA and Rec8-HA were visualized by DAPI and anti-HA staining, respectively. Synapsed chromosome V can be identified because these cells contain tetO operators on both copies of chromosome V and express tetR-GFP. The length of the Rec8 axis of chromosome V was measured in those spreads where chromosome V could be clearly distinguished from the other chromosomes as indicated by arrows. Enlarged views of the Rec8 axis of chromosome V are shown by Rec8-HA staining in the right. (B) Summary of average length of the Rec8 axis of chromosome V. Approximately 12 cells were scored for each strain. Standard error of the mean is shown in parentheses. t test shows the length of chromosome V from condensin mutants is significantly different from that of condensin wild-type cells (P < 0.001).
The other property of compaction is chromosome individualization, the resolution of chromosomes into distinct spatial domains. This property likely reflects a combination of overall chromatin compaction and the resolution of entanglements between homologues. In spread nuclei of wild-type meiotic prophase, DAPI stained homologues are individualized as discrete linear structures (Fig. 3 A). However, in condensin mutants homologues often fail to resolve and are fuzzy (Fig. 3 A). Thus, condensin is required for individualization as well as longitudinal compaction.
The function of condensin in these two aspects of meiotic chromosome compaction might be specific to condensin or a general property of any component of the meiotic core. To test this possibility, we monitored chromosome organization in hop1 ndt80 cells. The end to end length of chromosomes in these mutant cells is indistinguishable from wild-type suggesting normal axial organization and longitudinal compaction (Fig. 3, A and B). In contrast chromosomes are fuzzy and fail to separate in hop1 cells (Fig. 3 A), indicating a defect in individualization. Similar results were obtained from red1 ndt80 cells (not depicted). In summary, longitudinal compaction requires condensin but not Red1 or Hop1, whereas individualization requires all three components of the meiotic chromosome core.
Condensin is required for the proper assembly of the SC
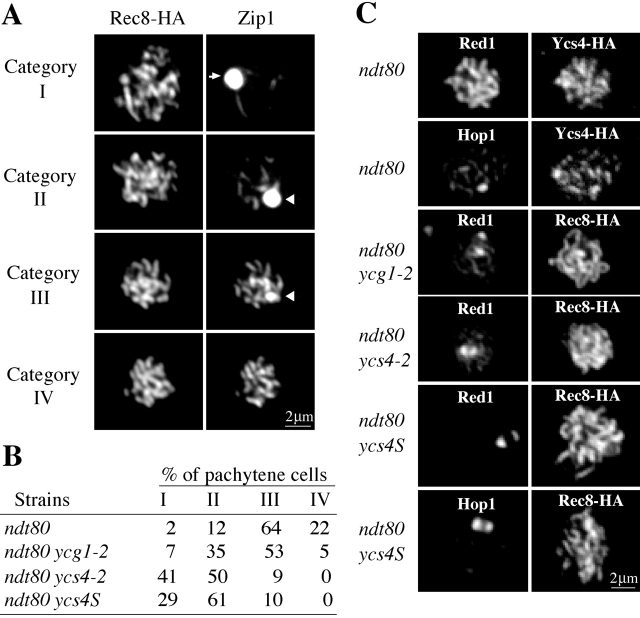
In addition to compaction, a second feature of meiotic chromosome structure is the SC. To assess the role of condensin in SC formation, wild-type and condensin mutants were placed in sporulation medium at 34°C and analyzed for Zip1 localization by indirect immunofluorescence at different time intervals. In synchronous cultures, chromosomal staining of Zip1 is dramatically reduced in condensin mutants (unpublished data). To better assess this phenotype, wild type and condensin mutants were staged at pachytene by inactivation of NDT80. As expected, the majority of cells with wild-type condensin exhibits staining with Zip1. A minor fraction possess small, but recognizable polycomplex (Fig. 4 A), which originates from the aggregation of SC components, and is indicative of defects in SC assembly (Sym and Roeder, 1995). Thus, high temperature in combination with ndt80 null causes very modest changes in Zip1 localization. In contrast, only 10% of ycs4-2 cells exhibit significant Zip1 chromosomal staining and even these still possess polycomplex (Fig. 4 A, category III). Localization of Zip1 is also perturbed in the ycg1-2 mutant although to a less extent (Fig. 4 B). The weaker phenotype of the ycg1-2 mutant is consistent with the fact that it has a significantly higher maximum permissive temperature for both mitosis and meiosis than the ycs4-2 mutant. Because both YCG1 and YCS4 are required for proper SC assembly, these genes most likely perform this common function in the context of condensin.
Figure 4.
Condensin is required for the assembly of axial elements and SC. (A) Nuclei from cells arrested at pachytene with ndt80 were spread and assigned to one of four categories based upon Zip1 localization: category I, absent from chromosomes and almost entirely in the polycomplex; category II, increased chromosome localization of Zip1 and persistence of large polycomplex; category III, significant chromosomal Zip1 localization and only residual polycomplex; and category IV, chromosome localization only. Arrows indicate the polycomplex. (B) Quantification of Zip1 localization in wild-type and condensin mutants arrested with ndt80. Strains harboring ndt80 (2880), ycg1-2 ndt80 (2879), ycs4-2 ndt80 (2881), and ycs4S ndt80 (2886) were harvested after 12 h of sporulation. For each strain spread nuclei were prepared, and ∼200 pachytene cells were categorized. (C) Condensin is required for the proper chromosome localization of Red1 and Hop1 at pachytene. Spread nuclei from cells arrested at pachytene by ndt80 (as in B) were processed for indirect immunofluorescence to detect Red1, Hop1, Ycs4-HA, and Rec8-HA. Both Red1 and Hop1 aggregate in condensin mutants.
Previous papers had shown that loading of Zip1 is dependent on Red1 and Hop1 which in turn are dependent on cohesin (Smith and Roeder, 1997; Klein et al., 1999). To position condensin in this hierarchy, we examined Rec8, Red1, and Hop1 localization in condensin mutants (Fig. 4 C). The chromosomal localization of Hop1 and Red1 was significantly altered in condensin mutants as evidenced by ∼2-fold reduction in the intensity of staining along the arms and the appearance of large polycomplex-like foci of Red1 and Hop1 (Fig. 4 C). In contrast, Rec8 staining was unaffected in condensin mutants (Fig. 3 A), and Ycs4-HA staining was unaffected by deletion of REC8 (not depicted). Together, these results suggest that cohesin and condensin load independently on meiotic chromosomes but are required for the proper loading of Red1 and Hop1, and subsequently Zip1.
The requirement for condensin in a meiotic specific process like SC assembly may reflect a meiosis-specific function for condensin. If so, then it should be possible to isolate meiosis-specific alleles of a condensin subunit. We identified such an allele called ycs4S (see Materials and methods). Vegetative cells with ycs4S as the only copy grow similarly to wild-type at all temperatures (unpublished data). Thus, this allele is competent for mitotic functions. In meiosis, ycs4S cells achieve longitudinal chromosome compaction of chromosome V (1.05 ± 0.05 μm, n = 14) that is very similar to wild-type. However, like condensin conditional alleles, the ycs4S allele fails to load Zip1 properly and forms polycomplex-like foci of Red1 and Hop1 (Fig. 4). Furthermore, the Red1 and Hop1 staining of the chromosome is reduced to background levels in ycs4S compared with only two- to threefold in the conditional alleles (Fig. 4 C) indicating an even more severe SC defect. From these data, we suggest that one function of condensin is required for the longitudinal compaction of both mitotic and meiotic chromosomes, and a second meiosis-specific function is required for proper recruitment of Red1 and Hop1 which helps to mediate chromosome individualization and SC assembly.
Condensin is required for efficient homologue pairing and proper DSB processing
The assembly of the SC results from a complex series of events, including pairing of homologues by a recombination-independent mechanism, formation of DSBs, presynaptic coalignment of homologues, and finally maturation of a subset of DSBs to crossovers with the concomitant nucleation of SC formation. A requirement for condensin in any one of these steps could explain the SC defect observed in condensin mutants. Therefore, we initiated studies to examine homologue pairing and the formation and processing of DSBs in condensin mutants.
Pairing at prophase I was assessed in both wild-type and mutant cells using a GFP-marked homologues of chromosome V (Fig. 5 A). This assay allows us to assess whether pairing has initiated but does not have the resolution to distinguish between recombination independent and later more intimate pairing. In more than 70% of wild-type cells, chromosome V homologues are paired after 6 h of sporulation at 34°C; whereas pairing in condensin mutant cells never exceeds 55% at any time. This reduced level of pairing in condensin mutants has several potential explanations, including a defect in the establishment of pairing, slower kinetics of pairing, poor culture synchrony, or an inability to maintain pairing. To begin to address these possibilities, we examined homologue pairing in condensin and other SC mutants that were arrested at pachytene by NDT80 inactivation. Both ycg1-2 ndt80 and ycs4-2 ndt80 cells achieve a level of pairing close to ndt80 cells (Fig. 5 B). By contrast, arrested hop1 ndt80 or red1 ndt80 cells show a significant lower pairing efficiency. The difference between condensin and red1 or hop1 mutants may reflect that the condensin conditional alleles only partially impede the chromosome association of Red1 and Hop1. Indeed, the ycs4S allele that shows a greater defect in the chromosome association of Red1 and Hop1 has a more severe pairing defect (Fig. 5 B). In summary, condensin is required for efficient homologue pairing.
Figure 5.
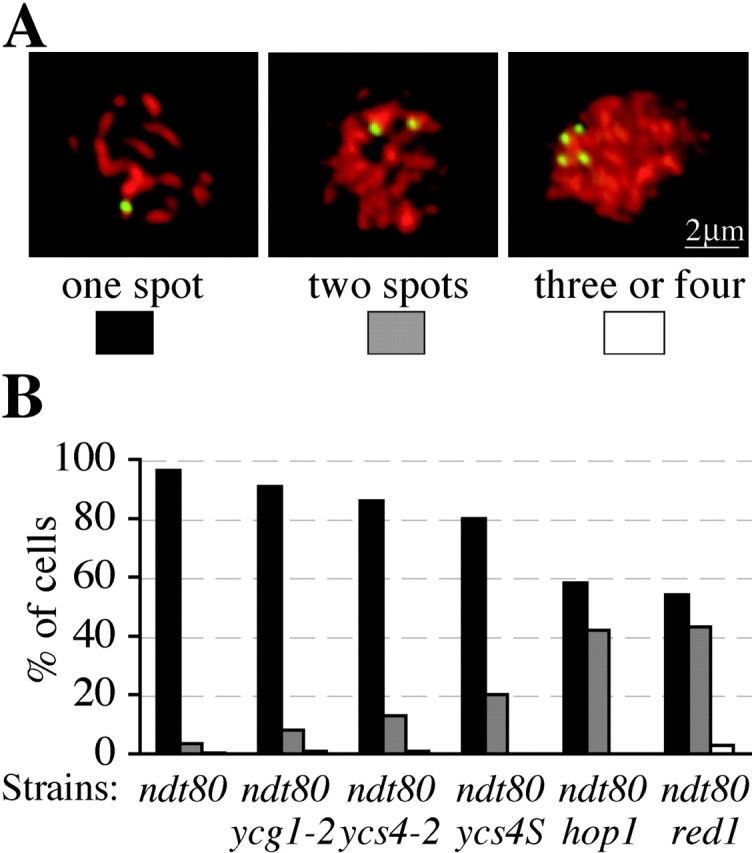
Condensin in homologue pairing. All strains shown were constructed with both homologues of chromosome V marked with GFP (Fig. 3). Spread nuclei were prepared and Rec8-HA and GFP were detected by indirect immunofluorescence. (A) Micrographs of spread nuclei at prophase I show examples with a single GFP spot (paired homologues), two GFP spots (unpaired homologues), and four spots (unpaired homologues and precociously separated sister chromatids). (B) Quantification of chromosome V pairing in cells arrested at pachytene by inactivation of NDT80. Strains harboring ndt80 (2880_), ycg1-2 ndt80 (2879),_ and ycs4-2 ndt80 (2881) were harvested after 12 h of sporulation at 34°C.
To assess the role of condensin in the formation DSBs, wild-type and condensin mutant cells were induced to sporulate and assayed for the presence of DSB at the recombination hotspot at the YCR047c/YCR048w locus (Fig. 6 A). Similar levels of DSBs are observed in wild-type (1.3%), ycs4-2 (1.1%), and ycs4S (0.7%). Although the appearance of DSBs is delayed ∼2 h in condensin mutants, this delay likely reflects slow progression to prophase (unpublished data). Furthermore, we monitored Rad51 foci as marker of DSB formation (Bishop, 1994). Consistent with previous reports, we observe ∼30 Rad51 foci in wild-type cells at prophase I (Fig. 6 B). A similar number of Rad51 foci are observed at prophase I in the ycs4S mutant (Fig. 6 B). These observations suggest that condensin is not essential for DSB production although the number or rate of formation of DSBs may be modestly affected.
Figure 6.
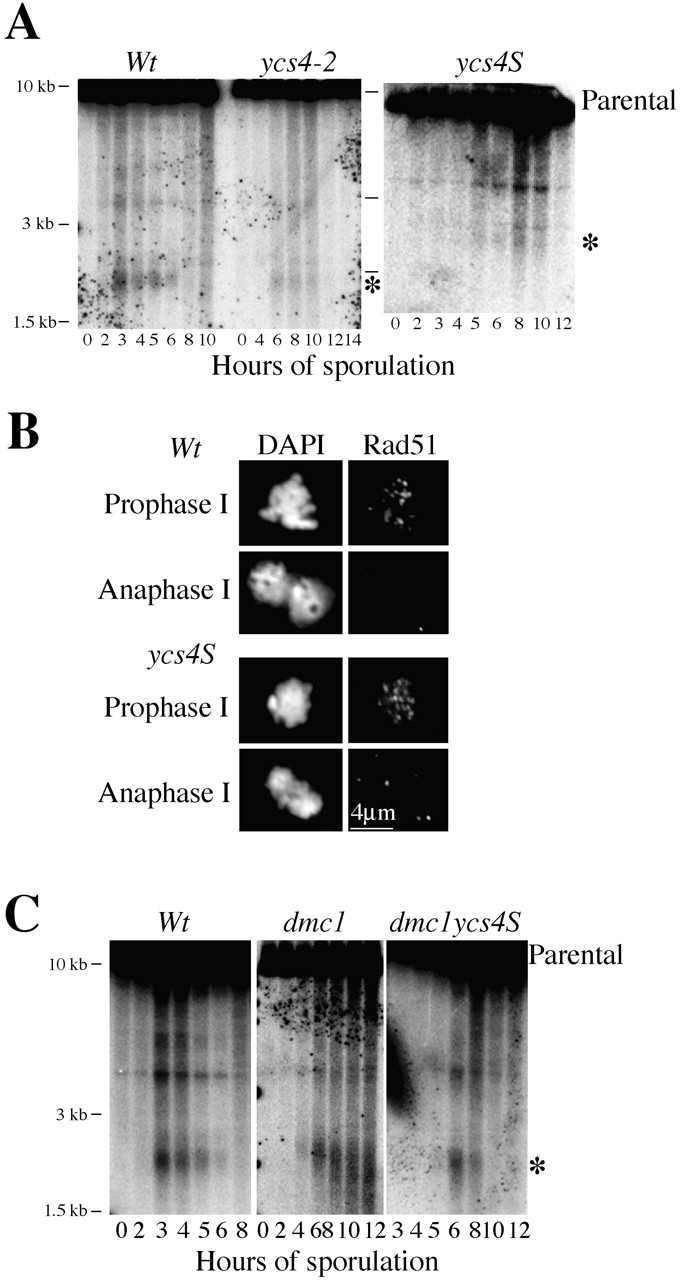
Analysis of condensin's role in meiotic DSB formation. (A) Genomic DNA was isolated from wild-type (2864), ycs4-2 (2975), and ycs4S (2922) strains at indicated time points, and processed to detect DSB at the YCR047c/YCR048w hot spot by Southern blot. The asterisks indicate the major DSB bands. (B) Spread nuclei from meiotic cultures of wild-type (2864) and ycs4S (2922) strains were processed for indirect immunofluorescence to detect chromosomal localization of Rad51. (C) As in A, genomic DNA was isolated from wild-type (2864), dmc1 (2924), and ycs4S dmc1 (2921) strains and processed to detect DSB at the YCR047c/YCR048w hot spot. Note that in dmc1 DSBs accumulate.
Furthermore, in condensin mutants DSBs appear to be processed as evidenced by the disappearance of Rad51 foci (Fig. 6 B) and the band corresponding to the DSB at the YCR047c/YCR048w locus (Fig. 6 A). Processing of DSBs occurs almost exclusively through a _DMC1_-dependent pathway, the major pathway for promoting recombination between homologues in meiosis (Roeder, 1997). Inactivation of DMC1 leads to the persistence of DSBs, and the activation of the meiotic recombination checkpoint which causes more than 98% of cells to arrest in prophase I (Fig. 6 C; Bishop et al., 1992). Similar phenotypes should be observed in a condensin dmc1 double mutant if DSBs in condensin mutants are processed solely by a _DMC1_-dependent pathway. However, the DSB at the YCR047c/YCR048w locus does not persist but rather disappears like the ycs4S single mutant (Fig. 6 C). Furthermore, 66% of ycs4S dmc1 cells fail to arrest and go on to sporulate. These data suggest that DSBs can be processed by a _DMC1_-independent mechanism in condensin mutants. Inactivation of the alternative processing in dmc1 ycs4S mutants should restore prophase arrest and/or prevent DSB processing. Interestingly, a dmc1 ycs4S rad54 triple mutant does undergo efficient prophase arrest (∼90%) but does not accumulate DSBs (unpublished data), indicating that another mechanism besides RAD54 also contributes to _DMC1_-independent DSB processing.
Condensin is required for meiosis I and II segregation and the resolution of recombination-dependent linkage between homologues
Our analyses of condensin had revealed meiotic functions that were likely to be critical for meiotic chromosome segregation. Therefore, we examined meiotic chromosome segregation in condensin mutants. At 34°C, less than 50% ycg1-2 and 30% ycs4-2 cells initiate meiotic I segregation. Furthermore, in these mutants, lagging chromosomes form a bridge between segregating chromosome masses during anaphase I and II. Eventually, condensin mutants, whether or not they attempt anaphase, form multiple DAPI-stained bodies and die (unpublished data). Cell death and nuclear fragmentation are observed in ycg1-2 and ycs4-2 but not ycs4S, red1, or hop1 mutants. The first two mutants are uniquely defective in axial compaction, suggesting that this defect is responsible for cell death and nuclear fragmentation.
Chromosome bridging in meiosis I is observed in the meiotic-specific ycs4S allele as well as in the condensin conditional mutants (Fig. 7 B), suggesting that this phenotype is a consequence of condensin's role in a meiotic process like meiotic recombination. To test this possibility, we examined bridging when the initiation of recombination was blocked by the elimination of SPO11 (Fig. 7 B). Indeed, anaphase chromosome bridging in ycs4-2 spo11 and ycs4S spo11 cells is reduced to levels similar to that of wild-type and spo11 cells (Fig. 7 B). The elimination of bridging by spo11 could result from a failure to form DSBs or to undergo recombination-independent homologue pairing (Weiner and Kleckner, 1994; Cha et al., 2000). To distinguish between these possibilities, we introduced the spo11-Y135F allele into condensin mutants (Fig. 7 B). This allele blocks only DSB formation but not recombination-independent homologue pairing (Cha et al., 2000). The spo11-Y135F allele is as competent as spo11 in reducing anaphase chromosome bridging in condensin mutants (Fig. 7 B). These observations suggest that the chromosome bridging in condensin mutants is the consequence of DSB induced recombination.
Figure 7.
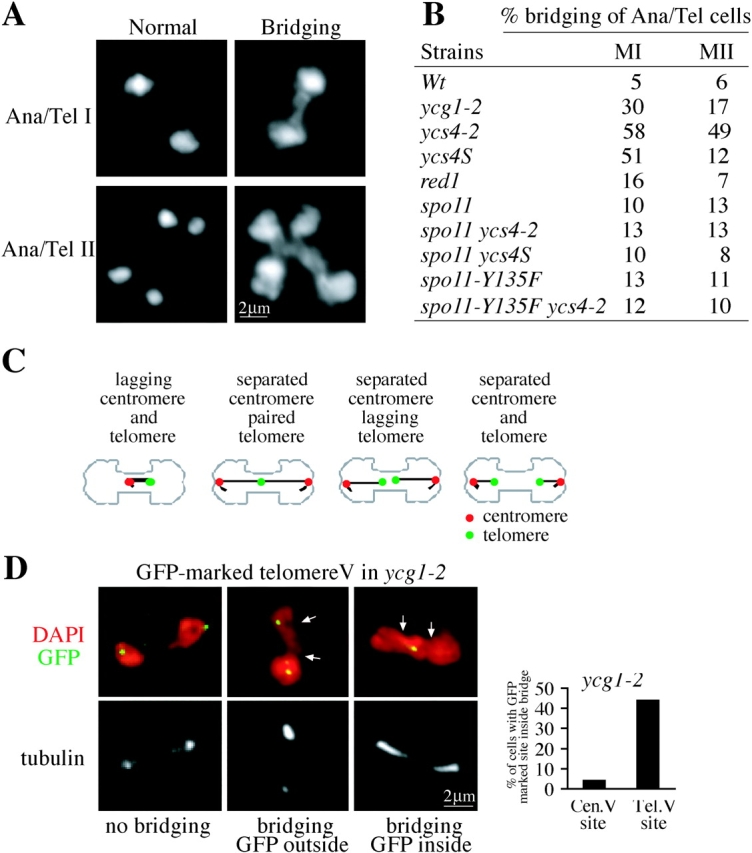
Condensin is required to resolve recombination-dependent chromosome linkages. (A) Chromosome bridging at anaphase I–telophase I and anaphase II–telophase II was detected by DAPI staining of spread nuclei (Materials and methods). (B) Quantification of chromosome bridging. Meiotic cultures of the listed strains were generated and assayed for chromosome bridging (as in A). In each strain at least 50 anaphase–telophase nuclei were scored for both meiosis I and II. (C) Illustration of potential centromere and telomere positions for chromosome V in bridged anaphase I cells. Four possible scenarios are shown. (D) Telomeres, but not centromeres, fail to resolve efficiently in condensin mutant ycg1-2. Nuclei were spread from ycg1-2 strains (2866 and 2888), containing centromere-proximal tetO (35 kb away from the centromere V; image not depicted) or telomere-proximal tetO (395 kb away form the centromere V). The arrows depict the bridging area. The position of GFP-marked centromere and telomere was assayed in nuclei with anaphase I chromosome bridges (graph).
A defect in recombination could cause chromosome bridging by several mechanisms including chromosome decondensation, lagging centromere movement to the poles, or failure to resolve recombination intermediates. To test these possibilities, we examined the position of the centromeres and telomeres of both chromosome V homologues in anaphase I nuclei that exhibit chromosome bridging (Fig. 7, C and D). In 96% of these cells centromeres are proximal to the spindle poles and outside of chromosome bridging area (Fig. 7 D), at a position expected for proper anaphase movement. In contrast, telomeres are frequently found in the bridging region (Fig. 7 D). Furthermore, 70% of cells with telomere trapped in the bridge show a single GFP-spot, indicating that links between homologues persists. This fact coupled with the spo11 results suggest that condensin is required to resolve recombination-dependent linkages between homologues. A significantly lower frequency of chromosome bridging is observed in red1 mutants (Fig. 7 B), and telomeres of chromosome V are not paired in the bridging region. These differences in quantity and quality of chromosome bridging suggests that the role of condensin in resolving homologue linkages is a distinct function from its role in Red1 and Hop1 loading. Finally, reciprocal crossovers are recovered among the viable spores of ycs4S suggesting that some recombination-dependent linkages between homologues can be resolved independently of condensin.
Discussion
Condensin function in meiotic chromosome compaction of budding yeast: shared and distinct features from mitotic compaction
In condensin mutants of budding yeast, the length of meiotic chromosomes is increased ∼50% and gross chromosome missegregation ensues (this paper). Condensin mutants cause similar changes in the length and segregation of mitotic chromosomes of budding yeast (Strunnikov et al., 1995; Lavoie et al., 2002). These phenotypic similarities strongly suggest that at least one activity of condensin is shared between mitosis and meiosis and this activity plays a common role in chromosome organization.
However, our experiments of condensin function in meiotic compaction revealed two features not observed from previous mitotic experiments of budding yeast. First, a highly organized albeit longer chromosome axis persists in meiotic cells severely compromised for condensin function, suggesting the existence of a condensin-independent pathway for axis formation. Second, we show that the individualization of meiotic chromosomes depends on the collaboration of condensin and meiosis-specific auxiliary proteins, such as Red1 and Hop1. Interestingly, meiotic chromosomes in budding yeast are better individualized and more compacted than their mitotic counterparts. In fact, compared with their mitotic counterparts, meiotic chromosomes in budding yeast are much more reminiscent of the meiotic and mitotic chromosomes in higher eukaryotes. One intriguing possibility is that alternative pathways for axis formation and auxiliary proteins for individualization are active in meiotic but not mitotic divisions of budding yeast, whereas they are active in both cell divisions of most other eukaryotic organisms. Recently several analyses have suggested the existence of condensin-independent pathways for mitotic condensation in these organisms (Bhat et al., 1996; Steffensen et al., 2001; Hagstrom et al., 2002; Kaitna et al., 2002). Thus, the study of condensin-independent axis formation and chromosome individualization during meiosis of budding yeast may prove a very powerful model for chromosome organization in both mitosis and meiosis of other organisms.
Meiotic functions of condensin in the structure and function of SC and resolution of recombination dependent chromosome linkages
Here, we show that condensin has a meiotic specific function that is essential for the proper assembly of the SC including axial–lateral (Red1 and Hop1) and central (Zip1) elements. It also promotes processing of DSBs by the DMC1 pathway by inhibiting alternative processing. These observations are consistent with previous analyses of SC assembly and function. Red1 and Hop1 are required for Zip1 chromosome localization and are also required to repress _RAD54_-dependent processing of DSBs (Smith and Roeder, 1997; Bishop et al., 1999).
Condensin could function in SC assembly at one of several steps given the complex prerequisites for SC assembly, including chromosome pairing, DSB formation and maturation, or loading of SC components. Condensin mutants do have detectable defects in pairing, DSB formation, and processing and these could account for the defect in SC assembly. However, these defects are relatively mild, and other mutants with similar mild defects can form SC.
We prefer an alternative structural model in which condensin organizes chromosome structure setting up a platform for the recruitment of SC components or assembly factors. In support of this model, the proper localization of these same SC components also requires cohesin (Klein et al., 1999), which has been shown to act coordinately with condensin to organize mitotic chromosomes in budding yeast (Guacci et al., 1997; Hartman et al., 2000; Lavoie et al., 2002). Indeed, precedent exists for the assembly of specialized chromosome structures using housekeeping chromosomal proteins as a platform. For example, the origin recognition complex not only directs the initiation of DNA replication but also serves to bind and therefore recruit factors necessary to build silent chromatin (Bell, 2002). To test whether condensin has an analogous recruitment function in SC assembly, we will need to gain a better understanding of the molecular basis for condensin dependence for the chromosomal localization of Red1 and Hop1.
A second meiosis-specific function of condensin in budding yeast is the resolution of _SPO11_-induced linkage between homologues (this paper). This resolution function of condensin is likely to be conserved in other eukaryotes because chromosome bridges in anaphase I have recently been observed in condensin mutants of Arabidopsis thaliana (Siddiqui et al., 2003). A role for condensin in meiotic recombination is also consistent with several previous observations in mitotic cells. Condensin is required for DNA damage repair in interphase in Schizosaccharomyces pombe (Aono et al., 2002) and can change the topology of DNA (Kimura and Hirano, 1997). This activity could be used for resolution of recombination intermediates. Alternatively, condensin could function in the resolution of protein-based homologue linkage, for example promoting the disassembly of chiasmata.
Novel functions for condensin in meiosis I, are they confined to meiotic cells?
Condensin regulates meiotic recombination by promoting the assembly of the SC and repressing alternative pathways for DSB processing. By analogy, during mitosis condensin could recruit chromosomal components to suppress sister chromatid exchange. In this way, sister chromatid exchange would be available to repair damage before M-phase but rendered inactive during M-phase to ensure that sister chromatid separation is not impeded in anaphase. Indeed, we have found that the ycs4-2 mutant stimulates sister chromatid exchange 50–100-fold in the tandem repeats of the rDNA (unpublished data). Therefore, condensin may well play a role in the repression of certain types of recombination in mitotic cells.
In addition, it will be interesting to reevaluate the mitotic function of condensin in light of its meiotic role in resolving recombination-dependent linkages between homologues. Bridging between sister chromatids in mitotic anaphase is the most shared phenotype of condensin inactivation in all organisms. It has been suggested that these sister chromatid bridges may be caused by catenation during DNA replication which are subsequently resolved by the compaction activity of condensin (Bhat et al., 1996; Koshland and Strunnikov, 1996; Steffensen et al., 2001). However, our results suggest that in meiosis I, linkages between homologues are generated by a recombination-dependent pathway. Thus, our experiments in meiosis raise the possibility that at least some mitotic linkages between sister chromatids may arise through recombination. It will be important to reassess the cause of mitotic linkages between sister chromatids and the function of condensin responsible for their resolution. Such a study may reveal a novel but universally conserved function of condensin in all mitotic cells.
Materials and methods
Yeast strains
Yeast strains used in this paper are congenic to SK1 (Table SI, available at http://www.jcb.org/cgi/content/full/jcb200308027/DC1). Condensin mutant alleles, smc2-8 (Freeman et al., 2000), ycg1-2, and ycs4-2 (Lavoie et al., 2002), were backcrossed to SK1 (strains RKY1145 and S2683) for more than eight times. SK1 strains with the following alleles: ndt80::KanMX4, hop1::LEU2, red1::LEU2, dmc1::KanMX4, and spo11-Y135F were constructed either previously (de los Santos and Hollingsworth, 1999; Cha et al., 2000) or by PCR-based gene knockout (Schneider et al., 1995). For epitope tagging of condensin subunits Ycs4 and Smc2, 3XHA and 8XMyc were incorporated in-frame at the COOH termini of the YCS4 and SMC2 genes, respectively, by PCR-based tagging strategy (Schneider et al., 1995). The Rec8–3HA strain was obtained from F. Klein (University of Vienna, Vienna, Austria; Klein et al., 1999). The ycs4S allele was generated by the addition of extra sequence at its COOH terminus, which includes 12Xmyc and linkers. Marking of chromosome V with GFP at the centromere-proximal locus and telomere-proximal locus was performed as described previously (Michaelis et al., 1997). Diploids homozygous for different combination of these markers and epitope tags (Table SI) were constructed by standard methods (Guthrie and Fink, 1991).
Culture methods
To induce synchronous sporulation, we adopted a method reported previously (Cha et al., 2000). In brief, a single colony was inoculated in 5 ml YEPD overnight at 30°C. This culture was diluted in YEPA medium at desired volume to reach OD (λ = 600) of 0.1–0.2. When the YEPA culture reached OD of 1.2–1.4 (∼12 h of incubation with vigorous shaking at 30°C), cells were harvested by centrifugation. Yeast cells were washed once in prewarmed water, and resuspended in 2% potassium acetate to induce sporulation at 30°C. For conditional mutants (ycg1-2, ycs4-2, and smc2-8) and their wild-type controls, vegetative growth was at 23°C (permissive). Cultures were grown in sporulation media at 23°C for 1.5 h and shifted to 34°C (nonpermissive temperature) to inactivate condensin. Time 0 of sporulation is defined as the point when yeast cultures were switched to sporulation medium. Sporulation efficiency (sporulated cells/total cells) was evaluated by phase-contrast microscopy after 24 h of sporulation. At least 500 total cells were counted for each strain. To monitor nuclear division during sporulation, aliquots of cells were withdrawn at indicated time intervals and fixed with 4% formaldehyde for 1 h at RT. Fixed cells were stained by DAPI and visualized under a fluorescence microscope.
Spread nuclei and immunofluorescence microscopy
Chromosome surface-spread was modified from a previous method (Dresser and Giroux, 1988; Engebrecht and Roeder, 1990). Cells at desired stages were harvested by centrifugation, and spheroplasted at 30°C with 10 μg/ml oxyliticase for ∼20 min. For conditional mutants and their wild-type controls, cells were spheroplasted at 34°C for ∼15 min. Nuclei spreads were fixed by 4% PFA with 0.25% Triton X-100 in PHEM buffer (60 mM Pipes, 25 mM Hepes, pH 6.95, 10 mM EGTA, and 4 mM MgCl2). Triton X-100 was omitted in nuclei spreads for detecting chromosome bridging. In the presence of detergent, nuclei/chromosomes spread greatly, potentially obscuring bridges. Indirect immunofluorescence of spread nuclei was performed as described previously (Sym et al., 1993). Primary antibodies used in this work—rabbit anti-Zip1, anti-Red1 (obtained from S. Roeder, Yale University, New Haven, CT), anti-Hop1 (obtained from N. Hollingsworth, SUNY Stony Brook, Stony Brook, NY), and anti-Rad51 (obtained from D. Bishop, University of Chicago, Chicago, IL)—were used at 1:100–200 dilution. Monoclonal anti-HA (12CA5; Roche) and anti-Myc (9E10; Roche) were used at 1–2 μg/ml. A polyclonal GFP antibody (CLONTECH Laboratories) was used as a primary antibody at 1:1,000 dilution to localize the GFP spots. An FITC-conjugated goat anti–rabbit antibody (Jackson ImmunoResearch Laboratories) and a rhodamine-conjugated goat anti–mouse antibody (Jackson ImmunoResearch Laboratories) were used as secondary antibodies at 1:200 dilution. Chromosomal DNA was counter-stained by DAPI. All images were acquired with an Axioplan 2 microscope (100× objectives, NA = 1.30; Carl Zeiss MicroImaging, Inc.) equipped with a cooled CCD camera. Images were processed with IP-Lab (Scanalytics) for contrast adjustment and pseudo-coloring.
Determining chromosome V length
Spread nuclei were prepared from strains with GFP marked chromosome V and Rec8-HA and processed for indirect immunofluorescence. The axial length of chromosome V was determined by tracking the Rec8-stained chromosome axis with the analyze tool supplied by IP-Lab. Chromosome V length was measured in spread nuclei only when its ends were clearly separated from the other chromosomes.
Chromosome bridging
Spread nuclei were prepared from meiotic cultures. Anaphase–telophase cells were identified by separated chromosome masses and tubulin staining (Lavoie et al., 2002), which revealed long or partially disassembled spindles. In spreads of anaphase–telophase nuclei, bridging is defined as a thick DAPI-stained band connecting the separating chromosome masses (Fig. 7). Occasionally, there was a very thin DAPI-stained thread connecting separated chromosomes (including in wild type), these cells were categorized as normal.
Immunoprecipitation and Western blots
Synchronous meiotic cultures were prepared as described in Culture methods. Protein extracts from these cultures and immunoprecipitation were performed as described previously (Freeman et al., 2000). Anti-HA (12CA5) or anti-Myc (9E10) antibodies were used at 10 μg/ml final concentration. Standard procedures for SDS-PAGE and blotting were followed (Sambrook et al., 1989). Primary anti-HA (12CA5) and anti-Myc (9E10) antibodies were used at 1–5 μg/ml. Binding of primary antibody to the blot was detected using an ECL kit (PerkinElmer). To determine sample loading, we used a rabbit anti yeast β-tubulin antibody (1:2,000 dilution) to detect the β-tubulin level on the same blot.
Physical analysis of DSBs
20 ml of synchronous meiotic culture was harvested at specified times after induction of sporulation. Genomic DNA was isolated as described by Woltering et al. (2000). Genomic DNA was digested with AseI at 37°C overnight before loaded onto a 1.0% agarose gel. Southern analysis was used to detect DSB fragments at the YCR047c/YCR048w locus (Smith et al., 2001).
Online supplemental material
Yeast strains used in this study are listed in Table S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200308027/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We are grateful to Nancy Hollingsworth, Shirleen Roeder, Doug Bishop, Franz Klein, Kim Nasmyth (Research Institute of Molecular Pathology, Vienna, Austria), and Nancy Kleckner (Harvard University, Cambridge, MA) for providing yeast strains and reagents. We also thank Nancy Kleckner, Nancy Hollingsworth, Agelika Amon, Judith Yanowitz, Yixian Zheng, and members of the Koshland laboratory for comments of the manuscript.
The online version of this article contains supplemental material.
Abbreviations used in this paper: DSB, double strand break; SC, synaptonemal complex.
References
- Aono, N., T. Sutani, T. Tomonaga, S. Mochida, and M. Yanagida. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 417:197–202. [DOI] [PubMed] [Google Scholar]
- Bell, S.P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659–672. [DOI] [PubMed] [Google Scholar]
- Bhalla, N., S. Biggins, and A.W. Murrray. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 13:632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, M.A., A.V. Philip, D.M. Glover, and H.J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 87:1103–1114. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K. 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 79:1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 69:439–456. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., Y. Nikolski, J. Oshiro, J. Chon, M. Shinohara, and X. Chen. 1999. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposed an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 4:425–443. [DOI] [PubMed] [Google Scholar]
- Cha, R.S., B.M. Weiner, S. Keeney, J. Dekker, and N. Kleckner. 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14:493–503. [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., and N.M. Hollingsworth. 1999. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J. Biol. Chem. 274:1783–1790. [DOI] [PubMed] [Google Scholar]
- Dong, H., and G.S. Roeder. 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser, M.E., and C.N. Giroux. 1988. Meiotic chromosome behavior in spread preparations of yeast. J. Cell Biol. 106:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht, J., and G.S. Roeder. 1990. MER1, a yeast gene required for chromosome pairing and genetic recombination is induced in meiosis. Mol. Cell. Biol. 10:2379–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 91:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods in Enzymology. Vol. 194. Academic Press, Inc., New York. 3–149. [PubMed]
- Hagstrom, K.A., V.F. Holmes, N.R. Cozzarelli, and B.J. Meyer. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, T., K. Stead, D. Koshland, and V. Guacci. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399–414. [DOI] [PubMed] [Google Scholar]
- Hirano, T., and T.J. Mitchison. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 79:449–458. [DOI] [PubMed] [Google Scholar]
- Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 89:511–521. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., P. Pasierbek, M. Jantsch, J. Loidl, and M. Glotzer. 2002. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798–812. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C.N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 88:375–384. [DOI] [PubMed] [Google Scholar]
- Kimura, K., and T. Hirano. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 90:625–634. [DOI] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S.B. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98:91–103. [DOI] [PubMed] [Google Scholar]
- Koshland, D., and A. Strunnikov. 1996. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 12:305–333. [DOI] [PubMed] [Google Scholar]
- Lavoie, B.D., K.M. Tuffo, S. Oh, D. Koshland, and C. Holm. 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell. 11:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, B.D., E. Hogan, and D. Koshland. 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Ouspenski, I., O.A. Cabello, and B.R. Brinkley. 2000. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell. 11:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600–2621. [DOI] [PubMed] [Google Scholar]
- Saka, Y., T. Sutani, Y. Yamashita, S. Saitoh, M. Takeuchi, Y. Nakaseko, and M. Yanagida. 1994. Fission yeast cut3 and cut14, members of the ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13:4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. SDS-PAGE. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 18.47–18.75.
- Schneider, B.L., W. Seufert, B. Steiner, Q.H. Yang, and A.B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 11:1265–1274. [DOI] [PubMed] [Google Scholar]
- Siddiqui, N., P.E. Stronghill, R.E. Dengler, C.D. Hasenkampf, and C.D. Riggs. 2003. Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development. 130:3283–3295. [DOI] [PubMed] [Google Scholar]
- Smith, A.V., and G.S. Roeder. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.N., A. Penkner, K. Ohta, F. Klein, and A. Nicolas. 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 11:88–97. [DOI] [PubMed] [Google Scholar]
- Stack, S.M., and L.K. Anderson. 2001. A model for chromosome structure during the mitotic and meiotic cell cycles. Chromosome Res. 9:175–198. [DOI] [PubMed] [Google Scholar]
- Steffensen, S., P.A. Coelho, N. Cobbe, S. Vass, M. Costa, B. Hassan, S.N. Prokopenko, H.J. Bellen, M.M.S. Heck, and C.E. Sunkel. 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11:295–307. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A.V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587–599. [DOI] [PubMed] [Google Scholar]
- Sym, M., and G.S. Roeder. 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym, M., J.A. Engebrecht, and G.S. Roeder. 1993. ZIP 1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 72:365–378. [DOI] [PubMed] [Google Scholar]
- Watrin, E., F. Cubizolles, H.B. Osborne, K.L. Guellec, and V. Legagneux. 2003. Expression and functional dynamics of the XCAP-D2 condensin subunit in Xenopus laevis oocytes. J. Biol. Chem. 278:25708–25715. [DOI] [PubMed] [Google Scholar]
- Weiner, B.M., and N. Kleckner. 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 77:977–991. [DOI] [PubMed] [Google Scholar]
- Woltering, D., B. Baumgartner, S. Bagchi, B. Larkin, J. Loidl, T. de los Santos, and N.M. Hollingsworth. 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 20:6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner. 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]