Pml Is Critical for Nd10 Formation and Recruits the Pml-Interacting Protein Daxx to This Nuclear Structure When Modified by Sumo-1 (original) (raw)
Abstract
Nuclear domain 10 (ND10), also referred to as nuclear bodies, are discrete interchromosomal accumulations of several proteins including promyelocytic leukemia protein (PML) and Sp100. In this study, we investigated the mechanism of ND10 assembly by identifying proteins that are essential for this process using cells lines that lack individual ND10-associated proteins. We identified the adapter protein Daxx and BML, the RecQ helicase missing in Bloom syndrome, as new ND10-associated proteins. PML, but not BLM or Sp100, was found to be responsible for the proper localization of all other ND10-associated proteins since they are dispersed in PML−/− cells. Introducing PML into this cell line by transient expression or fusion with PML-producing cells recruited ND10-associated proteins into de novo formed ND10 attesting to PMLs essential nature in ND10 formation. In the absence of PML, Daxx is highly enriched in condensed chromatin. Its recruitment to ND10 from condensed chromatin requires a small ubiquitin-related modifier (SUMO-1) modification of PML and reflects the interaction between the COOH-terminal domain of Daxx and PML. The segregation of Daxx from condensed chromatin in the absence of PML to ND10 by increased accumulation of SUMO-1–modified PML suggests the presence of a variable equilibrium between these two nuclear sites. Our findings identify the basic requirements for ND10 formation and suggest a dynamic mechanism for protein recruitment to these nuclear domains controlled by the SUMO-1 modification state of PML.
Keywords: nuclear structure, nuclear proteins, protein interaction, supramolecular regulation, knockout cells
Specific nuclear domains, ND101 (also referred to as PML nuclear bodies or promyelocytic oncogenic domain), but more specifically their constituent proteins have been intensively investigated, following the findings that some ND10-associated proteins are connected to certain disease processes. One of the proteins consistently present in ND10 is Sp100, autoantibodies to which are prevalent in primary billiary cirrhosis (Szostecki et al. 1990). Another protein in ND10 is the promyelocytic leukemia protein (PML), which is fused to the retinoic acid receptor α in acute promyelocytic leukemia (APL) (deThe et al., 1990; Goddard et al. 1991; Kakizuka et al. 1991; Pandolfi et al. 1991). The presence of wild-type PML in ND10 and the dispersion of this structure in APL suggested the involvement of these domains in the differentiation of promyelocytes (Dyck et al. 1994; Weis et al. 1994). Also, PML has recently become the center of attention in other contexts. It was identified as a tumor suppressor protein (Mu et al. 1994), involved in apoptosis (Quignon et al. 1998; Wang et al. 1998), a regulator of MHC expression (Zheng et al. 1998) and, together with Sp100, to be upregulated by interferon (Lavau et al. 1995; Grotzinger et al. 1996). Disparate information about ND10 as a nuclear structure is surreptitiously accumulating from several investigative directions. A concentrated approach to solve the question of how such a prominent nuclear domain is formed and maintained has been lacking. We focus here on how ND10 are assembled and how its formation and maintenance are controlled within the conceptual background that defines ND10 as a potential nuclear depot where certain proteins are regulated through recruitment and release.
ND10 are structurally defined by the immunohistochemical localization of certain proteins at specific punctate nuclear sites. New ND10-associated proteins are, therefore, often found accidentally when such patterns are discovered while working on a given protein in other contexts. Such was the case with two proteins that will feature in the present attempt to evaluate ND10 structure. One of them, Daxx, has been identified as a protein that binds with its COOH-terminal end to the Fas death domain (Yang et al. 1997), the centromeric protein CENP-C (Pluta et al. 1998), the transcription factor Pax3 (Hollenbach et al. 1999), and DNA methyltransferase I (Michaelson et al. 1999). None of these proteins are located predominantly at ND10. Daxx also has been found to interact with the steroidogenic acute regulatory protein gene promoter DNA sequence (Kiriakidou et al. 1997), and antibodies against Daxx showed a nuclear distribution like ND10. Therefore, it was further investigated in the context of ND10 structure. Similar to Daxx, BLM, the RecQ helicase missing in the Bloom syndrome patients, had a punctate distribution in most cells (Neff et al. 1999), which in the course of this investigation proved to be ND10. Therefore, BLM−/− fibroblasts could be used to test what effect lack of BLM has on the structure of ND10.
The stability, function, and localization of proteins can be changed by posttranscriptional modification. At least four ND10 proteins, Sp100, PML, CBP, and pRB, are phosphoproteins, and PML and Sp100 are covalently modified by the small ubiquitin-related modifier (SUMO-1) (Sternsdorf et al. 1997; Kamitani et al. 1998b; Muller et al. 1998). SUMO-1 modification of PML has been suggested to target this protein to ND10 (Muller et al. 1998), and removal of sites for SUMO-1 modification from PML seems to prevent PML from accumulating in ND10 (Kamitani et al. 1998a). In contrast, such a modification did not influence the localization pattern for Sp100 (Sternsdorf et al. 1997). Therefore, modification of PML as a determinant of its interaction with other proteins suggests itself as a potential control mechanism in the accumulation of proteins at ND10. The effect of SUMO-1 modification or the lack of such modification may be central to the structural integrity of ND10 and needs to be evaluated.
A remarkable feature of ND10 is the deposition of DNA viruses, such as herpes simplex virus type 1, cytomegalovirus, adenovirus 5, and SV40, at ND10 and that their transcription and replication begins associated with these sites (Ishov and Maul 1996; Maul et al. 1996; Ishov et al. 1997). Except for SV40, these viruses express proteins that modify ND10. Specifically, ICP0 of herpes simplex virus type 1 accumulates in ND10 and induces degradation of ND10-associated proteins (Maul et al. 1993; Everett and Maul 1994; Maul and Everett 1994; Everett et al. 1998, Everett et al. 1999; Chelbi-Alix and de The 1999). The degradation of PML involves desumofication followed by hydrolysis through the ubiquitin/proteosome pathway (Everett et al. 1998, Everett et al. 1999). This finding implicated desumofication and/or hydrolysis of PML in the destruction of ND10, although the advantage to the virus is as yet not apparent.
ND10 are modified by other effectors of nuclear functions. Stress in the form of heat shock or heavy metal exposure results in dispersion of ND10-associated proteins (Maul et al. 1995). Increases in the number and size of ND10 have been reported to be due to the interferon-induced upregulation of PML and Sp100 (Lavau et al. 1995; Grotzinger et al. 1996). Also, the earliest observed changes in ND10 were the size variations associated with hormonal changes in the uterine endometrium and the mammary epithelial cells (Padykula et al. 1981; Fitzgerald and Padykula 1983). Common to all the agents that induce modifications in ND10 are their global effects on the nuclear metabolism, although the signaling pathway that affects the changes at ND10 is wholly unexplored and can only be approached after we have knowledge on the recruitment and release mechanisms at this nuclear site.
ND10s are dispersed upon viral infection and in APL and are reassembled upon treatment of this disease by retinoic acid and arsenic (As2O3). This strongly suggests that this nuclear structure is important for a number of pathological processes. Therefore, investigating the assembly mechanism is an essential step toward understanding ND10 function in disease processes as well as in normal nuclear activities. Because the mechanism of ND10 assembly is likely to involve protein–protein interactions, we carried out studies to identify proteins that are essential for the formation of these domains. During the course of this study, we found the following: (1) PML, but not Sp100 nor the new ND10 component BLM, is essential for ND10 assembly; (2) the recently characterized Daxx (Kiriakidou et al. 1997; Yang et al. 1997; Pluta et al. 1998; Hollenbach et al. 1999; Michaelson et al. 1999) is a new ND10-associated protein and interacts with PML; (3) this interaction is necessary to recruit Daxx from heterochromatin to ND10; and (4) this recruitment is controlled by SUMO-1 modification of PML. Taken together, these results provide us with the basics of ND10 formation and suggest a mechanism for its dynamic control.
Materials and Methods
Cells and Growth Conditions
HEp-2 carcinoma human primary fibroblasts (HF) have been previously described (Ishov et al. 1997). Fibroblasts from Bloom syndrome patient (GM01492F) were obtained from the Coriell Institute for Medical Research; mouse primary embryonic fibroblasts (MPEF) and PML−/− mouse primary embryonic fibroblasts (PML−/− MPEF; Wang et al. 1998), N-myc-amplified neuroblastoma NGP cells (Zehnbauer et al. 1988), were maintained in DME supplemented with 10% FCS and antibiotics. Human embryonic NT2 cells and the retinoic acid–differentiated NT2 cells were provided by V. Lee (University of Pennsylvania) (Kleppner et al. 1995). All cells were grown at 37°C in a humidified 5% CO2 atmosphere. For immunohistochemical staining, cells were grown on round coverslips in 24-well plates (Corning Glass, Inc.) until ∼80% confluent before fixation. For cell fusion experiments, both cell types were plated on glass coverslips at a 1:1 ratio. The following day, they were treated with prewarmed 50% PEG-6000 (Serva Co.) for 2 min and washed three times with complete culture medium. Immunostaining was performed 16 h after fusion. Unfused cells in the PEG-6000–treated cells and unmixed fused cells were used as negative controls. During microscopic analysis, DNA staining (0.5 μg/ml Hoechst 33258; American Hoechst) was used to identify the nuclei of mouse and human cells.
Antibodies
ND10 were visualized using the following antibodies: mAb 138 labels NDP55 (Ascoli and Maul 1991), whereas mAb 5E10 reacts with PML (Stuurman et al. 1992). Isotype-matched mAbs of unrelated specificity were used as controls. Polyclonal rabbit antiserum against PML and rabbit antibodies against Sp100 were obtained from Dr. J. Frey (Korioth et al. 1996). A human antiserum specific for Sp100 from patients with primary billiary cirrhosis was found positive for mouse ND10. Human serum 602 recognized the centromeres in immunohistochemistry and the centromeric recombinant protein CENP-C in Western blots (Jarzabek-Chorzelska et al. 1990). Rabbit antibodies against human Daxx were produced using recombinant RGS-His-tagged human Daxx purified on NTX resin (Qiagen). Antibodies against RGS-HIS were purchased from Qiagen. A different antibody against hDaxx was produced in mouse against the hDaxx COOH-terminal 480–740 amino acid fragment. In addition, a rabbit anti–Daxx antibody was purchased from Santa Cruz Biotechnology, Inc. Antibodies against SUMO-1 were obtained from Dr. P. Freemont (I.C.R.F. London) (Boddy et al. 1996). Anti-CBP, antibodies were purchased from Santa Cruz Biotechnology, Inc., and those against RGS-His from Qiagen (Valencia, CA). Rabbit anti-BLM has been described (Neff et al. 1999).
Immunolocalization of Proteins
2 d after plating on round glass coverslips, cells were fixed at room temperature for 15 min with freshly prepared 1% paraformaldehyde in PBS and treated as previously described (Ishov and Maul 1996). Cells were analyzed using a Leica confocal laser scanning microscope. Leica image enhancement software was used to balance signal strength and eightfold scanning was used to separate signal from noise. Because of the variability among cells in any given culture, the most prevalent cells were photographed and are presented as small groups of nuclei or single nuclear images at high magnification.
Yeast Two-Hybrid Protein Interaction Assay
The interaction between Daxx and PML or Sp100 was measured in a yeast two-hybrid assay using β-galactosidase activity as a reporter of protein–protein interaction. Sp100 and PML were cloned in the pAS1 vector that contains the TRP1 gene (Durfee et al. 1993), so that these proteins are fused to GAL4-DBD (a gift from R. Evans, Salk Institut). The pAS1-PMLΔ coil deletion mutant was generated by digestion of pAS1-PML with endonuclease Bss HII and self-ligation, which resulted in a deletion of 214–329 amino acids of the PML protein. hDaxx cDNA and the respective mutants were produced by PCR using the high fidelity Vent polymerase (New England Biolabs) and pQE-30 hDaxx (Kiriakidou et al. 1997) as a template and cloned into the BamHI site downstream of NLS-VP16 driven by the ADH promoter into pVP16 (Vojtek et al. 1993), which carries the LEU2 gene. pSD5 (a gift from S. Berger, The Wistar Institute) containing the HIS2 gene and GAL4 binding site upstream of the bacterial lacZ gene was used as a reporter plasmid. Plasmids were transfected into the trp1 derivative of yeast strain PSY316 (MATα his 3-200 leu 2-3, 112 lys 2 ura 3-53) (Candau et al. 1996). β-galactosidase activity was assessed and normalized to protein concentration as described (Rose et al., 1998). The data represent results of three independent experiments.
Probes and Expression Plasmids
To analyze the intranuclear distribution of various Daxx mutants, we first constructed a pET plasmid, encoding GFP with a nuclear localization signal from SV40 large T-antigen (amino acid PKKKRKV). Two synthetic oligonucleotides (5′-AATTCTCCTAAGAAGAAGCGTAAGG-3′ and 5′-TCGACCTTACGCTTCTTCTTAGGAG-3′) were annealed and inserted into the COOH-terminal end of the GFP open reading frame between the EcoRI and SalI sites on pEGFP-C1 (CLONTECH Laboratories). Different deletion mutants of hDaxx were constructed by subcloning into BamHI cut pET vector. The respective fragments of BamHI cut PCR products were amplified with the sense primer 5′-CACACGGATCCGCCACCGCTAACAGC-3′ and the antisense primers: 5′-GTGGTGGATCCCTCCTCTGATTGCTTCCTGG-3′ for Daxx 1–595 amino acids; 5′-GTGGTGGATCCATCAGAGTCTGAGAGCACGATG-3′ for Daxx 1–740 amino acids; or BglII-BamHI cut PCR fragment amplified with 5′-CACACAGATCTGATTCTGGTCCCCCCTGC-3′ as the sense and antisense primers 5′-GTGGTGGATCCATCAGAGTCTGAGAGCACGATG-3′ for Daxx 624–740 amino acids using high fidelity Vent polymerase (New England Biolabs) and pQE-30hDaxx (Kiriakidou et al. 1997) as a template for the Daxx gene. All constructs were verified by sequencing. Plasmids PML-K65,160,490R (referred to here as PMLΔSUMO), PML-K65R, and PML-K65,160R (Kamitani et al. 1998a), based on the pcDNA3 plasmid (Invitrogen), expresses RGS-His-fused PML mutants with corresponding lysines substituted by arginines. Transient transfections were carried out using the DOSPER reagent (Boehringer) according to the manufacturer's recommendations.
Results
Identification of the Protein Essential for the Maintenance of ND10 by Using Cell Lines Lacking Specific ND10-associated Proteins
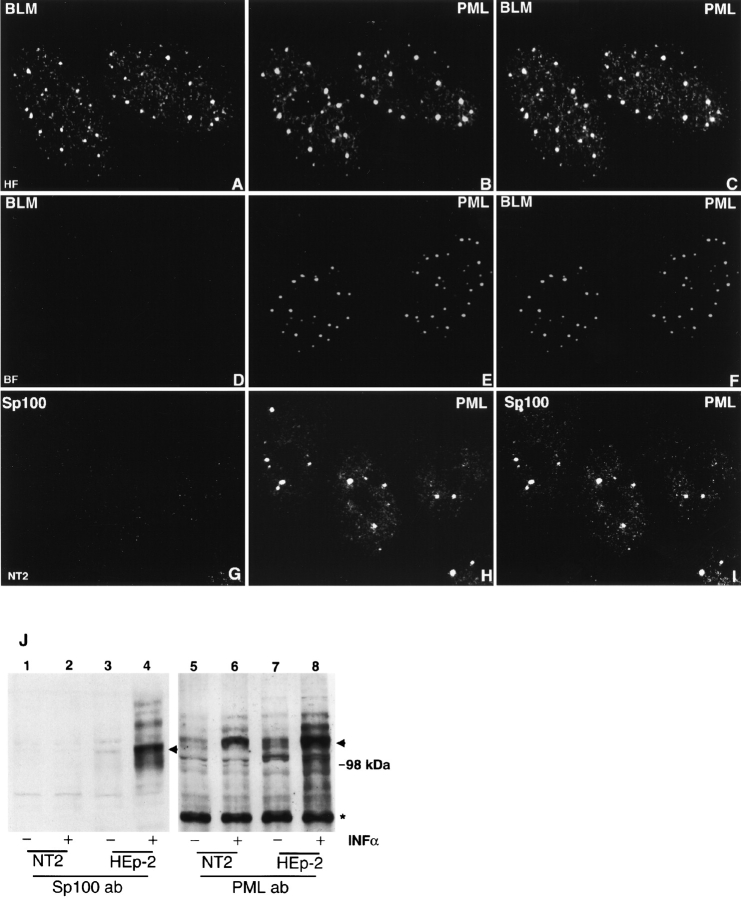
To address the question, which proteins are essential for the formation or maintenance of ND10, we identified cell lines that do not express certain ND10-associated proteins. The first cell line identified came from the observation that the BLM protein, a member of the DExH box containing DNA helicases, was located in discrete nuclear domains in most cells (Neff et al. 1999). Immunostaining of BML−/− fibroblasts derived from BLM syndrome patients did not reveal any discrete domain staining, confirming specificity of BML localization at such sites in normal cells. These patients are homozygous for the mutation in the BLM gene, which leads to the early truncation of the BLM helicase. Double labeling of primary human fibroblasts with antibodies against PML, a constitutive ND10-associated protein, and antibodies against BLM showed that the two antibodies labeled ND10 (Fig. 1, A–C). To determine whether the absence of BLM helicase affects ND10, BML−/− fibroblasts were double labeled with antibodies against several ND10-associated proteins (PML, Sp100, SUMO-1, and CBP). All were detected in ND10 (shown for PML in Fig. 1, D–F). Thus, BLM helicase is not essential for the formation of ND10.
Figure 1.
The new component of ND10, BLM helicase, as well as Sp100 are not necessary for ND10 maintenance. Confocal micrographs of various cells are presented with the labeled proteins in the upper corners. The cell type is indicated at the lower part of the image. (A–C) HF double labeled for BLM protein (A) and PML (B); the merged image shows that both proteins colocalize (C). (D–F) Bloom syndrome fibroblasts (BF) double labeled for BLM (D) and PML (E); there is no ND10 labeling with the BLM antibodies (F). (G–I) NT2 cells double labeled for Sp100 (G) and PML (H); there is no ND10 labeling for Sp100 (I). (J) Western blot analysis to determine the presence of Sp100 (lanes 1–4) and PML (lanes 5–8) in NT2 (lanes 1, 2, 5, and 6) and HEp-2 cells (lanes 3, 4, 7, and 8). IFNα + represents interferon α–treated cells. Asterisk marks unspecific protein recognized by anti–PML antibody. It indicates equal protein load. Arrowheads mark most abundant Sp100 and PML signals. Higher molecular mass polypeptides represent alternative splicing or posttranscriptional modification (by SUMO-1 or phosphorylation) of proteins. It is clearly seen that NT2 cells do not express Sp100 in normal and IFNα–upregulated conditions (lanes 1 and 2), whereas in HEp-2 cells IFNα upregulation results in a dramatic increase of Sp100 signal (lanes 3 and 4). Endogenous as well as IFNα–upregulated PML are present in both cell lines (lanes 5–8).
In previous studies, we found that nuclei of most brain cells do not contain Sp100 accumulations (Cho et al. 1998). Therefore, we checked for Sp100 expression in human NT2 embryocarcinoma cells, which can be induced to differentiate into nerve-like cells by retinoic acid (Dyck et al. 1995). Using HEp-2 cells as a control, we tested by Western blot analysis for the presence of Sp100 and PML in NT2 cells. Since Sp100, as well as PML, have interferon response elements in their promoter regions (Grotzinger et al. 1996), we stimulated the cells for 24 h with interferon α to upregulate a potentially undetectable level of expression. Both cell lines contained PML (Fig. 1 J, lanes 5 and 7), although the amount of PML was higher in the HEp-2 cells. Interferon treatment dramatically increased the amount of PML (compare lanes 5 and 6 with 7 and 8). The unspecific band marked by an asterisk indicates equal loading. No Sp100 was found in NT2 cells even upon interferon upregulation (lanes 1 and 2), whereas in HEp2 cells interferon treatment resulted in a massive enhancement of the Sp100 signal (lanes 3 and 4). The results of Western blot analysis were confirmed by immunostaining with two different anti–Sp100 antibodies of human and rabbit origin. No Sp100 accumulations were seen in NT2 cells with or without interferon activation (shown for rabbit Sp100 antibodies in Fig. 1, G–I). However, most NT2 cells contained ND10 as judged by the presence of PML at specific sites. Analysis using the entire panel of ND10-specific antibodies revealed the presence of all corresponding antigens at the PML-positive sites. The negative Sp100 phenotype was also retained after differentiation induced by retinoic acid treatment and in NGP neuroblastoma cells (data not shown). Thus, we concluded that Sp100 was absent in NT2 cells and, that the presence of Sp100 was not essential for the maintenance of ND10.
The question whether PML is important for ND10 assembly and maintenance was tested in PML−/− MPEF cells. To obtain a reasonable assurance that ND10 are present or absent in PML−/− MPEF cells, we needed a panel of antibodies that reacted with several mouse ND10-associated proteins. We had identified a rabbit antibody that reacted with human and mouse PML and labeled ND10 in mouse fibroblasts (Fig. 2, A–C). This antibody was used to identify antibodies that were specific for mouse proteins located in ND10. Screening several Sp100-positive human autoantibodies, we identified one that interacts with mouse Sp100 (Fig. 2A and Fig. D). Antibody specificity was confirmed by Western blotting using recombinant Sp100. In addition, the anti-NDP55 mAb labeled ND10 (Fig. 2B and Fig. E). Also, SUMO-1, which can modify both PML and Sp100, was detected in mouse ND10 (Fig. 2C and Fig. F). To show that Sp100, NDP55, and SUMO-1 are concentrated in ND10, they are presented as separate green images below the merged one in the upper row. This panel of four antibodies recognized mouse ND10 and was used to probe for the presence of ND10 in PML−/− MPEF.
Figure 2.
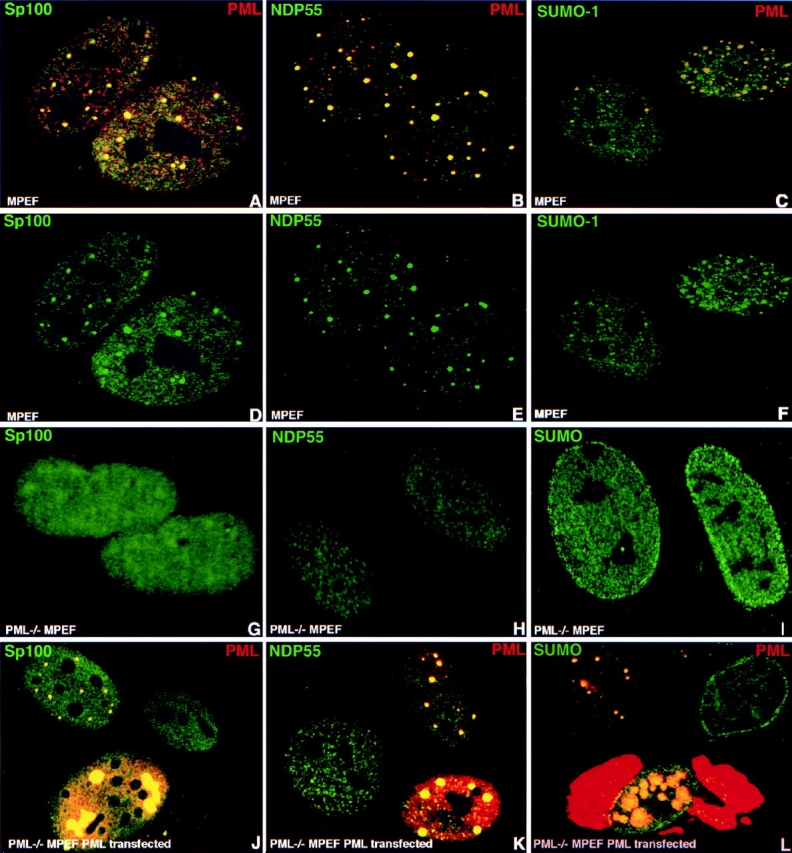
PML is responsible for the proper localization of all other ND10-associated proteins. Confocal micrographs of immunolabeled proteins are presented with the respective colors in the upper corners of the image. Cell type and overexpressed protein are indicated at the lower part of the image. (A) MPEF cells labeled for Sp100 and PML; the major ND10-associated proteins are present in mouse ND10. (B) MPEF cells double labeled for NDP55; mouse ND10 contain NDP55. C. MPEF cells double labeled for SUMO-1 and PML; mouse ND10 contain SUMO-1–modified proteins. D–H represent the same fields as in pictures above emphasizing the highest concentration of the corresponding proteins in ND10. (G) PML−/− MPEF cells labeled for Sp100 showing this protein only is dispersed throughout the nucleus in the absence of PML. (H) PML−/− MPEF cells labeled for NDP55; NDP55 is dispersed in the nucleus. (I) PML−/− MPEF cells labeled for SUMO-1; SUMO-1 is dispersed in the nucleus. (J) PML-transfected (left upper and lower cells) PML−/− MPEF labeled for Sp100 and PML showing that Sp100 is recruited to ND10-like sites. (K) PML transfected (right, upper and lower cells) PML−/− MPEF labeled for NDP55 and PML; NDP55 is recruited to ND10-like sites. (L) PML transfected PML−/− MPEF (left upper and lower cells) labeled for SUMO-1 and PML; SUMO-1 is recruited to ND10-like sites. Note exclusive nuclear localization of SUMO-1 even when PML is accumulated in the cytoplasm.
When PML−/− MPEF were tested for the distribution of the different ND10-associated proteins, neither Sp100, NDP55, nor SUMO-1 was seen in typical ND10. Instead, these proteins appeared throughout the nucleus without any ND10 accumulations (Fig. 2, compare D–F with G–I). The distribution pattern of three ND10-associated proteins, in the absence of PML, suggested PML as a likely candidate for establishing ND10 integrity. To test this possibility, we expressed PML in the PML−/− MPEF cells by transient transfection and probed for the location of Sp100, NDP55, and SUMO-1. At low expression levels, PML appeared in domains with the frequency and distribution of ND10 in wild-type cells (Fig. 2, J–L). Upon accumulation of PML, larger aggregates with a lower frequency appeared (Fig. 2 J, compare lower left and upper left cells) and sometimes cytoplasmic accumulations (Fig. 2 L, lower cell) were seen. All tested mouse ND10 proteins were segregated into PML-positive structures. These observations established that PML is essential for the assembly of ND10 and for the segregation and accumulation of ND10-associated proteins.
Daxx Is a New Component of ND10 and Interacts with PML
It has been shown before that PML cannot interact with Sp100 directly (Sternsdorf et al. 1997). However, it can recruit Sp100 into ND10 upon transient expression in PML−/− MPEF cells. We concluded that some adapter proteins must mediate this interaction and searched for new proteins that were part of ND10 and interacted with PML. In previous studies, we had cloned human Daxx through its ability to bind to the steroidogenic factor-1–like binding site in the human steroidogenic acute regulatory protein gene promoter DNA sequence (Kiriakidou et al. 1997). Rabbit antibodies produced against this protein showed an ND10-like nuclear distribution. When compared with PML, Daxx colocalized perfectly in ND10 (Fig. 3, A–C). Since Daxx interacts with the death domain of Fas (Yang et al. 1997) and, therefore, was anticipated to be a cytoplasmic protein, we confirmed the ND10 location of Daxx with two other independently generated antibodies to ensure that the antigen detected in ND10 was truly identical to Daxx. Both a commercial rabbit antibody and a mouse antibody produced against a recombinant hDaxx fragment reacted with the same structure as did PML antibodies. Specificity of Daxx antibodies were confirmed by Western blot analysis of in vitro translated hDaxx (not shown). These experiments strongly argued against the possibility that antibodies identified a spurious localization.
Figure 3.
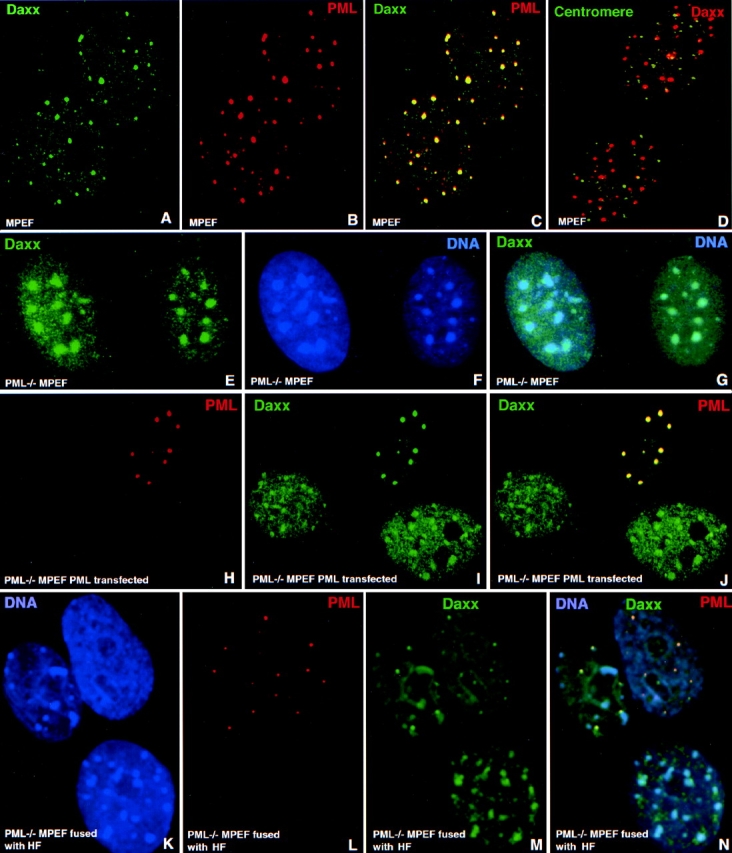
The new ND10-associated protein Daxx and its localization upon ND10 destruction and reconstitution. Confocal micrographs of immunolabeled proteins are presented with the respective colors in the upper corners of the image. Cell type and overexpressed proteins are indicated in the lower part of the image. (A–C) MPEF cells labeled for Daxx (A), PML (B), and both proteins (C) shown to colocalize at ND10. (D) PML−/− MPEF cells double labeled for centromeres and PML; some centromeres are associated with Daxx-positive sites, but mostly Daxx is not present in the same space as centromeres. (E–G) PML−/− MPEF cells double labeled for Daxx (E) and DNA (F), and together (G) show Daxx located at the condensed chromatin in the absence of PML. (H–J) PML−/− MPEF cells transfected to express PML (upper cell) and labeled for PML (H), Daxx (I), and both proteins (J); Daxx is located in PML-positive sites and contrary to the untransfected cells, very little is in the nucleoplasm and is unrecognizable at sites that may resemble condensed chromatin. (K–N) PML−/− MPEF cells fused to HF and stained for DNA (K) identifying the lower and left upper cell as of mouse origin by their DNA staining of condensed chromatin, and the right upper cell as human. Staining for PML is shown in L and for Daxx in M. In the merged image (N), the left upper mouse cell is shown to contain human PML and is, therefore, fused to HF. In the fused mouse cell, Daxx is present not only at condensed chromatin as in the lower mouse cell, but also in ND10-like domains.
In addition to Fas, Daxx was shown to interact with CENP-C and to be located at centromeres in a cell cycle–dependent fashion (Pluta et al. 1998). When we used human autoantibodies against centromeric proteins together with Daxx antibodies in wild-type mouse fibroblasts, we observed that a few sites did appear to colocalize. However, in most cases Daxx was situated beside centromeres or was not apparent at centromeres (Fig. 3 D). We concluded that Daxx is at its highest concentration in ND10 and is, therefore, a novel ND10-associated protein.
To investigate the possibility that the localization of Daxx reflects an interaction between Daxx and another ND10-associated protein, we employed the yeast two-hybrid assay. We determined whether Daxx could interact with Sp100 and/or PML fused to GAL4DB. Daxx was fused to VP16 and the VP16-producing plasmid was used as a negative control. To quantitate the strength of interaction, we used the liquid β-galactosidase assay. Using this assay, we did not observe any evidence of interaction between Daxx and Sp100. In contrast, Daxx strongly interacted with PML (Table ).
Table 1.
PML–Daxx and Sp100–Daxx Interaction in the Yeast Two-Hybrid System
To assess the specificity of this interaction, we mapped the region of Daxx that is required for interaction with PML (see Table for details). The PML coil-coiled region deletion mutant can still interact with Daxx, although it can also interact with Vp16 alone (see also Ahn et al. 1998). Unexpectedly, two of the Daxx NH2-terminal deletion mutants (amino acids 488–740 and 625–740) interacted with PML approximately threefold more strongly than the full-length molecule. The NH2-terminal region of Daxx can lower the strength of its interaction with PML, probably as a result of protein folding. In contrast, the COOH-terminal deletion mutant (amino acids 1–625) and the smallest COOH-terminal construct (amino acids 661–740) failed to interact at all. Another Daxx deletion mutant (amino acids 433–740) showed an ∼20-fold weaker interaction than the Daxx 488–740 amino acid mutant. The weaker signal might reflect the exposure of the acidic amino acid–rich region between amino acids 434 and 485, diminishing the interaction. Thus, the PML interaction domain, as defined by the yeast two-hybrid assay, lies between amino acids 625 and 740, and amino acids 625–661 are essential for this interaction. Together, these data demonstrate that Daxx interacts with PML and suggests that the spatial colocalization of PML and Daxx in ND10 reflects an interaction between these two proteins.
Daxx Location Is Mediated by PML
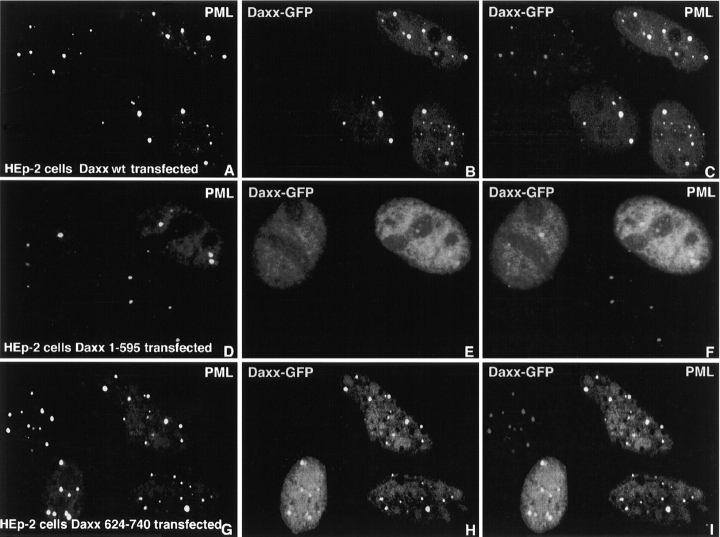
The results of the yeast two-hybrid assay indicated that Daxx and PML interacted, and the colocalization of the two proteins in ND10 was consistent with physiological association. To test whether various deletions in Daxx would affect Daxx localization in the context of mammalian cells, we fused the mutants with GFP-NLS. In agreement with indirect immunofluorescence results, the GFP-Daxx fusion protein was found to accumulate efficiently at PML-positive sites (Fig. 4, A–C). In contrast, the COOH-terminal deletion mutant (amino acids 1–595), which lacked the PML-interacting region, was diffusely present throughout the nucleus and was not accumulated at PML-positive sites (Fig. 4, D–F). The results from the yeast two-hybrid interaction were also confirmed by the finding that the Daxx COOH-terminal region (amino acids 624–740) alone was sufficient to localize GFP to ND10 (Fig. 4, G–I). These data suggest that Daxx interaction with PML is necessary for ND10 localization.
Figure 4.
Localization of GFP-fused wild-type Daxx and Daxx mutants upon transfection. Confocal micrographs presenting HEp-2 cells transiently transfected by plasmids expressing Daxx and Daxx mutants fused with GFP and stained by PML (A, D, and G) and GFP (B, E, and H). Merged images are presented in C, F, and I. Overexpressed proteins are indicated in the lower part of the image. (A–C) HEp-2 cells 16 h after transfection with wild-type Daxx fused to GFP-NLS; transiently expressed Daxx accumulates in PML-positive sites. (D–F) HEp-2 cells 16 h after transfection with Daxx 1-595 fused to GFP-NLS; this COOH-terminal deletion mutant does not accumulate in PML-positive sites. (G-I) HEp-2 cells 16 h after transfection with Daxx 624-740 fused to GFP-NLS; the transiently expressed Daxx COOH-terminal fragment accumulates in PML-positive sites but also floods the nucleoplasm.
If Daxx is accumulated in ND10 through interaction with PML, one would predict a different Daxx distribution in cells without PML. In normal mouse fibroblasts, Daxx colocalized specifically with PML in ND10 (Fig. 3, A–G). However, in the absence of PML (mouse PML−/− fibroblasts), we found the localization of Daxx to be quite different. Daxx was localized in patches which, when counterstained for DNA, proved to be condensed chromatin (Fig. 3, E–G). Therefore, in the absence of ND10, condensed chromatin is an alternative nuclear compartment of Daxx accumulation.
To investigate the relationship between PML and Daxx further, we tested whether PML could recruit endogenous Daxx into ND10 when transiently expressed in PML−/− MPEF cells. As shown in Fig. 3H–J, the untransfected PML−/− MPEF cells exhibited the patchy Daxx distribution characteristic of its condensed chromatin location. In PML-transfected cells (Fig. 3, H–J, upper cell), Daxx was not seen at condensed chromatin but, instead, now colocalized almost exclusively with PML. These PML-induced structures were found to also contain the other ND10-associated proteins (Fig. 2, J–L) and were, therefore, considered to be ND10.
To test if PML could restore Daxx accumulation at ND10 without the strong overexpression induced by transient transfection, we performed a cell fusion experiment where PML−/− MPEF cells were fused with human fibroblasts as a source of PML. Species specificity of the cells was determined by DNA distribution, which showed strongly condensed chromatin for mouse cells (Fig. 3 K, compare left and lower cells with the one in the upper right). In unfused mouse cells (Fig. 3, K–N, lower cell), Daxx appeared at its highest concentration only in condensed chromatin. But in another mouse cell (upper left cell), which became positive for human PML as a result of fusion with HF (upper right cell), Daxx started to appear in additional domains that colocalized with PML. In this experiment we could not distinguish between human and mouse Daxx. However, the finding that mDaxx, after hPML overexpression in PML−/− MPEF cells was recruited to ND10 (Fig. 3 J), made it likely that in the fusion experiment mDaxx could be accumulated into PML-positive structures. Therefore, we concluded that physiological quantities of PML can induce ND10 formation in nuclei that normally do not have them.
SUMO-1 Modification of PML Is Essential for Daxx Recruitment to ND10
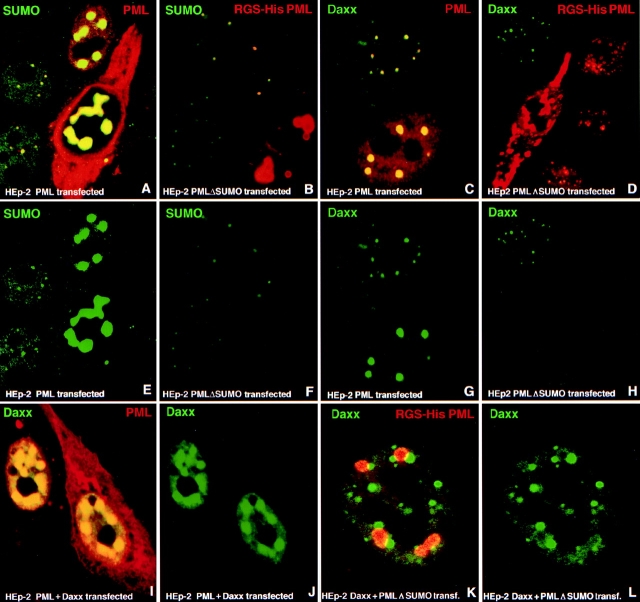
PML has been shown to be modified by SUMO-1 at least at three sites. The SUMO-1 modification seemed necessary for the deposition of PML at ND10 (Kamitani et al. 1998a; Muller et al. 1998). Therefore, we were interested in determining whether this modification affected interaction with Daxx. Double labeling of HEp-2 cells for SUMO-1 and overexpressed PML showed that PML in the nucleus colocalized with SUMO-1 in large aggregates, and that the cytoplasmic PML aggregates did not stain for SUMO-1. This suggested that only nuclear PML was modified by SUMO-1 (Fig. 5A and Fig. E, compare yellow nuclear domains resulting from the PML/SUMO-1 colocalization and red staining of cytoplasmic PML). We tested whether elimination of SUMO-1 modification sites in PML influenced SUMO-1 aggregation in PML-positive sites. SUMO-1 modification sites have been identified previously at lysines 65, 160, and 490 (Kamitani et al. 1998a). We transfected HEp-2 cells with RGS–His-tagged PML mutants containing successively decreasing numbers of SUMO-1–modified lysines and analyzed the localization of these overexpressed PML mutants and endogenous SUMO-1. We observed that a decrease of SUMO-1 accumulation in PML domains paralleled the number of mutated lysines. Shown in Fig. 5B and Fig. F, are cells transfected with the RGS–His-tagged mutant PMLΔSUMO, in which all three lysine residues are substituted with arginine. Upon high PMLΔSUMO accumulation, similar enlarged and later distorted ND10 appear as seen for the wild-type PML overexpression, except that these accumulations did not label with SUMO-1 antibodies like the wild-type PML accumulations (Fig. 5 A, compare upper right cell with lower right cell). Contrary to published reports, all PML mutants were accumulated at ND10 (recognized by the location of endogenous SUMO-1) at low level of expression (Fig. 5B and Fig. F, upper two cells are transfected, lower right is not transfected). This PMLΔSUMO accumulation indicates that either SUMO-1 modification is not necessary for ND10 targeting of PML and/or is due to potential dimer formation between PMLΔSUMO and wild-type PML, where the wild-type PML would serve as an ND10 targeting vehicle. We concluded that SUMO-1–modified PML appears only in the nucleus, and confirmed in situ that PMLΔSUMO is not SUMO-1–modified.
Figure 5.
SUMO-1 modification of PML is important for Daxx localization. Confocal micrographs of immunolabeled proteins are presented with the respective colors in the upper corners of the image. Cell type and overexpressed proteins are indicated in the lower part of the image. (A and E) PML-transfected HEp-2 cells double labeled for SUMO-1 and PML (A) or SUMO-1 only (E). SUMO-1 and PML colocalized only in the nucleus. (B and F) HEp-2 cells transfected to express the triple SUMO-1 site deletion PML mutant (PMLΔSUMO) double labeled for His-tagged PMLΔSUMO and SUMO-1 (B) or SUMO-1 only (F); PMLΔSUMO is accumulating in ND10-like structures in the upper two transfected cells; endogenous SUMO-1 indicating the location of ND10. There is no SUMO-1 in large nuclear PMLΔSUMO aggregates of the lower right cell as compared with the large aggregates of wild-type PML in A. The lower left cell is not transfected. (C and G) PML-transfected HEp-2 cells double labeled for Daxx and PML (C) or Daxx only (G); Daxx is accumulated in PML-positive sites. (D and H) HEp-2 cells transfected by His-tagged PMLΔSUMO and double labeled for Daxx and PMLΔSUMO (D) or Daxx only (H); there is no Daxx in the mutant PML accumulations as compared with the wild-type PML transfection in C. The upper left cell is not transfected. (I and J) HEp-2 cells double transfected to express PML and Daxx and double labeled for Daxx and PML (I) or Daxx only (J); Daxx and PML colocalized, but in the nucleus only. (K and L) Daxx and PMLΔSUMO double-transfected HEp-2 cells double labeled for Daxx and His-tagged PMLΔSUMO (K) or Daxx only (L); Daxx and PMLΔSUMO are deposited in different spaces.
We tested whether endogenous Daxx recruitment into ND10 was influenced by the level of PML SUMO-1 modification. HEp-2 cells were transfected with PML and PMLΔSUMO expression plasmids and tested for the location of endogenous Daxx. We found that Daxx is accumulated in domains formed by wild-type PML but not in those formed by PMLΔSUMO (Fig. 5, compare lower cell in C and G with lower left cell in D and H; the cell in the upper left of D and H is not transfected). These data demonstrate that Daxx accumulation at ND10, and potentially PML-Daxx interaction, depends on SUMO-1 modification of PML (Fig. 5, compare A and B with C and D). If PMLΔSUMO does not recruit endogenous Daxx into domains, both proteins should not colocalize upon overexpression. When we cotransfected Daxx and PML into HEp-2 cells, we found that the two proteins colocalized in the nucleus (Fig. 5I and Fig. J). However, PMLΔSUMO and Daxx formed separated aggregates upon overexpression (Fig. 5K and Fig. L). Taken together, these results show that the SUMO-1 modification of PML determines the ability of PML to segregate Daxx into ND10.
Discussion
The nucleus has been increasingly segmented into different domains that relate to the traditional nuclear functions of replication and transcription. These domains are defined by specific chromosomal territories (Lichter et al. 1988), replication sites (Wei et al. 1998), transcription sites (Jackson et al. 1993), or domains that contain excess splicing components (Spector et al. 1991). Like the nucleolus, coiled bodies have been suggested to reflect high rates of transcription because of the localization of certain genes at these sites (Gao et al. 1997; Smith et al. 1995). Nuclear domains such as ND10 and Gemini have gained attention through their connection to specific diseases (Szostecki et al. 1990; Maul et al. 1993; Dyck et al. 1994; Liu and Dreyfuss 1996; Liu et al. 1997) or viral infections (Maul et al. 1993, Maul et al. 1996; Maul and Everett 1994; Ishov and Maul 1996; Ishov et al. 1997). For ND10, a function in transcription also has been postulated (LaMorte et al. 1998). The role ND10 plays as a structure remains unclear, although we have suggested that these domains function as nuclear depots for a number of proteins (Maul 1998). Such a model dissociates the function of the respective ND10-associated proteins from their location at ND10. It also suggests that physiologically relevant interactions of these proteins with other proteins might occur at different locations. Recruitment of proteins from nucleoplasm to ND10, leading to the changes in the intranuclear protein balance, may affect cellular functions.
Modification of ND10 in a number of pathological processes strongly suggests that investigating the ND10 assembly mechanism is an essential step towards understanding the function of this nuclear structure. Using cells that lack either Sp100 or the newly described ND10-associated protein BML, we demonstrated that neither the lack of Sp100 nor the absence of BLM affected the structure of ND10. In contrast, cells lacking PML exhibited dispersion of all ND10-associated proteins. ND10 could be reconstructed by the introduction of PML into PML−/− cells either by transfection or, at more physiological concentrations of PML, through the fusion with PML-containing cells. This reconstruction includes the recruitment of all ND10 proteins, including Sp100, which does not interact with PML, suggesting the presence of mediator proteins. Our observation establishes that PML is the essential protein for ND10 assembly under physiological conditions.
The destruction of ND10 induced by the herpes virus immediate early gene products (IE1 of HCMV and ICP0 of HSV; Everett and Maul 1994; Kelly et al. 1995; Ishov et al. 1997; Ahn et al. 1998; Everett et al. 1998) supports the notion that PML plays a central part in the maintenance of ND10. Particularly the direct interaction of IE1 with PML (Ahn et al. 1998) may deprive ND10 of PML and so lead to their destruction. Consistent with this effect, constitutive expression of IE1 in astrocytoma cells and human fibroblasts also results in the loss of detectable ND10 (Ishov, A.M., unpublished observation).
The formation of ND10 has been found to accompany APL remission in promyelocytes (Dyck et al. 1995). After retinoic acid treatment of APL-derived NB4 cells, the dominant negative PML–retinoic acid receptor α (PML-RARα) fusion protein is selectively hydrolyzed through the proteosome pathway, releasing wild-type PML (Muller et al. 1998). This, in turn, might be the reason for ND10 formation in RA-treated NB4 cells, which normally have only dispersed ND10-associated proteins. Therefore, the recovery of ND10 may be a consequence of PML availability, which leads to the segregation of other ND10 proteins.
The central role of PML in ND10 formation suggests the presence of proteins that are accumulated at ND10 through interaction with PML. We found that the recently cloned DNA-binding protein Daxx (Kiriakidou et al. 1997; Yang et al. 1997) was highly concentrated in ND10. Moreover, we found that Daxx interacted with PML in the yeast two-hybrid assay, mapped the interaction domain of Daxx, and demonstrated that Daxx localization at ND10 depended on the presence of a PML interaction domain as well as SUMO-1 modification of PML. The discrepancy between previously reported Daxx interactions with Fas (Yang et al. 1997), CENP-C (Pluta et al. 1998), and ND10 localization of this protein suggests that Daxx does not accumulate together with all interaction partners, but is preferentially accumulated at ND10.
In the absence of PML in PML−/− MPEF cells, ND10 are destroyed. Therefore, ND10-associated proteins are expected to be found at their alternative binding locations. Most of these proteins were dispersed throughout the nucleus and, thus, were not amenable to microscopic analysis. Only Daxx was detected at higher concentrations in the areas of condensed chromatin. Daxx was removed from these chromatin regions through the introduction of PML by transient transfection concomitant with the formation of ND10. Therefore, these chromatin regions have a substantial amount of unsaturated Daxx binding sites. We propose that Daxx acts at sites other than ND10 by interactions with DNA (Kiriakidou et al. 1997) or other proteins (Yang et al. 1997; Pluta et al. 1998; Hollenbach et al. 1999) at a variety of cellular locations. Whether the balance of Daxx between ND10 and condensed chromatin can be modified under physiological conditions is not yet conclusively established; however, such a balance may constitute a potential control mechanism.
A key finding of our study was that the Daxx accumulation in ND10 is dependent on SUMO-1 modification of PML. In the yeast two-hybrid system, SMT3, the yeast homologue of SUMO-1 (Lapenta et al. 1997), may have facilitated the PML–Daxx interaction. Moreover, conjugation of SMT3 to other yeast proteins is facilitated by Ubc9 (Johnson and Blobel 1997; Schwarz et al. 1998), the yeast homologue of human Ubc9, which is involved in the SUMO-1 modification of PML (Duprez et al. 1999). Failure to coimmunoprecipitate in vitro translated Daxx with recombinant PML (not shown) is consistent with the idea that SUMO-1 modification of PML mediates this interaction.
The SUMO-1 modification level of PML might be a determinant of the amount of Daxx recruited and the avidity with which Daxx is retained at ND10. Therefore, the regulated posttranslational modification of PML may balance the amount of Daxx available in the nucleus. Contrary to previous reports (Kamitani et al. 1998a; Muller et al. 1998), we found that SUMO-1 modification may not be essential for the deposition of PML at ND10, but rather results in the Daxx accumulation at this domain. The central role of PML in ND10 formation suggests the presence of a protein network where some adapter proteins can mediate recruitment of non-PML–interacting proteins to ND10. Daxx may act as such an adapter and recruit other ND10-associated proteins that do not bind directly to PML (Negorev, D., unpublished results).
An emergent hierarchical model for ND10 formation is presented schematically in Fig. 6. The appearance of ND10 after mitosis must result from a nucleation event possibly through homo- or heteromultimerization of PML. This event may take place at specific nuclear deposition sites, as postulated earlier (Maul 1998). Transcriptional activation, for instance by interferon, can upregulate PML expression (Lavau et al. 1995), nucleating additional aggregation sites. SUMO-1 modification-demodification of PML (third level) may lead to a reversible accumulation of Daxx to ND10 (fourth level), increasing or decreasing the availability of this protein for alternative binding partners (DNA, CENP-C, Fas, Pax3, DNA methyltransferase), and thus regulate corresponding functions. The complexity and plasticity of such a supramolecular regulatory mechanism are evident and envisioned structurally as a network of interacting proteins with PML at its core.
Figure 6.
Hierarchical scheme of ND10 formation and maintenance. PMLS indicates PML conjugated to SUMO-1; DNA M-Tase, DNA methyltransferase.
Acknowledgments
We thank Qinwu Lin (The Wistar Institute) for technical assistance, P. Freemont, J. Frey, R. Evans, and N. Stuurman (University of Amsterdam), for providing the antibodies and plasmids, and Dr. P. Pandolfi for the PML−/− cells. We also thank S. Berger for providing yeast strains and vectors for the yeast assays, and G. Prendergast (The Wistar Institute) and J. Dyck (Salk Institut) for critically reading the manuscript.
This study was supported by funds from the National Institutes of Health (AI 41136; NIH GM 57599, NIH HD 34612, NSF MCB9728398) and the Mathers Foundation. The NIH Core grant CA-10815 is acknowledged for the support of the microscopy facility.
Footnotes
1.used in this paper: APC, acute promyelocytic leukemia; BSF, human Bloom syndrome–derived primary fibroblasts; Daxx, Fas death domain–associated protein; HF, human primary fibroblasts; MPEF, embryonic primary mouse fibroblasts; ND10, nuclear domain 10; NGP, human neuroblastoma cells; PML, promyelocytic leukemia protein; PML−/− MPEF, SV40 large T-antigen transformed cells derived from embryos of PML−/− knockout mouse; Sp100, autoantigen in primary billiary cirrhosis; SUMO-1, small ubiquitin-related modifier
References
- Ahn J.H., Brignole E.J., III, Hayward G.S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli C.A., Maul G.G. Identification of a novel nuclear domain. J. Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Howe K., Etkin L.D., Solomon E., Freemont P.S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Candau R., Moore P.A., Wang L., Barlev N., Ying C.Y., Rosen C.A., Berger S.L. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix M.K., de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- Cho Y., Lee I., Maul G.G., Yu E. A novel nuclear substructure, ND10Distribution in normal and neoplastic human tissues. Int. J. Mol. Med. 1998;1:717–724. doi: 10.3892/ijmm.1.4.717. [DOI] [PubMed] [Google Scholar]
- de The H., Chomienne C., Lanotte M., Degos L., Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- Duprez E., Saurin A.J., Desterro J.M., Lallemand-Breitenbach V., Howe K., Boddy M.N., Solomon E., de The H., Hay R.T., Freemont P.S. SUMO-1 modification of the acute promyelocytic leukaemia protein PMLimplications for nuclear localisation. J. Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Dyck J.A., Maul G.G., Miller W.H., Jr., Chen J.D., Kakizuka A., Evans R.M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Dyck J.A., Warrell R.P., Jr., Evans R.M., Miller W.H., Jr. Rapid diagnosis of acute promyelocytic leukemia by immunohistochemical localization of PML/RAR-alpha protein. Blood. 1995;86:862–867. [PubMed] [Google Scholar]
- Everett R.D., Maul G.G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R.D., Freemont P., Saitoh H., Dasso M., Orr A., Kathoria M., Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R.D., Meredith M., Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Padykula H.A. Differing functional responses of simple and complex nuclear bodies in uterine luminal epithelial cells following estrogenic stimuli. Anat. Rec. 1983;205:131–141. doi: 10.1002/ar.1092050204. [DOI] [PubMed] [Google Scholar]
- Gao L., Frey M.R., Matera A.G. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard A.D., Borrow J., Freemont P.S., Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- Grotzinger T., Sternsdorf T., Jensen K., Will H. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur. J. Biochem. 1996;238:554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- Hollenbach A.D., Sublett J.E., McPherson C.J., Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Maul G.G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Stenberg R.M., Maul G.G. Human cytomegalovirus immediate early interaction with host nuclear structuresdefinition of an immediate transcript environment. J. Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Hassan A.B., Errington R.J., Cook P.R. Visualization of focal sites of transcription within human nuclei. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzabek-Chorzelska M., Blaszczyk M., Kolacinska-Strasz Z., Chorzelski T., Jablonska S., Maul G.G. Antikinetochore and antitopoisomerase I antibodies in systemic sclerodermacomparative study using immunoblotted recombinant antigens, immunofluorescence, and double immunodiffusion. Arch Dermatol. Res. 1990;282:76–83. doi: 10.1007/BF00493462. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Kakizuka A., Miller W.H., Jr., Umesono K., Warrell R.P., Jr., Frankel S.R., Murty V.V., Dmitrovsky E., Evans R.M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Kamitani T., Kito K., Nguyen H.P., Wada H., Fukuda-Kamitani T., Yeh E.T. Identification of three major sentrinization sites in PML J. Biol. Chem. 273 1998. 26675 26682a [DOI] [PubMed] [Google Scholar]
- Kamitani T., Nguyen H.P., Kito K., Fukuda-Kamitani T., Yeh E.T. Covalent modification of PML by the sentrin family of ubiquitin-like proteins J. Biol. Chem. 273 1998. 3117 3120b [DOI] [PubMed] [Google Scholar]
- Kelly C., Van Driel R., Wilkinson G.W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J. Gen. Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M., Driscoll D.A., Lopez-Guisa J.M., Strauss J.F., III. Cloning and expression of primate Daxx cDNAs and mapping of the human gene to chromosome 6p21.3 in the MHC region. DNA Cell Biol. 1997;16:1289–1298. doi: 10.1089/dna.1997.16.1289. [DOI] [PubMed] [Google Scholar]
- Kleppner S.R., Robinson K.A., Trojanowski J.Q., Lee V.M. Transplanted human neurons derived from a teratocarcinoma cell line (NTera-2) mature, integrate, and survive for over 1 year in the nude mouse brain. J. Comp. Neurol. 1995;357:618–632. doi: 10.1002/cne.903570410. [DOI] [PubMed] [Google Scholar]
- Korioth F., Maul G.G., Plachter B., Stamminger T., Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- LaMorte V.J., Dyck J.A., Ochs R.L., Evans R.M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenta V., Chiurazzi P., van der Spek P., Pizzuti A., Hanaoka F., Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- Lavau C., Marchio A., Fagioli M., Jansen J., Falini B., Lebon P., Grosveld F., Pandolfi P.P., Pelicci P.G., Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Maul G.G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Everett R.D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Guldner H.H., Spivack J.G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J. Gen. Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Yu E., Ishov A.M., Epstein A.L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Ishov A.M., Everett R.D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- Michaelson J.S., Bader D., Kuo F., Kozak C., Leder P. Loss of daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Z.M., Chin K.V., Liu J.H., Lozano G., Chang K.S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Matunis M.J., Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff N.F., Ellis N.A., Ye T.Z., Noonan J., Huang K., Sanz M., Proytcheva M. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of Bloom syndrome cells. Mol. Biol. Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padykula H.A., Fitzgerald M., Clark J.H., Hardin J.W. Nuclear bodies as structural indicators of estrogenic stimulation in uterine luminal epithelial cells. Anat. Rec. 1981;201:679–696. doi: 10.1002/ar.1092010412. [DOI] [PubMed] [Google Scholar]
- Pandolfi P.P., Grignani F., Alcalay M., Mencarelli A., Biondi A., LoCoco F., Pelicci P.G. Structure and origin of the acute promyelocytic leukemia myl/RAR alpha cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- Pluta A.F., Earnshaw W.C., Goldberg I.G. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J. Cell Sci. 1998;111:2029–2041. doi: 10.1242/jcs.111.14.2029. [DOI] [PubMed] [Google Scholar]
- Quignon F., De Bels F., Koken M., Feunteun J., Ameisen J.C., de The H. PML induces a novel caspase-independent death process. Nat. Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- Rose M., Winson F., Hieter P. Methods in Yeast Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Schwarz S.E., Matuschewski K., Liakopoulos D., Scheffner M., Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.P., Carter K.C., Johnson C.V., Lawrence J.B. U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Spector D.L., Fu X.D., Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N., de Graaf A., Floore A., Josso A., Humbel B., de Jong L., van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Szostecki C., Guldner H.H., Netter H.J., Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg S.M., Cooper J.A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P.P. PML is essential for multiple apoptotic pathways. Nat. Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- Wei X., Samarabandu J., Devdhar R.S., Siegel A.J., Acharya R., Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Weis K., Rambaud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Yang X., Khosravi-Far R., Chang H.Y., Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnbauer B.A., Small D., Brodeur G.M., Seeger R., Vogelstein B. Characterization of N-myc amplification units in human neuroblastoma cells. Mol. Cell Biol. 1988;8:522–530. doi: 10.1128/mcb.8.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Guo Y., Niu Q., Levy D.E., Dyck J.A., Lu S., Sheiman L.A., Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]