Genetic identification of a novel NeuroD1 function in the early differentiation of islet α, PP and ε cells (original) (raw)
. Author manuscript; available in PMC: 2008 Dec 15.
Summary
Nkx2.2 and NeuroD1 are vital for proper differentiation of pancreatic islet cell types. Nkx2.2-null mice fail to form β cells, have reduced numbers of α and PP cells, and display an increase in ghrelin-producing ε cells. NeuroD1-null mice display a reduction of α and β cells after embryonic day (e) 17.5. To begin to determine the relative contributions of Nkx2.2 and NeuroD1 in islet development, we generated Nkx2.2−/−;NeuroD1−/− double knockout (DKO) mice. As expected, the DKO mice fail to form β cells, similar to the Nkx2.2-null mice, suggesting that the Nkx2.2 phenotype may be dominant over the NeuroD1 phenotype in the β cells. Surprisingly, however, the α, PP and ε phenotypes of the Nkx2.2-null mice are partially rescued by the simultaneous elimination of NeuroD1, even at early developmental time points when NeuroD1 null mice alone do not display a phenotype. Our results indicate that Nkx2.2 and NeuroD1 interact to regulate pancreatic islet cell fates, and this epistatic relationship is cell-type dependent. Furthermore, this study reveals a previously unappreciated early function of NeuroD1 in regulating the specification of α, PP and ε cells.
Keywords: Nkx2.2, NeuroD1, islet development, α cells, cell differentiation, glucagon, ghrelin, genetic analysis
Introduction
The pancreas is a multifunctional organ consisting of exocrine and endocrine components, which is essential for maintaining glucose homeostasis and energy metabolism in the body. In developing mice, the pancreatic islets consist of five major endocrine cell types; insulin-producing β cells, glucagon-producing α cells, somatostatin-producing δ cells, pancreatic polypeptide (PP)-producing PP cells, and ghrelin-producing ε cells. In addition, there exists a small population of islet cells that co-express ghrelin and glucagon and appear to be distinct from either the α cell or ε cell lineage (Heller et al., 2005). Several mouse models have demonstrated that individual transcription factors, such as Pdx1, Hnf6, Ngn3, NeuroD1, Nkx2.2, Nkx6.1, Pax4, and Pax6 are essential for islet cell differentiation, maintenance and proper organization (Collombat et al., 2003; Gannon et al., 2000; Gradwohl et al., 2000; Jacquemin et al., 2000; Jonsson et al., 1994; Naya et al., 1997; Offield et al., 1996; Sander et al., 2000; Sosa-Pineda et al., 1997; St-Onge et al., 1997; Sussel et al., 1998). Elimination of each of these transcription factors gives rise to a pancreatic defect at a distinct developmental stage, thus allowing for the organization of these factors into a genetic regulatory cascade (Gasa et al., 2004; Habener et al., 2005; Schwitzgebel, 2001). More recently, sophisticated genetic approaches have been used to show complex temporal requirements and novel functions for several of these regulatory factors. Inducible activation of Pdx1 at discrete embryonic time points demonstrates that it functions at multiple stages of pancreas development, as well as in the adult to regulate pancreatic differentiation and function (Holland et al., 2005; Holland et al., 2002). Furthermore, double knockout studies have revealed novel roles for Pax4, Arx, Nkx6.1 and Nkx6.2 regulatory factors in islet cell type specification (Collombat et al., 2005; Henseleit et al., 2005).
In this study, we focus on two essential islet regulatory proteins, Nkx2.2 and NeuroD1/Beta2 (hereafter referred to as NeuroD1). Nkx2.2 is a homeodomain-containing protein that is expressed in the central nervous system, pancreas and intestine. Nkx2.2 is present in the earliest progenitor cells of the developing pancreas and gradually becomes restricted to β cells and a subset of α and PP cells (Sussel et al., 1998).
In the absence of Nkx2.2, β cells fail to differentiate and α and PP cell numbers are reduced. Interestingly, the ghrelin-producing ε cell population appears to replace these islet cell types, implicating Nkx2.2 as a major regulator of the initial islet cell fate decisions (Prado et al., 2004). NeuroD1 is a basic helix-loop-helix (bHLH) transcription factor, which is also expressed in the central nervous system, pancreas, and intestine. In the pancreas, NeuroD1 expression begins at the onset of pancreatic bud formation and by e10.5, NeuroD1 is restricted to the endocrine cell population (Chu et al., 2001; Naya et al., 1997). In NeuroD1−/− mice, the pancreas develops without any discernable phenotypes until e15.5; however, islets fail to form and increased islet cell death is apparent by e17.5 (Naya et al., 1997). These analyses suggest that NeuroD1 may not be required for initial islet cell specification, but may play a predominant role in islet cell differentiation and survival. The phenotypes of the individual Nkx2.2 null and NeuroD1 null mice suggest that Nkx2.2 functions upstream of NeuroD1 in the genetic cascade; however it remains unclear what role each of these transcription factors plays in the early pancreatic primordia and whether they genetically interact to control islet cell development.
To clarify the relative roles of Nkx2.2 and NeuroD1 in the development of the individual islet cell populations, we generated Nkx2.2−/−;NeuroD1−/− double knockout (DKO) mice. We found that the Nkx2.2−/−;NeuroD1−/− DKO mice failed to form insulin-producing β cells, similar to what was observed in the single Nkx2.2−/− mice. Unexpectedly, however, we observed a significant recovery of α cells and PP cells in the Nkx2.2−/−;NeuroD1−/− DKO mice, compared to the Nkx2.2−/− single knockout mice. Furthermore, there appeared to be a corresponding decrease of the ghrelin-producing ε cells in Nkx2.2−/−;NeuroD1−/− DKO mice compared to Nkx2.2−/− mice. Interestingly, this partial restoration of normal α, PP, and ε cell ratios was evident during a major wave of islet cell differentiation, termed the secondary transition (Pictet and Rutter, 1972), which is prior to an observed NeuroD1 single knockout phenotype. These results suggest that NeuroD1 has a previously unknown role in regulating islet cell fate specification prior to e15.5. In addition, these studies reveal a complex genetic interaction between Nkx2.2 and NeuroD1 that is responsible for the specification of α, PP, and ε cell fates.
Materials and Methods
Animals
Nkx2.2 and NeuroD1 heterozygous mice were generated by homologous recombination as previously described (Miyata et al., 1999; Sussel et al., 1998). Nkx2.2+/− and NeuroD1 +/− animals were maintained in a Swiss Black (Taconic) background. All mice that were homozygous for the NeuroD1 null allele on this genetic background died shortly after birth and displayed no variability in phenotype. Double heterozygous mice were mated to obtain DKO mice. Genotyping of mice and embryos was performed by PCR analysis. The Nkx2.2 wild type locus was identified with primers as previously described (Sussel et al, 1998). In order to detect the Nkx2.2 null allele, we used a forward primer (5′-cagcgatgggaaggagagatttca-3′) to detect sequence within the 5′ Nkx2.2 promoter and a reverse primer (5′-gatcagcagcctctgttccacata-3′) homologous to sequences within the PGK1-neo gene. To genotype NeuroD1, we used a forward primer (5′-accatgcactctgtacgcatt-3′) that recognized sequences within the 5′ NeuroD1 promoter and two reverse primers, one within the LacZ gene (5′-gagaactgagacactcatgtg-3′) and one within the NeuroD1 locus (5′-aaacgccgagttaaagccatc-3′) to identify the knockout and wild type alleles, respectively. Animals were housed and treated according to UCHSC Institutional Review Board approval protocols.
Immunofluorescence
Immunofluorescence was performed on e12.5, e15.5, e16.5 and P0 cyropreserved tissue fixed with 4% paraformaldehyde at 4°C for 3 hours (MafA) or overnight (Arx, ghrelin, glucagon, insulin, somatostatin, PP). A subset of P0 pancreatic tissue was dissected and fixed with 10% formalin for 1.5 hours (Nkx6.1, Pdx1, and Pax6). Antibodies used were rabbit anti-amylase (1:1000, Sigma), rabbit anti-Arx (1:1000, P. Collombat, Goettingen, Germany), guinea-pig anti β-galactosidase (1:1000, T. Finger, University of Colorado Health Science Center), rabbit anti-ghrelin (1:200, Phoenix), guinea-pig anti-glucagon (1:3000, Linco), guinea-pig anti-insulin (1:1000, Linco), rabbit anti-MafA (1:1000, Bethyl), rabbit anti-Nkx6.1 (1:800, Beta Cell Biology Consortium (BCBC)), rabbit anti-Pax6 (1;1000, Chemicon), rabbit anti-Pdx1 (1:1000, Chemicon), guinea-pig anti-pancreatic polypeptide (1:500, Linco), and rabbit anti-somatostatin (1:200, Phoenix). Secondary antibodies used were Alexafluor-488 anti-rabbit (1:400), Alexafluor-488 anti-guinea pig (1:400), Alexafluor-594 anti-guinea pig (1:400), and Alexafluor-594 anti-rabbit (1:400). Confocal images were processed with a Zeiss Microscope, LSM 510 META at 25X magnification.
Morphometric analysis
For each genotype (wild type, Nkx2.2−/−, NeuroD1−/−, and Nkx2.2−/−;NeuroD1−/−), e16.5 embryos and P0 mice were processed as above and every 10 sections were collected and stained by immunofluorescence for ghrelin and glucagon. For morphometric analysis, n = 3 at each time point. Non-overlapping images of the total pancreatic area were taken at 10x magnification for counting of fluorescent cells and total pancreatic area on a Leica CTR 5000. Immunofluorescent cell area on glucagon+, ghrelin+, and glucagon+ghrelin+ sections was counted and then divided by total pancreatic area using morphometric analysis in Image Pro Plus 5.1 software. Results are shown as percent fluorescent area per total pancreatic area. Significance was calculated using the two-tailed student t-test with unequal variance. A p value of <0.05 was considered statistically significant.
Quantification of mRNA by Real Time Taqman PCR
Total RNA was isolated with the RNeasy Micro Kit (Qiagen) from whole pancreas and small intestine at P0. For each tissue, mRNA (2ug) was converted into cDNA using superscript III (Invitrogen). Quantitative PCR (qPCR) for Brn4 (Mm00447171_S1), cholecystokinin (Mm00446170_m1), gapdh (Mm99999915_g1), ghrelin (Mm00445450_m1), glucagon (Mm00801712_m1), insulin II (Mm00731595_gH), Irx2 (Mm01340316_m1), NeuroD1 (Mm01280117_m1), pancreatic polypeptide (Mm00435889_m1), Pdx1 (Mm00435565_m1), secretin (Mm00441235_g1), and somatostatin (Mm00436671_m1) was performed using Assay on Demand primer/probe sets (Applied Biosystems). Primer/probe sets were designed for Nkx2.2, Nkx6.1, MafA, Pax4, and Pax6 (Doyle et al., 2007). The probe, forward primer and reverse primer sequences for Arx were FAM-ctaccgcaccacgttcaccagt-MGB, 5′gaagcgcaaacagagg3′, and 5′gctccagttcctccag3′, respectively. All qPCR was performed on the ABI PRISM 7000 Sequence Detection System. An n=5–6 was obtained for wild type, Nkx2.2−/−, NeuroD1−/−, Nkx2.2−/−;NeuroD1−/−, Nkx2.2−/−NeuroD1+/−, and Nkx2.2+/−NeuroD1−/−. A p value of <0.05 were considered statistically significant.
RNA in situ hybridization
RNA in situ hybridization was performed as previously described (Prado et al., 2004) using antisense riboprobes transcribed from linearized plasmids on processed tissue (described in immunofluorescence section of materials and methods). Riboprobes for Nkx2.2 and Ngn3, were made from PCR-cloned full-length cDNAs (Decker et al., 2006; Sussel et al., 1998). The riboprobe for Brn4 was generated from full length Brn4 cDNA PCR amplified from a pancreatic cDNA library (Prado et al., 2004) and cloned into the TOPO blunt vector (Invitrogen). NeuroD1, Arx and Irx2 riboprobes were generated from the plasmids pCS2:MTmNeuroD1 (J. Lee.), pYX-Asc-Arx (Open Biosystems) and pYX-Asc-Irx2 (Open Biosystems), respectively. RNA in situ hybridization was performed on tissue from Nkx2.2−/−, NeuroD1−/−, and Nkx2.2−/−;NeuroD1−/− embryos and a corresponding number of wild type littermates at e12.5, e16.5, and P0.
Results
The Nkx2.2−/−;NeuroD1−/− DKO mice fail to form insulin-producing β cells
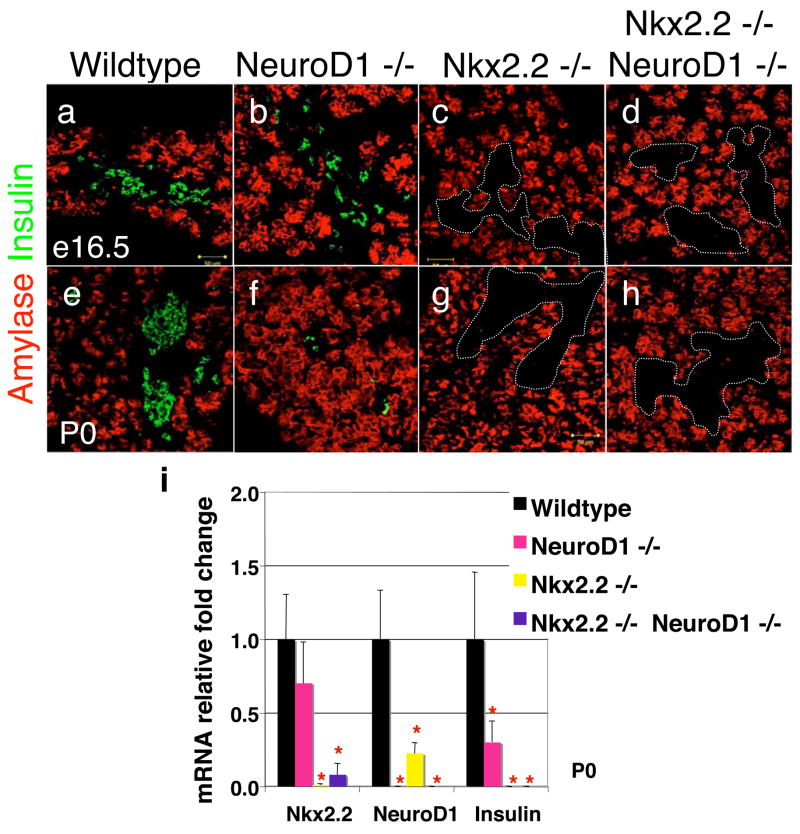
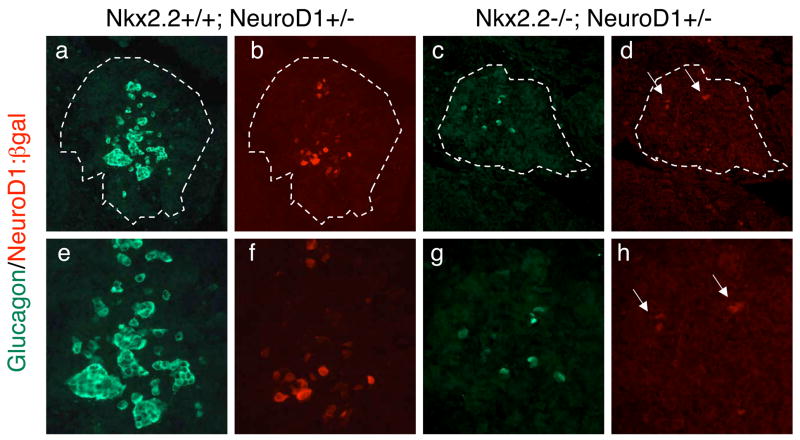
To determine the potential molecular and genetic interactions between Nkx2.2 and NeuroD1 during islet development, we generated Nkx2.2−/−;NeuroD1−/− DKO mice. Since the presence of the NeoR gene in both the Nkx2.2 and NeuroD1 mutant alleles was previously used for genotyping, we developed new PCR primer sets to distinguish each of the alleles (Materials and methods) and verified the genotypes using quantitative real time PCR for detection of the Nkx2.2 and NeuroD1 mRNA (Figure 1). This analysis confirmed the efficacy of our new genotyping primers. Interestingly, the assessment of gene expression also demonstrated that NeuroD1 is down regulated in the islets of Nkx2.2−/− mice (Figure 1i). We further explored the reduction in NeuroD1 expression in the Nkx2.2−/− mice by comparing the expression of β-galactosidase (the lacZ gene is inserted into the NeuroD1 locus (Miyata et al., 1999)) in wild type and Nkx2.2−/− mice at e12.5 and e18.5 (Figure 2 and data not shown). The immunohistochemical analysis of β-galactosidase expressioin confirms that NeuroD1 is expressed in reduced numbers of cells in the Nkx2.2−/− mice, possibly partially due to the loss of specific islet cell types. Nkx2.2 mRNA expression is not significantly changed in the NeuroD1−/− knockout at each stage of development tested (Figure 1 and data not shown).
Figure 1. Insulin-producing β cells are absent in Nkx2.2−/−;NeuroD1−/− DKO mice.
Immunofluorescence of insulin (green) and amylase (red) in e16.5 (A–D) and P0 (E–H) pancreas of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice. Islet clusters are outlined by the dotted lines with no insulin staining. Quantitative PCR results for mRNA of Nkx2.2, NeuroD1 and insulin in P0 pancreas of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice (I). N = 3–6 pancreas per genotype. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an * compared to wild type phenotype.
Figure 2. NeuroD1 expression in Nkx2.2−/− islets.
Immunofluorescence of glucagon (green) and β-galactosidase (red) in e12.5 tissue. The β-galactosidase signal reflects NeuroD1 expression since the cytoplasmic LacZ gene has been knocked into the NeuroD1 genomic locus. Nkx2.2+/+; NeuroD1+/− at 20x (a, b) and 40x (e, f) magnification. Nkx2.2−/−; NeuroD1+/− at 20x (c, d) and 40x (g, h) magnification.
We next examined Nkx2.2−/−;NeuroD1−/− DKO mice at e12.5, e15.5, e16.5 and P0 and compared them to the single knockout phenotypes. Similar to the Nkx2.2-null mice, Nkx2.2−/−;NeuroD1−/− DKO mice do not contain insulin-producing cells at any developmental time point (Figure 1a-h and data not shown). The complete absence of insulin expression was confirmed by quantitative PCR at P0 (Figure 1i). The single knockout phenotypes suggest that Nkx2.2 is required for β cell fate determination and specification, whereas NeuroD1 appears to be necessary for the subsequent differentiation and survival of β cells at subsequent developmental stages. The Nkx2.2−/−;NeuroD1−/− DKO phenotype supports the single knockout data; however, the complete absence of β cells in the Nkx2.2−/− mice precludes our ability to make additional conclusions about the relative roles of Nkx2.2 and NeuroD1 in the β cell.
The aberrant ratios of glucagon-producing cells and ghrelin-producing cells found in Nkx2.2−/− mice are partially restored in Nkx2.2−/−;NeuroD1−/− DKO mice
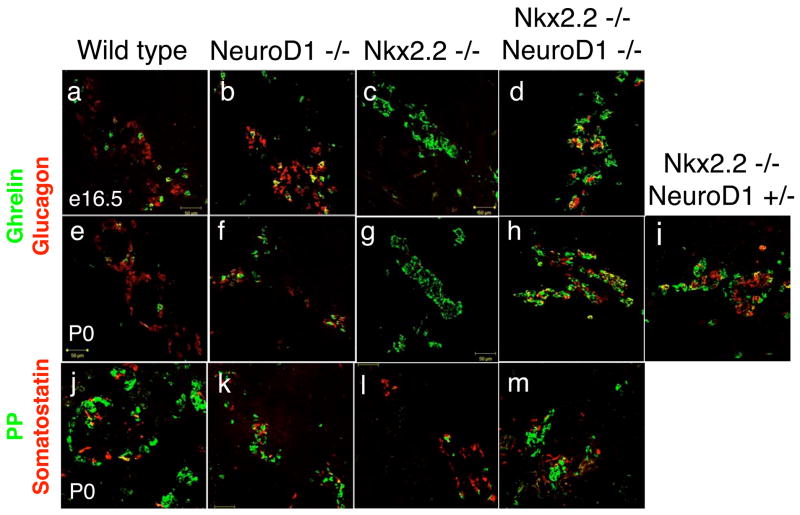
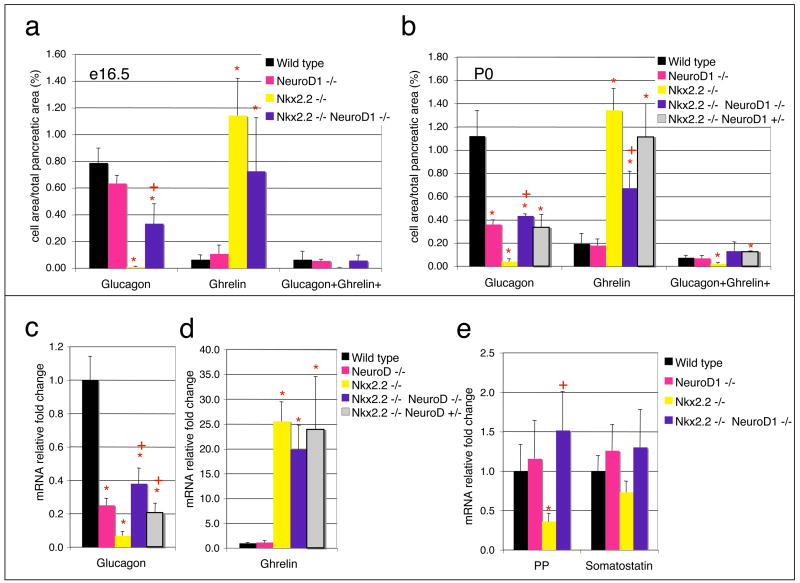
In the Nkx2.2−/− mice, only a small fraction of glucagon-producing α cells form (Sussel et al., 1998). In the NeuroD1−/− mice, α cells form in their normal numbers; however, there is a 50% reduction in α cells at e16.5, the molecular basis of which is unknown (Naya et al., 1995). Surprisingly, at e12.5 and e15.5, which is prior to any apparent NeuroD1−/− single knockout phenotype, α cell numbers are partially restored in the islets of the Nkx2.2−/−;NeuroD1−/− DKO mice. These results would suggest that the absence of NeuroD1 in the Nkx2.2−/− genetic background allowed for partial recovery of the α cell population (Figure 3, Figure 4 and data not shown). In the newborn Nkx2.2−/−;NeuroD1−/− DKO mice, the recovered α cell population is present at approximately 50% of wild type levels (Figure 3a-h, and Figure 4), which is significantly higher than observed in the Nkx2.2−/− mice. On the other hand, the amount of α cells that are recovered is similar to the numbers observed in the late gestation NeuroD1−/−mice, suggesting that NeuroD1 function may still be required in the Nkx2.2−/−;NeuroD1−/− DKO for full maintenance of the α cell population. Quantitative PCR analyses confirm that glucagon mRNA expression levels corresponded to the relative numbers of glucagon-producing cells in each genotypic background (Figure 3). Interestingly, an intermediate number of α cells are restored in the Nkx2.2−/−;NeuroD1+/− mice (Figure 3i and 4c–d), suggesting that the dosage of NeuroD1 activity may play an important role in determining the number of α cells recovered in the absence of Nkx2.2. Similar dosage effects were not observed in Nkx2.2+/−;NeuroD1−/− mice, which had an α cell phenotype indistinguishable from that of the NeuroD1−/− mice (data not shown). These findings suggest that Nkx2.2 and NeuroD1 have interrelated functions in the differentiation of glucagon-producing α cells and the absence of Nkx2.2 uncovers novel NeuroD1 activities in α cell specification.
Figure 3. Glucagon-producing α cells and Ghrelin-producing ε cells partially restored in Nkx2.2−/−;NeuroD1−/− DKO mice.
Immunofluorescence of glucagon (red) and ghrelin (green) in e16.5 (A–D) and P0 (E–I) tissue of wild type, NeuroD1−/−, Nkx2.2−/−, Nkx2.2−/−;NeuroD1−/− DKO, and Nkx2.2−/−NeuroD1+/− mice. Immunofluorescence of PP (green) and somatostatin (red) in P0 (J–M) tissue of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice.
Figure 4. Quantification of mRNA and islet hormone-positive area.
Graphs of the percent immunofluorescent glucagon, ghrelin, and glucagon+ghrelin+ cell area per total pancreatic area from e16.5 (A) and P0 (B) tissues. Every ten sections from 100 sections for each genotype, wild type, NeuroD1−/−, Nkx2.2−/−, Nkx2.2−/−;NeuroD1−/−, and Nkx2.2−/−NeuroD1+/− mice were counted. N=3 for each genotype in cell area counts. Quantitative PCR results for fold change of glucagon (C) and ghrelin (D) mRNA in P0 pancreas of wild type, NeuroD1−/−, Nkx2.2−/−, Nkx2.2−/v;NeuroD1−/−, and Nkx2.2−/−NeuroD1+/− mice. Real time PCR results for fold change of PP and somatostatin mRNA in P0 pancreas of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice (E). n = 3–6 pancreas per genotype in all qPCRs. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an * compared to wild type and + compared to Nkx2.2 knockout phenotype.
In the Nkx2.2−/− mice, the loss of glucagon-producing α cell formation is accompanied by a corresponding increase in ghrelin-producing ε cell numbers and a loss of the minor glucagon+ghrelin+ co-expressing cell population (Prado et al., 2004). In the NeuroD1−/− mice, ghrelin appears unchanged at both the mRNA and protein levels in all ages examined (Figure 3 and 4). To determine the status of the ghrelin-producing and ghrelin+glucagon+ populations in the Nkx2.2−/−;NeuroD1−/− DKO mice, we assessed ghrelin mRNA and protein expression at e16.5 and/or P0. Interestingly, although not reduced to wild type levels, ε cell numbers were decreased approximately 50% in the Nkx2.2−/−;NeuroD1−/− DKO mice compared to the Nkx2.2−/− mice (Figure 4b). The decrease in ε cell numbers corresponded with the observed increase in the α cell numbers. We confirmed that the alteration in the relative glucagon- and ghrelin-producing cell numbers was not due to an abnormal increase in the glucagon+ghrelin+ double positive cell population; however, this minor cell population was fully recovered to wild type levels compared to the Nkx2.2 null mice (Figure 4a–b). These results suggest that the glucagon-producing α cell population has been restored at the expense of the ghrelin-producing ε cell population and further implicates an important role for NeuroD1 in these early islet cell fate decisions.
The Nkx2.2−/− mice also display a partial loss of PP cells (Sussel et al., 1998), while the PP cell population appears to be unaffected in the NeuroD1 null mice at all ages (Figure 3k). Interestingly, PP cells are restored to wild type levels in the Nkx2.2−/−; NeuroD1−/− DKO mice (Figure 3m and 4e), suggesting that Nkx2.2 and NeuroD1 also genetically interact in this cell fate determination pathway. The somatostatin-producing δ cell populations are unaffected in both single knockouts and remained unchanged in the Nkx2.2−/−;NeuroD1−/− DKO mice (Figure 4e).
The rescued glucagon-producing cell population in the Nkx2.2−/−;NeuroD1−/− DKO mice express the α cell-specific factor, Brn4
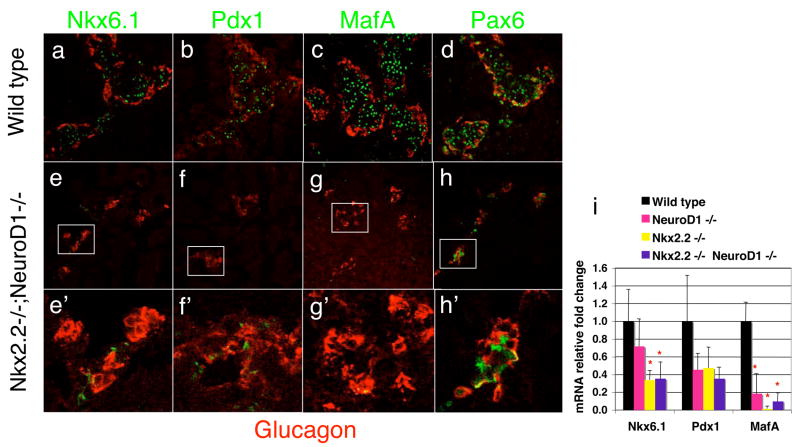
In the Nkx2.2−/−;NeuroD1−/− DKO mice, approximately 50% of the glucagon-producing cells are recovered compared with the Nkx2.2−/− mice and we have demonstrated that the majority of these cells do not co-express either ghrelin or the other islet hormones (Figure 4 and data not shown). To further characterize the glucagon-expressing cells recovered in the Nkx2.2−/−;NeuroD1−/− DKO, we assessed the expression of Brn4, a well-characterized α cell specific marker (Heller et al., 2004; Jensen et al., 2000). Similar to the recovery of glucagon expression in these mice, Brn4 expression was partially restored in the Nkx2.2−/−;NeuroD1−/− DKO (Figure 5 and Supplemental Figure 2). We also analyzed the expression of Arx and Irx2, two recently described α cell factors (Collombat et al., 2003; Heller et al., 2005). Unexpectedly, we observed a significant upregulation of Arx expression and relatively little change of Irx2 expression in the Nkx2.2−/− pancreata, in which α cells are almost completely absent and the majority of endocrine cells produce ghrelin (Supplemental Figure 1 and 2). The expression of both these factors is moderately reduced in the Nkx2.2−/−;NeuroD1−/− DKO compared to the Nkx2.2−/− mice, which again does not correlate with the comparative increase in α cells we observe compared to either single knockout. To better understand these surprising results, we assessed the cell types expressing Arx protein (antibodies against Irx2 were unavailable). Interestingly, in the Nkx2.2−/− mice, the majority of Arx expressing cells did not co-express glucagon (Supplemental figure 3g and 3k) and were located in the region of the islet containing large numbers of ghrelin-producing cells (Supplemental figure 3c and 3g; the Arx and ghrelin antibodies are both generated in rabbit, which precluded double labeling of the two proteins). In the Nkx2.2−/−;NeuroD1−/− DKO, there still remained a number of Arx+, glucagon- cells present (Supplemental figure 3m); however, significant numbers of the recovered glucagon-positive cells now co-expressed Arx (Supplemental figure 3h and 3l). While the unexpected upregulation of Arx and unchanged Irx2 expression in the Nkx2.2−/− pancreata somewhat impair the use of these markers to assess the recovered α cell population in the Nkx2.2−/−;NeuroD1−/− DKO, it is clear that the newly recovered α cells do co-express Arx. Futhermore, the recovered α cells co-express Pax6, which is also highly expressed in α cells, and do not inappropriately co-express the β cell transcription factors Nkx6.1, Pdx1 and MafA (Figure 6). Consistent with the immunohistochemical analysis, quantitative PCR analysis indicated that Nkx6.1, Pdx1 and MafA expression is not recovered in Nkx2.2−/−;NeuroD1−/− DKO mice compared to the Nkx2.2−/− mice (Figure 6i). Therefore, it appears that a characteristic glucagon-producing α cell population expressing Brn4, Arx and Pax6 is recovered in the Nkx2.2−/−;NeuroD1−/−DKO mice. Further analysis of the ghrelin cell population in the Nkx2.2−/− mice will be necessary to assess why they apparently express the α cell marker, Arx.
Figure 5. Brn4 expression in recovered glucagon cells in Nkx2.2−/−;NeuroD1−/− DKO mice.
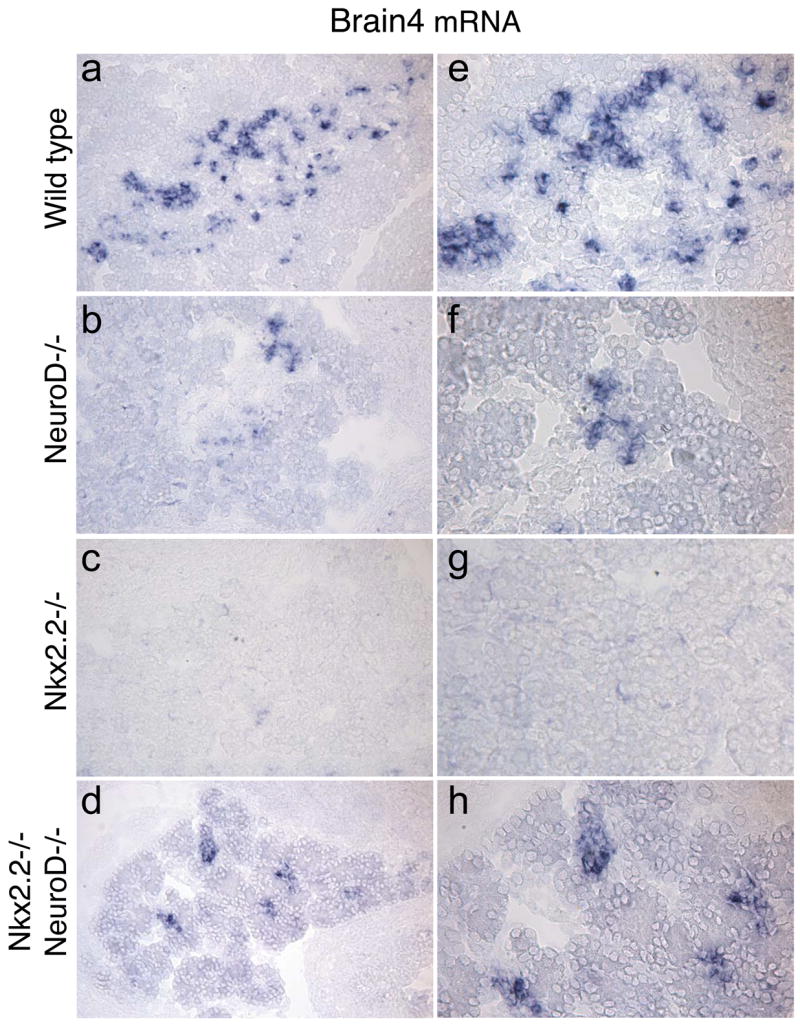
RNA in situ analysis for Brn4 expression on wild type (A, E), NeuroD1−/− (B, F), Nkx2.2−/− (C, G) and Nkx2.2−/−;NeuroD1−/− DKO (D, H) tissue. E – H are higher magnification images of staining in A–D.
Figure 6. Transcription Factor Profile of Nkx2.2−/−;NeuroD1−/− DKO mice.
Immunofluorescence images at P0 of wild type (A–D) and Nkx2.2−/−;NeuroD1−/− DKO (E–H) mice for glucagon (red: A–H), and Nkx6.1 (green: A, E), Pdx1 (green: B, F), MafA (green: C, G) and Pax6 (green: D, H). An enlarged image of panel D shows in wild type islets some alpha cells do not normally express Pax6 (arrow). Panels A–H are images taken on a Zeiss LSM 5 confocal microscope at 25x. E′–H′ are 40x images of the islets shown in E–H. Quantitative PCR results for fold change of Nkx6.1, Pdx1, and MafA in P0 pancreas of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice (I). n = 3–6 pancreas per genotype in qPCR. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an * compared to wild type and + compared to Nkx2.2 knockout phenotype.
Proglucagon-producing cells in the intestine of Nkx2.2−/− are also recovered in Nkx2.2−/−;NeuroD1−/−DKO mice
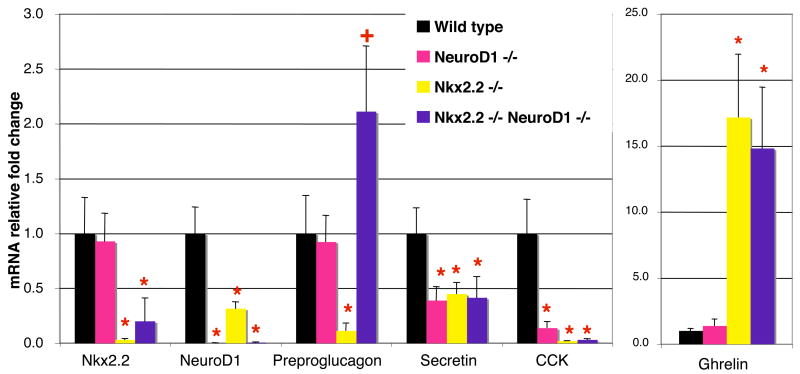
The differentiation of the enteroendocrine cell population in the intestine is regulated by many of the same transcription factors that regulate islet cell development. Ngn3−/− mice fail to generate both intestinal and pancreatic endocrine progenitor cells (Gradwohl et al., 2000; Jenny et al., 2002; Lee et al., 2002), and Pax4 and Pax6 affect the formation of subsets of endocrine cells in both organs (Larsson et al., 1998). Several enteroendocrine cell populations are also reduced in the Nkx2.2−/− and NeuroD1−/− mice (Figure 7, Desai et al, submitted, and (Naya et al., 1995). Similar to the islet phenotype, the reduction of intestinal endocrine cell populations in Nkx2.2−/− mice corresponds with an increase in ghrelin-producing cells (Figure 7 and Desai et al, submitted). On the other hand, NeuroD1−/− mice only lack cholecystokinin (CCK) and secretin-producing enteroendocrine cells in the intestine (Naya et al., 1995). Given the regulatory conservation that appears to exist between the intestinal and pancreatic endocrine populations, we assessed whether a functional interaction between Nkx2.2 and NeuroD1 also existed in the formation of the enteroendocrine cell populations both affected by Nkx2.2 and NeuroD1. We examined mRNA expression levels of CCK, secretin, proglucagon, and ghrelin in the Nkx2.2−/−;NeuroD1−/− DKO mice (Figure 7). In each single knockout, expression of CCK and secretin is reduced, but there is no additional reduction or rescue observed in the DKO mice (Figure 7 and data not shown). In contrast, proglucagon expression is significantly reduced and ghrelin is upregulated approximately 20 fold in the Nkx2.2−/−mice. These hormones are unaffected in the NeuroD1−/− mice (Figure 7). Strikingly, proglucagon expression is completely restored and ghrelin levels are decreased in the Nkx2.2−/−;NeuroD1−/− DKO, similar to what we observed in the pancreas. Our results indicate that Nkx2.2 and NeuroD1 genetically interact to regulate the differentiation of both islet and intestinal proglucagon- and ghrelin- expressing endocrine populations.
Figure 7. Preproglucagon-producing cells in the intestine also recovered in Nkx2.2−/−;NeuroD1−/−DKO mice.
Quantitative PCR of Nkx2.2, NeuroD1, preproglucagon, secretin, CCK and ghrelin mRNA in P0 intestine of wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice. N = 3–6 pancreas per genotype in qPCR. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an * compared to wild type phenotype.
Discussion
In this study, we created NeuroD1 and Nkx2.2 DKO mice to understand the relative roles of these two essential transcription factors in islet development. Surprisingly, we demonstrated that there is a unique genetic interaction between Nkx2.2 and NeuroD1 during α, PP and ε cell differentiation that is only revealed when Nkx2.2 and NeuroD1 are simultaneously deleted from the developing pancreas. These observations provide the first evidence that these two essential regulatory factors can form cell type-dependent functional interactions to differentially regulate the individual islet cell fates. Interestingly, it also appears that Nkx2.2 and NeuroD1 functionally cooperate to specify the fate decision between L cells and ghrelin cells in the intestine. In addition, these studies have uncovered a novel role for NeuroD1 in directing early α, PP and ε cell fate decisions that is distinct from its role in later stages of α cell development and β cell differentiation and survival.
The majority of recent pancreatic islet research has focused on β cell development due to the importance of β cells in islet function, as well as their relative abundance compared to the other islet cell types. As a result of this extensive body of research, a more defined picture of β cell differentiation is beginning to emerge. Specifically, many β cell regulators and markers have been identified and distinct stages of β cell differentiation have been characterized (Cerf et al., 2005; Doyle et al., 2007; Holland et al., 2005; Lantz and Kaestner, 2005; Nishimura et al., 2006; Sosa-Pineda, 2004). The Nkx2.2 and NeuroD1 single knockout β cell phenotypes suggest that Nkx2.2 and NeuroD1 function at different stages of β cell differentiation; Nkx2.2 is required for the initial specification of all β cells, while NeuroD1 appears to be required for late stage differentiation and maintenance. Furthermore, NeuroD1 expression is reduced in the Nkx2.2 knockout mice, whereas Nkx2.2 expression is unchanged in the NeuroD1 mutant, supporting the idea that Nkx2.2 functions upstream of NeuroD1 in β cells. Consistent with this, we do observe a significant reduction of NeuroD1-expressing cells as early as e12.5 (prior to the formation of many of the individual islet cell types) and there appear to be fewer cells expressing high levels of NeuroD1. At this point, however, we cannot rule out that the down regulation of NeuroD1 is merely secondary to the loss of specific islet cell types, especially late in gestation. The fact that the Nkx2.2−/−;NeuroD1−/− DKO mice display an Nkx2.2 KO phenotype also suggests that Nkx2.2 is epistatic to NeuroD1, the caveat being that the complete absence of β cells may preclude observing phenotypes associated with the NeuroD1 deletion. A conditional mutation of Nkx2.2 is being developed to resolve these issues and to further explore the relationship between NeuroD1 and Nkx2.2 specifically in the β cell.
In contrast to the β cells, much less is understood about the regulation of the non-β cell lineages. Although glucagon-producing α cells are also essential for proper islet function, little is known about the regulation of α cell formation and the stages of α cell differentiation. The restoration of the glucagon-producing α cell population in the Nkx2.2−/−;NeuroD1−/− DKO mice, compared to the Nkx2.2−/− mice implicates NeuroD1 as an important regulator of α cell determination. While these studies are beginning to clarify the role of NeuroD1 in α cell fate determination, the molecular mechanism by which NeuroD1 operates remains unclear. NeuroD1 is present in glucagon-producing α cells throughout embryogenesis; however, the NeuroD1−/− mice do not display an α cell phenotype until late in gestation (Chu et al., 2001; Naya et al., 1997). In contrast, NeuroD1 activity appears to be required to maintain repression of the α cell fate in the absence of Nkx2.2. Furthermore, incremental reductions in NeuroD1 activity, as seen in the NeuroD1 heterozygous mice, can restore to an intermediate level the α cell defects associated with loss of Nkx2.2. We propose that Nkx2.2 may be responsible for the normal repression of NeuroD1 function in early α cells, possibly through activation of a negative regulator of NeuroD1, such as an Id protein (Norton, 2000). Alternatively, Nkx2.2 and NeuroD1 may function in parallel to modulate the formation of α cells during islet development.
An unexpected finding emerged from our characterization of the glucagon-positive cell population in Nkx2.2−/−;NeuroD1−/− DKO mice with regards to α cell marker expression in the Nkx2.2−/− ghrelin cell population. While Brn4 expression levels corresponded to the presence and absence of glucagon-producing α cells in each of the genotypes analyzed, Arx and Irx2 expression levels corresponded more closely with the Nkx2.2−/− ghrelin cell population. In our previous analyses of the Nkx2.2−/− ghrelin cells, these two transcription factors had not yet been identified as α cell markers and were not assessed. It is possible, that the ghrelin-producing cells in the Nkx2.2−/− mice are α cells that merely misexpress ghrelin, however none of the ghrelin cells in the Nkx2.2−/− islets co-express glucagon or Brn4. It is also possible that Arx and Irx2 are also expressed in wild type ghrelin cells, a hypothesis we could not test due to the lack of antibodies (Irx2) or incompatibility of antibodies (Arx); however, this explanation is not supported by the phenotype of the Arx null mice in which there is a loss of glucagon cells and the glucagon+ ghrelin+ population, but no effect on the ghrelin ε cells (Collombat et al., 2003; Heller et al., 2005). An intriguing possibility is that the ghrelin-producing cells that form in the Nkx2.2−/− mice represent a novel bipotential intermediate cell that expresses Arx, but in the absence of Nkx2.2 cannot inactivate ghrelin and initate the expression of Brn4 and glucagon to allow mature α cell formation. If this is the case, then the simultaneous loss of NeuroD1 in the Nkx2.2−/− mice may function to partially suppress this block and allow normal differentiation of α cells to proceed. While we have yet to resolve this issue and the mechanism underlying the phenotypes we observe, it does appear that the loss of NeuroD1 in the Nkx2.2−/− mice allows the restoration of apparently normal α cells that express Brn4 and Arx.
Previous studies of the NeuroD1−/− mice have suggested that NeuroD1 is required for the terminal differentiation of α and β cells; at birth both these cell populations are significantly depleted compared to wild type mice (Chu 2001, Naya 1997). While the absence of normal numbers of β cells appears to be caused by increased levels of apoptosis, the mechanism underlying the decrease in α cell numbers was not determined, possibly due to the relatively small number of α cells available for analysis (Naya et al., 1997). We have demonstrated that the elimination of NeuroD1 in the Nkx2.2−/− mice restores α cell formation during the secondary transition; however, α cell numbers never return to wild type levels. Based on the previously characterized role for NeuroD1 in the terminal differentiation of α cells (Naya et al., 1997), we hypothesize that NeuroD1 also plays a later role in α cell survival that is independent of Nkx2.2. Alternatively, an additional complex relationship may exist between NeuroD1 and Nkx2.2 in maintaining the α cell population, and the loss of Nkx2.2 may partially rescue the survival of NeuroD1-deficient α cells. The generation of temporal and cell type specific knockout alleles of Nkx2.2 and NeuroD1 will allow us to explore these questions further.
In summary, a complex and overlapping regulatory network that requires Nkx2.2 and NeuroD1 determines the cell fate decisions in the islet and the intestine. A large number of mutational studies from many labs have uncovered many of the regulatory factors that contribute to these processes, however it is likely that additional regulatory factors and/or novel roles for known factors have yet to be identified. Using genetic epistasis analysis, we have demonstrated that Nkx2.2 and NeuroD1 functionally cooperate during islet cell differentiation and we have uncovered a novel function for NeuroD1 in the regulation of α, PP and ε cell fate determination suggesting there exists a unique interaction between Nkx2.2 and NeuroD1 in these early cell fate decisions. The corresponding decrease in ε cell numbers that occurs with the increase in α cells in the Nkx2.2−/−;NeuroD1−/− DKO mice provides further evidence that Nkx2.2 regulates the cell fate choice between these two islet lineages and implicates NeuroD1 as mediator of these important developmental fate decisions. NeuroD1 and Nkx2.2 also appear to interact in their regulation of PP cell fate determination. It is clear from this analysis that simple single gene mutations may not uncover the full functional repertoire of known transcription factors or the full complexities of these regulatory pathways. Additional studies will be required to determine epistatic relationships between known and novel islet factors to gain a complete understanding of their relative functions in directing pancreatic islet cell fates.
Supplementary Material
01. Supplemental Figure 1. RNA in situ analysis of Arx and Irx2.
RNA in situ analysis of e16.5 embryos. a–d) Arx mRNA expression in wild type (a), NeuroD1−/− (b), Nkx2.2−/− (c), and Nkx2.2−/−;NeuroD1−/− DKO mice (d). e–h) Irx2 mRNA expression in wild type (e), NeuroD1−/− (f), Nkx2.2−/− (g), and Nkx2.2−/−;NeuroD1−/− DKO mice (h). Tissue in e-h are co-stained with anti-glucagon antibody; α cells are only apparent in the wild type and Nkx2.2−/−;NeuroD1−/− DKO mice. 40x magnification..
02. Supplemental Figure 2. Real time PCR analysis of α cell marker expression.
Real time PCR results for fold change of Brn4, Arx and Irx2 mRNA in e18.5 pancreas of wild type (n=5), NeuroD1−/− (n=6), Nkx2.2−/− (n=5), Nkx2.2−/−;NeuroD1−/−, and Nkx2.2−/−NeuroD1+/− (n=8) mice. All samples were normalized to Gapdh. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an ** compared to wild type.
03. Supplemental Figure 3. Immunofluorescence analysis of Arx protein expression.
A–d) Co-staining of glucagon (blue) and ghrelin (red) on tissue isolated from pancreas of newborn (P0) wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice. E–h) Adjacent sections from each genotype co-stained with Arx (green) and glucagon (blue). The boxes outline regions shown in higher magnification below (panels i-m). l) shows a cluster of glucagon+, Arx+ cells. m) shows a cluster of glucagon-, Arx+ cells.
Acknowledgments
We thank Morgan Singleton for her help in maintaining the mouse colony, Tanya McNeal for initial help in breeding and harvesting of the double knockout embryos, and Angela Minic for her expertise and assistance with the real time PCR. We thank Dr. Lee Niswander for use of the Zeiss confocal microscope. We would also like to thank Min Han for helpful discussions regarding the interpretation of the double knockout phenotypes and Kristin Artinger, Michelle Doyle, Teresa Mastracci, and Keith Anderson for critical reading of this manuscript. This work was supported by the NIDDK: U01 DK072504 (LS and JL), ADA RRG (LS), and NIGMS F31-GM75456-01 (CC). We would also like to acknowledge support from the DERC (NIH P30 DK57516) and the UCHSC Cancer center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cerf ME, et al. Transcription factors, pancreatic development, and beta-cell maintenance. Biochem Biophys Res Commun. 2005;326:699–702. doi: 10.1016/j.bbrc.2004.10.217. [DOI] [PubMed] [Google Scholar]
- Chu K, et al. BETA2 and pancreatic islet development. Recent Prog Horm Res. 2001;56:23–46. doi: 10.1210/rp.56.1.23. [DOI] [PubMed] [Google Scholar]
- Collombat P, et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–80. doi: 10.1242/dev.01870. [DOI] [PubMed] [Google Scholar]
- Collombat P, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K, et al. Gata6 is an important regulator of mouse pancreas development. Dev Biol. 2006;298:415–29. doi: 10.1016/j.ydbio.2006.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MJ, et al. Nkx2.2-repressor activity is sufficient to specify {alpha}-cells and a small number of {beta}-cells in the pancreatic islet. Development. 2007;134:515–23. doi: 10.1242/dev.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, et al. Persistent expression of HNF6 in islet endocrine cells causes disrupted islet architecture and loss of beta cell function. Development. 2000;127:2883–95. doi: 10.1242/dev.127.13.2883. [DOI] [PubMed] [Google Scholar]
- Gasa R, et al. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci U S A. 2004;101:13245–50. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, et al. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habener JF, et al. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–34. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- Heller RS, et al. Genetic determinants of pancreatic epsilon-cell development. Dev Biol. 2005;286:217–24. doi: 10.1016/j.ydbio.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Heller RS, et al. The role of Brn4/Pou3f4 and Pax6 in forming the pancreatic glucagon cell identity. Dev Biol. 2004;268:123–34. doi: 10.1016/j.ydbio.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Henseleit KD, et al. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132:3139–49. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Holland AM, et al. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54:2586–95. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- Holland AM, et al. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–41. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–54. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M, et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. Embo J. 2002;21:6338–47. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, et al. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49:163–76. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- Jonsson J, et al. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Lantz KA, Kaestner KH, et al. Winged-helix transcription factors and pancreatic development. Clin Sci (Lond) 2005;108:195–204. doi: 10.1042/CS20040309. [DOI] [PubMed] [Google Scholar]
- Larsson LI, et al. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mech Dev. 1998;79:153–9. doi: 10.1016/s0925-4773(98)00182-8. [DOI] [PubMed] [Google Scholar]
- Lee CS, et al. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–97. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, et al. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–52. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–34. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, et al. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–19. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–39. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JD, et al. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–95. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Petri A, et al. The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J Mol Endocrinol. 2006;37:301–16. doi: 10.1677/jme.1.02096. [DOI] [PubMed] [Google Scholar]
- Pictet R, Rutter WJ. Development of the embryonic endocrine pancreas. Williams and Wilkins; Washington, DC: 1972. [Google Scholar]
- Prado CL, et al. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–9. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–40. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM, et al. Programming of the pancreas. Mol Cell Endocrinol. 2001;185:99–108. doi: 10.1016/s0303-7207(01)00628-1. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, et al. The gene Pax4 is an essential regulator of pancreatic beta-cell development. Mol Cells. 2004;18:289–94. [PubMed] [Google Scholar]
- Sosa-Pineda B, et al. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- St-Onge L, et al. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–9. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Sussel L, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–21. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. Supplemental Figure 1. RNA in situ analysis of Arx and Irx2.
RNA in situ analysis of e16.5 embryos. a–d) Arx mRNA expression in wild type (a), NeuroD1−/− (b), Nkx2.2−/− (c), and Nkx2.2−/−;NeuroD1−/− DKO mice (d). e–h) Irx2 mRNA expression in wild type (e), NeuroD1−/− (f), Nkx2.2−/− (g), and Nkx2.2−/−;NeuroD1−/− DKO mice (h). Tissue in e-h are co-stained with anti-glucagon antibody; α cells are only apparent in the wild type and Nkx2.2−/−;NeuroD1−/− DKO mice. 40x magnification..
02. Supplemental Figure 2. Real time PCR analysis of α cell marker expression.
Real time PCR results for fold change of Brn4, Arx and Irx2 mRNA in e18.5 pancreas of wild type (n=5), NeuroD1−/− (n=6), Nkx2.2−/− (n=5), Nkx2.2−/−;NeuroD1−/−, and Nkx2.2−/−NeuroD1+/− (n=8) mice. All samples were normalized to Gapdh. Significance was calculated using the two-tailed student t-test with unequal variance. P value of <0.05 is indicated by an ** compared to wild type.
03. Supplemental Figure 3. Immunofluorescence analysis of Arx protein expression.
A–d) Co-staining of glucagon (blue) and ghrelin (red) on tissue isolated from pancreas of newborn (P0) wild type, NeuroD1−/−, Nkx2.2−/−, and Nkx2.2−/−;NeuroD1−/− DKO mice. E–h) Adjacent sections from each genotype co-stained with Arx (green) and glucagon (blue). The boxes outline regions shown in higher magnification below (panels i-m). l) shows a cluster of glucagon+, Arx+ cells. m) shows a cluster of glucagon-, Arx+ cells.