T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant β-catenin (original) (raw)
Abstract
N-terminal mutations in β-catenin that inhibit β-catenin degradation are found in primary tumors and cancer cell lines, and increased β-catenin/T cell factor (TCF)-activated transcription in these cells has been correlated with cancer formation. However, the role of mutant β-catenin in cell transformation is poorly understood. Here, we compare the ability of different N-terminal mutations of β-catenin (ΔN131, ΔN90, ΔGSK) to induce TCF-activated transcription and anchorage-independent growth in Madin–Darby canine kidney epithelial cells. Expression of ΔN90 or ΔGSK β-catenin increased TCF-activated transcription but did not induce significant anchorage-independent cell growth. In contrast, deletion of the α-catenin-binding site in ΔN131 β-catenin reduced TCF-activated transcription, compared with that induced by ΔN90 or ΔGSK β-catenin, but significantly enhanced anchorage-independent cell growth.
The cytoplasmic protein β-catenin has important structural and signaling functions (1–3). β-Catenin binds to the cytoplasmic domain of the cadherin family of Ca2+-dependent, homophilic cell–cell adhesion proteins and to α-catenin, which, in turn, mediates the interaction of the cadherin/catenin complex with the actin cytoskeleton (4–6). This linkage to the actin cytoskeleton is essential for the adhesion function of cadherins (7, 8). β-Catenin is also a component of the Wnt signaling pathway and activates gene expression in response to Wnt signaling by binding to transcription factors of the T cell factor (TCF) family (2, 9–12). Wnt signaling inhibits the degradation of cytosolic β-catenin, resulting in an increase in the cytosolic level of β-catenin and, thereby, the amount of β-catenin available to bind TCF and activate transcription (13–15). Thus, degradation of cytosolic β-catenin is an essential regulatory mechanism to keep the cellular β-catenin/TCF activity low. β-catenin degradation involves the ubiquitin–proteasome pathway (16) and is mediated by the adenomatous polyposis coli (APC) protein (17), the product of a tumor-suppressor gene mutated in somatic and hereditary colorectal cancer (18).
Abnormalities in the regulation of structural and signaling functions of β-catenin have been implicated in tumorigenesis. Loss of cadherin function or a β-catenin mutation that decreases cadherin-mediated adhesion have been correlated with cell dedifferentiation and high invasiveness (19, 20). In addition, mutations in APC protein or mutations in β-catenin that inhibit β-catenin degradation have been found in a variety of solid tumors (21–24). However, mutations in APC protein, which lead to an increase in β-catenin/TCF activity, are also found in well differentiated, benign adenomas (18, 25). Thus, it appears that mutations that cause deregulation of β-catenin/TCF-activated transcription define early events in tumorigenesis, whereas those that cause a loss of cadherin function define later stages in tumorigenesis (18, 20, 25, 26). These observations and conclusions are based on analysis of different tissues and cells as well as on heterogeneous levels of β-catenin mutants. Here, we have taken a systematic approach to this problem by examining the ability of equivalent levels of specific β-catenin mutants to induce TCF-activated transcription and transformation within the same epithelial background.
MATERIALS AND METHODS
Cell Lines and cDNA Constructs.
The human colon cancer cell line HCT116 was obtained from the American Type Culture Collection (CCL 247). Parental Madin–Darby canine kidney (MDCK) cell line T23 and MDCK clones ΔN90-A and ΔN131-D have been described previously (27). Cultures were passaged without doxycycline (Dox) or expression of mutant proteins was repressed for the times indicated by the addition of 20 ng/ml Dox (Sigma) to the culture medium. The construction of the expression vectors for ΔN90 and ΔN131 β-catenin has been described previously (27). A short N-terminal sequence for ΔGSK (glycogen synthase kinase) β-catenin was constructed by PCR. The oligonucleotide defining the start site of the cDNA, 5′-CCGATATCGAATTCCCGCGGCCACCATGGCTACTCAAGCTGACCTGATG, contained a Kozak consensus sequence for translation initiation (28). The mutations Ser-33, Ser-37, Thr-41, and Ser-45 → Ala were introduced with the oligonucleotide 5′-CATCTTCTTCCTCAGGGTTGCCCTTGCCACTCAGGG_C_AGGAGCTGTGG_C_GGTGGCACCAG_C_ATGGATTCCAG_C_ATCCAAG. Underlined sequences are derived from the β-catenin cDNA, and introduced point mutations are shown in italics. The N-terminal _Sac_II/_Bsu_36I fragment of the full-length KT3-tagged wild-type β-catenin (β-catenin*) cDNA in pUDH10–3 (27) was replaced with the N-terminal ΔGSK β-catenin fragment to obtain the expression vector pUDH10–3/ΔGSK. The original start site in the full-length KT3-tagged β-catenin* cDNA (27) was modified by PCR, with the oligonucleotide defining the start site in ΔGSK β-catenin to introduce the Kozak consensus sequence. All PCR-derived sequences were confirmed by sequencing. Expression vectors pUDH10–3/ΔGSK and pUDH10–3/β-catenin* were transfected into MDCK line T23 as described previously (27).
Antibodies.
Mouse mAbs against the carboxyl-terminal 212 aa of β-catenin (β-cat.C) and the cytoplasmic domain of E-cadherin were obtained from Transduction Laboratories (Lexington, KY). A mouse mAb KT3 against a simian virus 40 large T antigen epitope was kindly provided by G. Walter (University of California, San Diego) and has been described previously (29). Polyclonal antisera against α-catenin and the cytoplasmic domain of E-cadherin have been described previously (30, 31). Polyclonal antisera against APC protein were kindly provided by I. Näthke (University of Dundee, Dundee, U.K.) and P. Polakis (ONYX Pharmaceuticals, Richmond, CA) (32). Affinity-purified antisera were used as indicated.
Immunoprecipitation and Immunoblotting.
Cell lysates and immunoprecipitations were prepared as described previously (27). Protein concentrations were determined by using the BCA protein assay reagent kit (Pierce), and equal amounts of protein were used for immunoprecipitation or separated by SDS/PAGE in 7.5% polyacrylamide gels. Proteins were immunoblotted and immunoblots were analyzed as described previously (27).
Indirect Immunofluorescence Microscopy.
For KT3/APC protein double-immunofluorescence, 2 × 105 cells were plated onto collagen-coated coverslips in 35-mm tissue culture dishes for 48 hr. Cells were washed once in Dulbecco’s PBS, fixed for 5 min at −20°C in precooled methanol, and processed as described previously (27).
Size-Exclusion Chromatography.
Cells were extracted and processed as described previously with minor modifications (33). Briefly, cells were extracted with DHE buffer (10 mM Hepes, pH 7.5/100 mM NaCl/0.5% Nonidet P-40) containing protease and phosphatase inhibitors and loaded on a Superose 6 HR 10/30 column (Pharmacia) equilibrated with DHE buffer. Proteins were eluted at a flow rate of 0.25 ml/min at 4°C, and 0.5-ml fractions were collected. Fractions were analyzed by Western blotting, and blots were quantitated as described previously (33).
Reporter Gene Assay.
Parental MDCK cells and MDCK clones expressing mutant β-catenin were cultured 3 days without or with Dox. Cells (2 × 105) were plated into 35-mm culture dishes without or with Dox. Sixteen hours after plating, cells were cotransfected with 1 μg of pTOPFLASH/0.5 μg of pSV-β-galactosidase and 10 μl of Lipofectamine (GIBCO/BRL). The vector pSV-β-galactosidase (Promega) was used to estimate transfection efficiencies. Cultures were kept without or with Dox during and after transfection and were extracted 48 hr after transfection. Luciferase and β-galactosidase activities were determined on a Turner TD-20e Luminometer by using the Dual-Light System (Tropix, Bedford, MA) as described by the manufacturer. Luciferase activities were corrected for differences in transfection efficiencies estimated from galactosidase activities in the same samples. For comparison of TCF-activated transcription in HCT116 and MDCK cells, cells were treated as described above but without addition of Dox to the culture medium. Cells were transfected with 1 μg of pTOPtkFLASH or 1 μg of control vector pFOPtkFLASH/0.5 μg of pSV-β-galactosidase and 10 μl of Lipofectamine. In control vector pFOPtkFLASH, the TCF-binding sites are replaced by a mutant motif (12). pTOPFLASH and pTOPtkFLASH/pFOPtkFLASH were kindly provided by M. van de Wetering and H. Clevers (University of Utrecht, Utrecht, The Netherlands).
Soft Agar Assay.
MDCK clones were passaged for a period of 2–4 weeks without Dox in DMEM supplemented with 10% FCS and selection drug, 300 μg/ml hygromycin. Parental cells and HCT116 were passaged for the same period of time without hygromycin. Expression of β-catenin mutant proteins was repressed in part of the cultures for 2 days before plating into soft agar. Five thousand cells were resuspended in 1 ml of DMEM/10% FCS/0.4% Sea Plaque GTG agarose (FMC) without or with 20 ng/ml Dox and plated on top of 1 ml of DMEM/10% FCS/0.8% agarose without or with 20 ng/ml Dox in 35-mm dishes. Samples were processed in triplicate for every experiment. After cells were incubated for 2 weeks at 37°C/5% CO2, colonies were stained overnight with 1 mg/ml _p_-iodonitrotetrazolium violet (Sigma), and four random areas of 11 mm2 were photographed from each dish by using a Nikon SMZ-U dissection microscope and a Nikon FX-35DX camera. Images were analyzed and colony size was measured with nih image.
RESULTS AND DISCUSSION
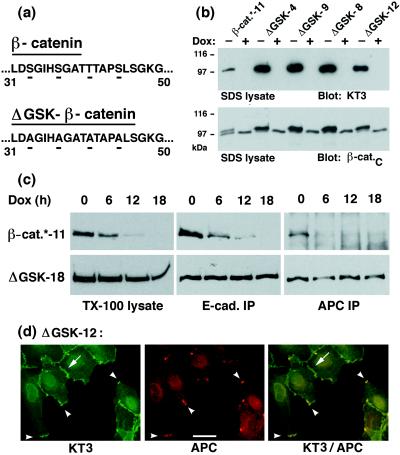
We showed previously that N-terminal deletions in β-catenin (the first 90 aa in ΔN90 or the first 131 aa in ΔN131 β-catenin) stabilized the protein in MDCK cells (27). The N terminus of β-catenin contains a recognition site for the serine/threonine kinase GSK-3β, which regulates β-catenin stability in Drosophila and Xenopus (34, 35). GSK-3β has been shown to bind to the APC/β-catenin complex in mammalian cells and to phosphorylate APC and β-catenin (35, 36). Furthermore, mutations in the potential GSK-3β phosphorylation site in β-catenin have been shown to stabilize β-catenin in Xenopus and in mammalian cells (23, 24, 35). To examine whether this site regulates β-catenin stability in MDCK cells, four serine and threonine residues in this site were changed to alanine (ΔGSK β-catenin; Fig. 1a), and MDCK cells were transfected stably with KT3-tagged ΔGSK β-catenin or with KT3-tagged β-catenin* under control of the tetR (Dox)-repressible transactivator (27, 37). The level of mutant β-catenin in clones expressing ΔGSK β-catenin was higher than that of wild-type β-catenin* in control clones (Fig. 1b). To examine whether the stability of ΔGSK β-catenin was increased compared with wild-type β-catenin*, expression of KT3-tagged β-catenin was repressed by addition of Dox to the cultures for 0, 6, 12, or 18 hr and the amount of protein remaining at subsequent times was determined (Fig. 1c). In Triton X-100 lysates, the amount of wild-type β-catenin* was reduced to 12% of the original level after 12-hr incubation with Dox, whereas the amount of ΔGSK β-catenin was reduced to 81% (Fig. 1c). A significant increase in relative stability of ΔGSK β-catenin compared with wild-type β-catenin* also was detected in the E-cadherin and APC protein complexes. For example, wild-type β-catenin* was removed from the APC protein complex within 6 hr of Dox addition, whereas there was little decrease in the amount of bound ΔGSK β-catenin after 18 hr (Fig. 1c). Immunofluorescence microscopy revealed that ΔGSK β-catenin localized to cell–cell contacts and colocalized with APC protein in punctate aggregates at the tip of membrane extensions (Fig. 1d; see also ref. 27). In summary, these results show that serine/threonine residues in the N terminus of β-catenin regulate both overall β-catenin stability and β-catenin stability in E-cadherin and APC protein complexes.
Figure 1.
Expression, stability, and subcellular localization of ΔGSK β-catenin in MDCK cells. (a) Illustration of the amino acid changes in the sequence of ΔGSK β-catenin. Serine/threonine residues Ser-33, Ser-37, Thr-41, Ser-45 in the putative GSK-3β phosphorylation site of β-catenin were changed to Ala (for details, see Material and Methods). (b) Dox-repressible expression of tagged wild-type β-catenin* and ΔGSK β-catenin in independently isolated MDCK clones (designated by numbers). Cells were cultured for 4 days without or with Dox (−/+ Dox) and extracted with 1% SDS. Fifteen micrograms of the protein lysates were subjected to SDS/PAGE and immunoblotted with the tag-antibody KT3 or mAb β-cat.C. Molecular mass standards are indicated in kDa. (c) MDCK clones were cultured for 0, 6, 12, or 18 hr with Dox and extracted with 1% Triton X-100 lysis buffer. Protein lysates were divided: one part was subjected to SDS/PAGE and immunoblotted with mAb KT3 (TX-100 lysates), another was immunoprecipitated with E-cadherin antiserum (E-cad IP), and another was immunoprecipitated with APC antiserum (APC IP). Equivalent fractions of the immunoprecipitates were subjected to SDS/PAGE and immunoblotted with mAb KT3. Three times more of the APC immunoprecipitates from β-catenin*-11 lysates were used than from the ΔGSK-18 lysates, and the blot for β-catenin* was exposed 10 times longer. The rate and efficiency of Dox repression of gene expression is very similar in different MDCK clones (27). Therefore, differences in the amounts of protein remaining after addition of Dox indicate the relative stability of each protein. (d) MDCK clones were double-stained with mAb KT3 against the epitope tag in ΔGSK β-catenin and with affinity-purified antiserum against APC protein. ΔGSK β-catenin localized to sites of cell–cell contact (arrow) and colocalized with APC protein in clusters at the tip of membrane extensions (arrowheads). (Bar = 20 μm.)
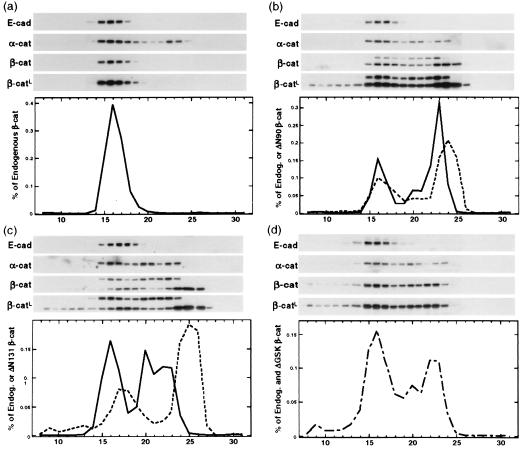
Wnt signaling during embryogenesis stabilizes the cytosolic pool of β-catenin, and this pool has been implicated in TCF-activated transcription (11–14) and induction of cell transformation (24, 33). To examine different β-catenin pools, we used size-exclusion chromatography to separate β-catenin complexes from MDCK clones expressing N-terminal-deleted (ΔN90, ΔN131) or ΔGSK β-catenin. In parental MDCK cells (33) and in MDCK clones in which expression of mutant β-catenins was repressed (+Dox), >99% of endogenous wild-type β-catenin coeluted with cadherin in a high-molecular-mass complex (Fig. 2a, fractions 14–19; shown for clone ΔN90-A/+Dox; see also ref. 33). ΔN90 and ΔN131 β-catenin eluted in two major peaks (Fig. 2 b and c); 35% coeluted with cadherin in a high-molecular-mass peak (fractions 14–19 for ΔN90 and 15–21 for ΔN131), and 55% (ΔN90) to 60% (ΔN131) eluted in a cadherin-free, low-molecular-mass peak (fractions 21–26 for ΔN90 and 22–27 for ΔN131). Expression of mutant β-catenins also resulted in the redistribution of endogenous β-catenin from the cadherin-associated pool into the low-molecular-mass pool (Fig. 2 a_–_c). In these cells, 55% (ΔN90-A) and 30% (ΔN131-D) of endogenous β-catenin was redistributed into the low-molecular-mass pool (fractions 21–27). Likewise, expression of ΔGSK β-catenin caused accumulation of 30% total β-catenin in the low-molecular-mass pool (Fig. 2d). However, the distributions of endogenous β-catenin and ΔGSK β-catenin could not be distinguished because of their similar electrophoretic mobilities. In longer exposures of the β-catenin immunoblots, mutant β-catenins, but not endogenous β-catenin, were detected in very high molecular mass fractions 8–10 (Fig. 2, β-catL), which contain APC protein (33), which correlates with high stability of mutant β-catenins in the APC complex (see Fig. 1 c and d for ΔGSK and ref. 27 for ΔN90 and ΔN131). In control cells, the major pool of α-catenin (75%) coeluted with E-cadherin and β-catenin in fractions 14–19, and the remainder eluted in low-molecular-mass fractions 22–25 (Fig. 2a, ΔN90/+Dox; see also ref. 33). This cadherin/β-catenin-free pool of α-catenin could be available to bind additional β-catenin because some α-catenin redistributed with β-catenin into a third, intermediate-molecular-mass peak (fractions 19–21) in cells expressing mutant β-catenin (Fig. 2 b_–_d). This third peak was found in the α-catenin blots from lysates of mutant cells (Fig. 2 b_–_d) and in the elution profiles of endogenous β-catenin (solid line in Fig. 2b) and of ΔN90 β-catenin (dotted line in Fig. 2b) in cell line ΔN90-A, of endogenous β-catenin in cell line ΔN131-D (solid line in Fig. 2c), and of total (endogenous and ΔGSK) β-catenin in cell line ΔGSK-4 (dashed line in Fig. 2d). This third peak was not found in the elution profile of ΔN131-β-catenin (dotted line in Fig. 2c), which is consistent with our previously published data that this mutant does not bind α-catenin (27). In summary, expression of any of the stabilized β-catenin mutants caused the accumulation of both endogenous and mutant β-catenin in cadherin-free, low-molecular-mass pools.
Figure 2.
Accumulation of low-molecular-mass β-catenin pools in MDCK clones expressing mutant β-catenin. Proteins extracted from MDCK clones ΔN90-A/+Dox (a) ΔN90-A/-Dox (b), ΔN131-D/-Dox (c), and ΔGSK-4/-Dox (d) were fractionated by Superose 6 size-exclusion chromatography as described in Materials and Methods. Equal amounts of fractions 8–31 were separated by SDS/PAGE and immunoblotted with antibodies specific for E-cadherin, α-catenin, and β-catenin; a second, longer exposure of the β-catenin immunoblots (β-catL) is shown to visualize small amounts of mutant β-catenin in high-molecular-mass fractions 8–10. The β-catenin antibody detected both endogenous β-catenin (single band in a and higher-molecular-mass bands in b and c) and ΔN90 or ΔN131 β-catenins (lower-molecular-mass bands in b and c). The single β-catenin band in d represents the total of both endogenous and ΔGSK β-catenin because the electrophoretic mobility of endogenous and ΔGSK β-catenin was very similar (see Fig. 1). β-Catenin immunoblots were quantified, and protein concentrations in each fraction were measured as percentage of all endogenous (solid lines in a_–_c), percentage of all mutant (dotted lines in b and c), and percentage of total (sum of endogenous and ΔGSK β-catenin; dashed line in d).
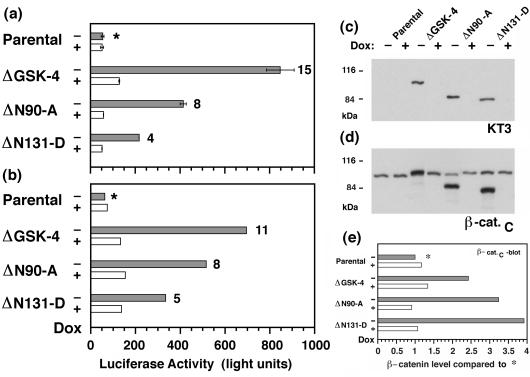
Increased amounts of β-catenin in a cadherin-free pool have been correlated with TCF-induced activation of transcription (24). To determine whether β-catenin in the mutant cell lines formed a complex with a TCF-like transcription factor, MDCK clones were transiently transfected with a luciferase reporter plasmid in which luciferase expression was controlled by a promoter containing multiple TCF-binding sites. TCF alone does not activate transcription from this promoter, but the β-catenin/TCF complex does (12). Expression of ΔGSK-, ΔN90-, or ΔN131-β-catenin increased TCF-activated transcription in MDCK cells compared with that in parental cells or clones in which expression of mutant β-catenins had been repressed (Fig. 3 a and b). Compared with parental cells, transcription was increased 11- to 15-fold in clone ΔGSK-4, 8-fold in clone ΔN90-A, and 4- to 5-fold in clone ΔN131-D (Fig. 3 a and b). Expression of mutant β-catenin caused the relocalization of endogenous β-catenin into cadherin-free, low-molecular-mass pools (Fig. 2) that may participate in activating transcription (38, 39). Therefore, we compared the levels of β-catenin/TCF activity in the different clones (Fig. 3 a and b) with the levels of total (sum of endogenous and mutant) β-catenin (Fig. 3 d and e). The hierarchy of transcriptional activation in the mutant cell lines (ΔGSK-4 > ΔN90-A > ΔN131-D; Fig. 3 a and b) was inverse to the total level of β-catenin in these cell lines (ΔN131-D > ΔN90-A > ΔGSK-4; Fig. 3 d and e). These data indicate that ΔN131 β-catenin is a weaker transcriptional activator than ΔGSK or ΔN90 β-catenin. These data are consistent with previously published results that the N-terminal region of β-catenin may contain a transcriptional activation domain and thus may contribute to the activity of the β-catenin/TCF complex (40).
Figure 3.
Increased TCF reporter activity in MDCK clones expressing mutant β-catenin. (a and b) Parental MDCK cells and MDCK clones expressing mutant β-catenin were preincubated for 3 days without or with Dox (−/+Dox) and then cotransfected with pTOPFLASH/pSV-β-galactosidase. Luciferase activities (relative light units) were corrected for differences in transfection efficiencies, which were estimated by β-galactosidase activities in the same samples. Data from two independent experiments are summarized in a and b; bars represent mean values from two independent samples (a) or one sample (b). Numbers at the head of the bars represent “fold activation” compared with luciferase activity in parental cultures without (−) Dox (∗) for each experiment. TCF-activated transcription of the luciferase reporter was higher in clones expressing the mutant β-catenins (−Dox, shaded bars) compared with cultures in which expression of mutant β-catenins was repressed (+Dox, open bars) or with parental cells. (c and d) Parallel cultures to those reported in a were extracted with 1% SDS, and 15-μg protein lysates were subjected to SDS/PAGE and immunoblotted with mAb KT3 (c) or mAb β-cat.C (d). Molecular mass standards are indicated in kDa. (e) Levels of total (sum of endogenous and mutant) β-catenin were quantified from the β-cat.C immunoblot. x axis represents “fold expression” compared with the level of endogenous β-catenin in the parental culture/−Dox (∗).
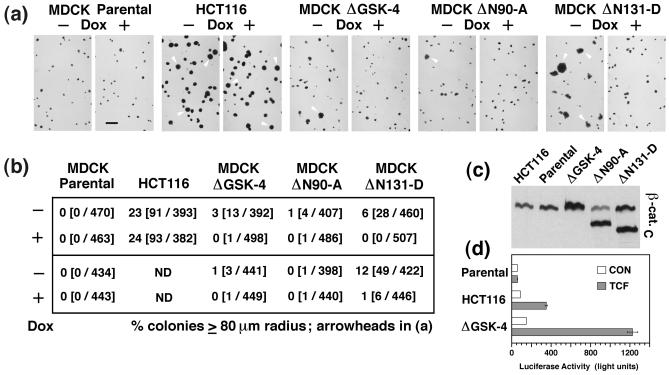
β-catenin mutants similar to the ones characterized here are found in a variety of tumors and cancer cell lines, and high β-catenin/TCF-mediated transcriptional activity in these cells has been correlated with cancer formation (21–24). To examine the transforming capacity of different β-catenin mutant proteins, we compared anchorage-independent growth of MDCK clones expressing mutant β-catenin with those of parental MDCK cells and the colorectal cancer cell line HCT116 (Fig. 4). HCT116 cells have a heterozygous β-catenin mutation that removes Ser-45 in the GSK-3β phosphorylation site (23), similar to the ΔGSK β-catenin mutation described here (Fig. 1a), have constitutive β-catenin/TCF-activated transcription, and are highly transformed (23). Approximately 25% of the HCT116 cells, but none of the parental MDCK cells, formed colonies with a radius ≥80 μm after 2 weeks of growth in soft agar (Fig. 4 a and b). MDCK clones ΔGSK-4 and ΔN90-A accumulated two to three times as much total β-catenin as HCT116 cells (Fig. 4c), and 30 and 55%, respectively, of their total β-catenin fractionated in the signaling active pool compared with only 15% in HCT116 cells (Fig. 2 and ref. 33). In addition, MDCK cells expressing ΔGSK β-catenin had a higher level of TCF-activated reporter transcription than HCT116 cells (Fig. 4d). Despite these high β-catenin levels and transcriptional activities, expression of ΔN90 or ΔGSK β-catenin only slightly increased the number of colonies with a radius ≥80 μm from 0% in control cultures (+Dox) to 1 and 3%, respectively, in cultures without Dox (Fig. 4). In contrast to ΔGSK and ΔN90 β-catenin, expression of ΔN131 β-catenin caused a lower level of TCF-activated transcription (Fig. 3 a and b) but an increase in the number of colonies with a radius ≥80 μm from ≈1% in control cultures (+Dox) to 6–12% in cultures without Dox (Fig. 4b).
Figure 4.
MDCK cells expressing ΔN131 β-catenin exhibit increased growth in soft agar compared with cells expressing ΔN90 or ΔGSK β-catenin. (a) Photomicrographs of colonies 15 days after plating in soft agar. Parental MDCK cells, the human colon cancer cell line HCT116, and MDCK clones ΔGSK-4, ΔN90-A, and ΔN131-D were preincubated for 2 days −/+Dox and cultured 15 days −/+Dox in soft agar. Colonies were stained overnight with _p_-iodonitrotetrazolium violet. Representative images of two independent experiments are shown, and some of the colonies with a radius of ≥80 μm in these images are marked with arrowheads. (Bar = 500 μm.) (b) Percentage of colonies that had a radius of ≥80 μm in the culture; numbers in parentheses are the numbers of colonies with ≥80-μm radius per total numbers of measured colonies. Results from two independent experiments are shown. (c) HCT116 cells, MDCK parental cells, and MDCK clones were extracted with 1% SDS, and 10-μg protein lysates were subjected to SDS/PAGE and immunoblotted with mAb β-cat.C. (d) In parallel with SDS extraction shown in c, part of the cultures of MDCK parental cells, HCT116 cells, or MDCK clone ΔGSK-4 were transfected with control vector pFOPtkFLASH (CON, open bars) or pTOPtkFLASH (TCF, shaded bars). Bars represent values from one sample (CON) or mean values from two independent samples (TCF).
We conclude that β-catenin/TCF-activated transcription is not sufficient to induce anchorage-independent growth in epithelial cells. We suggest that in the presence of ΔGSK or ΔN90 β-catenin and high β-catenin/TCF-activated transcription, MDCK cells have a mechanism to suppress anchorage-independent cell growth (see below). However, this mechanism is defective or overridden in HCT116 cells and in MDCK cells expressing ΔN131 β-catenin. HCT116 cells contain mutations in the DCC and Ki-ras genes (41, 42), in addition to the β-catenin mutation, which may promote anchorage-independent cell growth. In MDCK cells expressing ΔN131 β-catenin, ΔN131 β-catenin itself may cause additional defects. ΔN131 β-catenin binds E-cadherin but not α-catenin (27) and cannot mediate linkage of the cadherin/β-catenin complex to the actin cytoskeleton that is required for strong cell–cell adhesion (8). Thus, cadherin-mediated cell–cell adhesion, such as in cells expressing ΔN90 or ΔGSK β-catenin, may help to suppress anchorage-independent growth even in the presence of increased TCF-activated transcription. We have shown previously that cells expressing either ΔN90- or ΔN131-β-catenin have a scattered morphology when plated at low cell density (27). Preliminary results from time-lapse analysis show that single MDCK cells expressing ΔN90- or ΔN131-β-catenin are more motile than control cells (A.I.M.B., unpublished results). Taken together, these data suggest that the scattered morphology in low-density cultures may be a combination of changes in both intercellular adhesion and cell motility. In collagen gels, these mutant cell lines formed polarized cysts that were indistinguishable from wild-type cysts. However, hepatocyte growth factor (HGF)-induced tubulogenesis, which requires cell extension from the cyst wall, was inhibited in cysts derived from cells expressing mutant β-catenins (43). Significantly, cysts expressing ΔN131-β-catenin also formed polyp-like cell aggregates in the cyst lumen after prolonged culture with HGF (43). This result indicates that ΔN131 β-catenin induced deregulated cell growth in these cysts, which is consistent with the increased anchorage-independent growth of cells expressing ΔN131 β-catenin observed in the present study.
The expression of stabilized mutant forms of β-catenin similar to the ones described here has been correlated previously with cell transformation and tumor formation. A β-catenin mutant with an N-terminal truncation of 173 aa was isolated in a functional screen for oncogenes and characterized as a moderate oncogene (44). This mutant form of β-catenin was missing the α-catenin-binding site. Forced expression of ΔN87 β-catenin in the basal layer of the murine skin epidermis and in the outer root sheath of the hair follicle induced hair follicle tumors (45), indicating that some type of skin cells are susceptible to neoplastic transformation by a mutant β-catenin similar to the ΔN90 β-catenin mutant described here. However, forced expression of ΔN89 β-catenin in the murine intestinal epithelium did not induce neoplastic transformation (46). These results indicate that, similar to MDCK cells expressing ΔGSK- or ΔN90–β-catenin, intestinal epithelial cells may have mechanisms to suppress deregulated cell growth induced by mutant β-catenin and that these mechanisms may be overridden in colon cancer cells such as HCT116 cells or other cancer cells expressing mutant β-catenin. Taken together, these data suggest that the role of β-catenin in cellular transformation is complex and probably depends on other negative and/or positive interacting factors.
Acknowledgments
We thank Dr. R. Kemler for providing the β-catenin cDNA, Dr. H. Bujard for providing the plasmids for the tetracycline-repressible expression system, Drs. P. Polakis and I. Näthke for antiserum to APC, Dr. G. Walter for mAb KT3, Drs. van de Wetering and H. Clevers for providing the TCF reporter plasmids, and Dr. D. Boffelli for helpful advice for the reporter gene assay. This work was supported by grants to W.J.N. from the National Institutes of Health. A.I.M.B. was supported by a North Atlantic Treaty Organization fellowship from the Deutscher Akademischer Austauschdienst and an American Heart Association postdoctoral fellowship. D.B.S. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
ABBREVIATIONS
TCF
T cell factor
APC
adenomatous polyposis coli
MDCK
Madin–Darby canine kidney
GSK
glycogen synthase kinase
Dox
doxycycline
−/+Dox
without or with Dox
References
- 1.Gumbiner B M. Curr Opin Cell Biol. 1995;7:634–640. doi: 10.1016/0955-0674(95)80104-9. [DOI] [PubMed] [Google Scholar]
- 2.Peifer M. Trends Cell Biol. 1995;5:224–229. doi: 10.1016/s0962-8924(00)89015-7. [DOI] [PubMed] [Google Scholar]
- 3.Barth A I, Näthke I S, Nelson W J. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 4.Aberle, H., Butz, S., Stappert, J., Weissig, H., Kemler, R. & Hoschuetzky, H. (1994) J. Cell Sci. 3655–3663. [DOI] [PubMed]
- 5.Rimm D L, Koslov E R, Kebriaei P, Morrow J L. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jou T S, Stewart D B, Stappert J, Nelson W J, Marrs J A. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozawa M, Ringwald M, Kemler R. Proc Natl Acad Sci USA. 1990;87:4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angres B, Barth A, Nelson W J. J Cell Biol. 1996;134:549–557. doi: 10.1083/jcb.134.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 10.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 11.Brunner E, Peter O, Schweizer L, Basler K. Nature (London) 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 12.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen F, Samos C H, Nusse R. Nature (London) 1994;368:342–344. doi: 10.1038/368342a0. [DOI] [PubMed] [Google Scholar]
- 14.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polakis P. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 19.Behrens J, Mareel M M, Van R F, Birchmeier W. J Cell Biol. 1989;108:2435–2447. doi: 10.1083/jcb.108.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oyama T, Kanai Y, Ochiai A, Akimoto S, Oda T, Yanagihara K, Nagafuchi A, Tsukita S, Shibamoto S, Ito F, et al. Cancer Res. 1994;54:6282–6287. [PubMed] [Google Scholar]
- 21.Robbins P F, El-Gamil M, Li Y F, Kawakami Y, Loftus D, Appella E, Rosenberg S A. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 23.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 24.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 25.Powell S M, Zilz N, Beazer-Barclay Y, Bryan T M, Hamilton S R, Thibodeau S N, Vogelstein B, Kinzler K W. Nature (London) 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 26.Behrens J. Breast Cancer Res Treat. 1993;24:175–184. doi: 10.1007/BF01833258. [DOI] [PubMed] [Google Scholar]
- 27.Barth A I, Pollack A L, Altschuler Y, Mostov K E, Nelson W J. J Cell Biol. 1997;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacArthur H, Walter G. J Virol. 1984;52:483–491. doi: 10.1128/jvi.52.2.483-491.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinck L, Näthke I S, Papkoff J, Nelson W J. J Cell Biol. 1994;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marrs J A, Andersson-Fisone C, Jeong M C, Cohen-Gould L, Zurzolo C, Nabi I R, Rodriguez-Boulan E, Nelson W J. J Cell Biol. 1995;129:507–519. doi: 10.1083/jcb.129.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain S H, Masiarz F R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 33.Stewart D B, Nelson W J. J Biol Chem. 1997;272:29652–29662. doi: 10.1074/jbc.272.47.29652. [DOI] [PubMed] [Google Scholar]
- 34.Peifer M, Sweeton D, Casey M, Wieschaus E. Development (Cambridge, UK) 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- 35.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 36.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 37.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J R, Moon R T. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze’ev A. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fearon E R, Cho K R, Nigro J M, Kern S E, Simons J W, Ruppert J M, Hamilton S R, Preisinger A C, Thomas G, Kinzler K W, et al. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 42.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 43.Pollack A L, Barth A I M, Altschuler Y, Nelson W J, Mostov K E. J Cell Biol. 1997;137:1651–1662. doi: 10.1083/jcb.137.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitehead I, Kirk H, Kay R. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 46.Wong M H, Rubinfeld B, Gordon J I. J Cell Biol. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]