Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends (original) (raw)
Abstract
Ku is a conserved DNA end-binding protein that plays various roles at different kinds of DNA ends. At telomeres, Ku is part of the structure that protects the chromosome end, whereas at broken DNA ends, Ku promotes DNA repair as part of the nonhomologous end-joining (NHEJ) pathway. Here, we present evidence of a new role for Ku that impacts both telomere-length maintenance and DNA repair in Saccharomyces cerevisiae. We show that Ku binds TLC1, the RNA component of telomerase. We also describe a novel separation-of-function allele of Ku that is specifically defective in TLC1 binding. In this mutant, telomeres are short and the kinetics of telomere addition are slow, but other Ku-dependent activities, such as chromosome end protection and NHEJ, are unaffected. At low frequency, yeast will use telomerase to heal DNA damage by capping the broken chromosome with telomeric DNA sequences. We show that when Ku's ability to bind TLC1 is disrupted, DNA repair via telomere healing is reduced 10- to 100-fold, and the spectrum of sequences that can acquire a telomere changes. Thus, the interaction between Ku and TLC1 RNA enables telomerase to act at both broken and normal chromosome ends.
Keywords: Telomerase, Ku, telomere, DNA repair, Saccharomyces cerevisiae, RNA-protein interaction
Telomeres are the specialized protein-DNA structures found at the ends of linear chromosomes (for review, see Greider 1991; McEachern et al. 2000). In most eukaryotes, the telomeric DNA consists of tandem repeats of a short, TG-rich sequence. These repeats are bound by a host of proteins that protect the chromosome end from undergoing degradation, fusion, or recombination. Thus, telomeres enable chromosome ends to be stably maintained, rather than being recognized and processed as double-strand breaks.
Telomeric DNA is typically synthesized by telomerase, a reverse transcriptase that has an essential RNA and an essential protein component (McEachern et al. 2000). In the budding yeast Saccharomyces cerevisiae, these components are encoded by TLC1 and EST2, respectively (Singer and Gottschling 1994; Lingner et al. 1997a). The TLC1 RNA contains a template sequence that Est2p uses to add irregular repeats of TG1-3 residues onto a DNA end; together, TLC1 RNA and Est2p are necessary and sufficient for telomerase activity in vitro (Cohn and Blackburn 1995; Lingner et al. 1997b). However, three additional proteins, Est1p, Est3p, and Cdc13p, are required for telomerase to act in vivo (Lundblad and Szostak 1989; Lendvay et al. 1996). Est1p and Est3p associate with the telomerase holoenzyme (Steiner et al. 1996; Hughes et al. 2000), whereas Cdc13p is a sequence-specific telomeric DNA-binding protein (Lin and Zakian 1995; Nugent et al. 1996). Interactions between Cdc13p and Est1p have been implicated in recruiting telomerase to the telomere (Evans and Lundblad 1999; Pennock et al. 2001); the role of Est3p is unclear.
TLC1 was first identified, not through its role in telomere synthesis, but from its impact on telomeric silencing. Genes located near telomeres are transcriptionally silent, due to the heterochromatic structure that initiates at telomeric DNA and spreads inward (Renauld et al. 1993). A screen for high-copy disruptors of telomeric silencing (the DOT screen) was conducted to identify factors that affect this telomeric chromatin, and TLC1 was one of the genes that emerged (Singer et al. 1998). Deletion analysis mapped the domain of TLC1 responsible for the DOT phenotype to a 48-nucleotide region of the RNA that folds into a stem-loop structure (Peterson et al. 2001). RNA stem-loops are often sites for protein binding, and it was hypothesized that overexpression of the TLC1 stem-loop titrates away one of the components of silent chromatin, or otherwise interferes with its function.
Genetic analysis suggested that Ku is the factor affected by TLC1 overexpression (Peterson et al. 2001). Ku is an abundant DNA end-binding protein that is conserved among eukaryotes (Tuteja and Tuteja 2000). The functional unit is a heterodimer, with the two subunits in yeast being encoded by YKU70 and YKU80. Ku binds chromosome ends and is involved in maintaining normal telomere length and structure, in addition to participating in the formation of silent chromatin at telomere-proximal genes (Porter et al. 1996; Boulton and Jackson 1998; Gravel et al. 1998; Nugent et al. 1998; Polotnianka et al. 1998; DuBois et al. 2002). Overexpression of TLC1 mirrors the phenotypes of a yku deletion, and overexpression of YKU70 + YKU80 can partially compensate for overexpression of TLC1 (Peterson et al. 2001). We interpreted these results to suggest that, in addition to all of its other roles, Ku may also interact with telomerase RNA.
Insights into how Ku binds DNA and might interact with other factors come from the crystal structure of human Ku bound to DNA (Walker et al. 2001). The Ku70 and Ku80 subunits interlock to form a ring-shaped molecule, with a large central cavity for binding DNA and globular domains on the outside of the structure that are potentially available to interact with other factors. The structure of Ku is the same when crystallized with or without DNA, suggesting that the ring is preformed, and there are no base-specific contacts in the structure, thus explaining why Ku can bind DNA ends without sequence specificity. Moreover, biochemical experiments demonstrate that Ku can bind a wide variety of DNA end structures, including flush ends and both 5′ and 3′ overhangs (Falzon et al. 1993). These properties enable Ku to bind not only chromosome ends but also broken DNA ends that arise from DNA damage.
S. cerevisiae has many mechanisms to cope with DNA damage, including homologous recombination, nonhomologous end-joining (NHEJ), and break-induced replication (Haber 2000). At low frequency, cells also use telomerase to heal DNA damage, by capping the broken DNA end with a telomere (Kramer and Haber 1993; Myung et al. 2001). Ku is involved in NHEJ; in this work, we provide evidence that Ku is involved in telomere healing as well.
Results
Ku binds the TLC1 stem-loop RNA in vitro
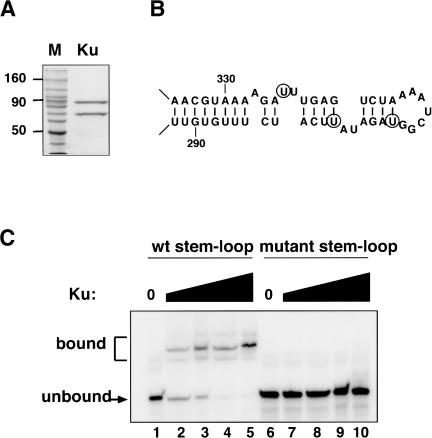
To determine whether Ku can physically bind the TLC1 stem-loop, a biochemical approach was taken. A TAP tag was placed on the Ku80 subunit, and Ku was isolated from yeast by two rounds of affinity purification (Rigaut et al. 1999). Both subunits of the heterodimer were recovered stoichiometrically (Fig. 1A). The purified Ku was incubated with a radiolabeled RNA substrate consisting of three tandem copies of the 48-nucleotide TLC1 stem-loop (Fig. 1B). The triple stem-loop had been found previously to most closely match the activity of full-length TLC1 in disrupting telomeric silencing (Peterson et al. 2001), perhaps because the stem-loop can fold most stably in this construct. Potential binding complexes were resolved on a nondenaturing polyacrylamide gel (Fig. 1C). In the presence of Ku, the mobility of the TLC1 RNA substrate was reduced, indicating that Ku had bound the TLC1 RNA, and the fraction of the RNA present in these shifted species increased with higher concentrations of Ku.
Figure 1.
Ku binds the TLC1 RNA stem-loop in vitro. (A) Purification of Ku from yeast. A Coomassie-stained protein gel is shown, containing a protein molecular weight ladder and 1 μg of the pooled eluate from the second round of affinity purification. (B) Secondary structure of the TLC1 RNA stem-loop. Nucleotides 288-335 of the 1.3-kb TLC1 RNA are shown. Circles indicate the residues that were changed (U301A, U307G, U324G) in the mutant RNA substrate used in C. Figure adapted from Peterson et al. (2001), and reproduced with permission from Nature Publishing Group. (C) Gel-shift analysis of Ku's RNA-binding activity. Reactions contained 12 pM wild-type stem-loop RNA (lanes 1-5) or 13 pM mutant RNA (lanes 6-10). Ku was present at 0 nM (lanes 1,6), 0.2 nM (lanes 2,7), 1 nM (lanes 3,8), 5 nM (lanes 4,9), and 25 nM (lanes 5,10).
To evaluate the specificity of this binding, a second RNA substrate containing three tandem copies of a mutated version of the 48-nucleotide RNA was tested. Three residues that affect the base-pairing of the stem and the sequence of the bulge (indicated by circles in Fig. 1B) were changed. Overexpression of a tlc1 allele containing these point mutations does not disrupt telomeric silencing (Peterson et al. 2001), suggesting that Ku does not interact with the mutant RNA in vivo. Ku did not bind this mutant RNA substrate in vitro, even at high protein concentrations (Fig. 1C, lanes 6-10), consistent with our interpretation of the in vivo results.
Thus, Ku can physically associate with the TLC1 RNA stem-loop. The simplest explanation for this result is that Ku directly binds the RNA. However, we cannot rule out the possibility that there is a bridging protein that mediates the interaction between Ku and TLC1 RNA. Attempts to detect in vivo binding of Ku to TLC1 RNA by coimmunoprecipitation have not been successful (data not shown). However, similar experiments with Cdc13p, a well-established regulator of telomerase in vivo, have also been unsuccessful (Hughes et al. 2000). These interactions may be transient, or may involve only a small fraction of the Ku or Cdc13p population in the cell, and, therefore, be difficult to detect by coimmunoprecipitation. Below, we present several lines of evidence that Ku impacts telomerase activity in vivo, making it likely that Ku's physical association with TLC1 RNA in vitro reflects a functional interaction with telomerase in vivo.
Isolation of a Ku allele that suppresses the effect of TLC1 overexpression
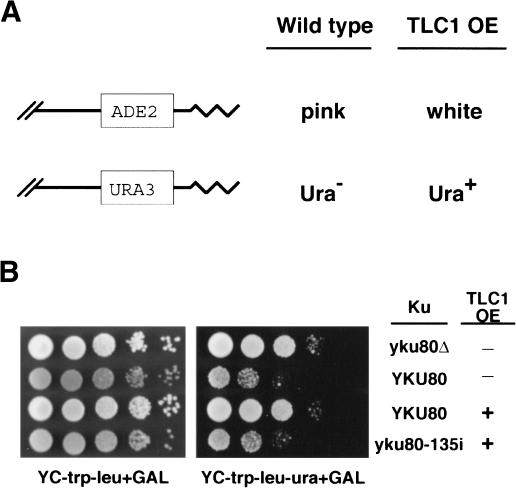
How does Ku bind TLC1 RNA, and what is the biological significance of the interaction? To address these questions, we sought to isolate an allele of Ku that is defective in TLC1 RNA binding, by taking advantage of the design of the original DOT screen (Singer et al. 1998). In that screen, ADE2 and URA3 serve as reporters of telomeric silencing. In wild-type strains, these genes are transcriptionally repressed when placed near telomeres; colonies are red with white sectors (and thus look pink on synthetic medium) and grow poorly in the absence of uracil (Fig. 2A,B). However, if silencing is disrupted, the reporters are expressed; colonies are white and Ura+. We reasoned that if high levels of TLC1 RNA negatively affect Ku, then it might be possible to identify alleles of Ku that would suppress the effects of TLC1 overexpression and give rise to colonies that maintain telomeric silencing (i.e., remain pink and Ura-).
Figure 2.
Isolation of a new yku80 allele that suppresses the effect of TLC1 overexpression on telomeric silencing. (A) Schematic of the reporters and phenotypes of telomeric silencing. ADE2 and URA3 are located near the telomeres (jagged lines) of chromosomes V and VII, respectively. (B) Observed telomeric silencing of UCC6058 strains containing the plasmids indicated below. Tenfold serial dilutions of each strain were plated onto YC-trp-leu + GAL medium to evaluate colony color (gray = pink) and number, and onto YC-trp-leu-ura + GAL medium to evaluate growth in the absence of uracil. (Row 1) pRS315 + pTCG. (Row 2) pRS315-YKU80 + pTCG. (Row 3) pRS315-YKU80 + pTCG-3X stem. (Row 4) pRS315-yku80-135i + pTCG-3X stem.
YKU70 and YKU80 were mutagenized using an in vitro transposition method that inserts a 15-bp sequence randomly throughout a plasmid containing the gene of interest (Biery et al. 2000). The mutagenized plasmids were then transformed into _yku70_Δ or _yku80_Δ strains that also contained a plasmid with three tandem copies of the TLC1 stem-loop motif adjacent to a galactoseinducible promoter. Thus, colony color phenotypes could be examined in the absence and presence of TLC1 overexpression. Approximately 11,000 yku70 transformants and 7,000 yku80 transformants were analyzed. 11%-12% of the transformants were white, even in the absence of TLC1 overexpression, presumably because those 15-bp insertions caused a complete loss of Ku function. However, most of the transformants were pink, and thus retained their competence for silent telomeric chromatin formation.
The transformants were then replica-plated to galactose-containing medium to induce the overexpression of the 48-nucleotide TLC1 stem-loop RNA. All of the yku70 transformants, like strains containing wild-type YKU70, became white. However, one of the yku80 transformants was pink on both glucose and galactose medium, indicating that telomeric silencing in this mutant was not disrupted by high levels of TLC1 RNA (Fig. 2B; data not shown). Similarly, the telomeric URA3 gene remained silenced in this mutant (Fig. 2B). Sequencing revealed that the only change in this allele was the 15-bp insertion, which fell between nucleotides 135 and 136 of YKU80; therefore, we have named this allele yku80-135i.
MutantKu binds DNA but not TLC1 RNA in vitro
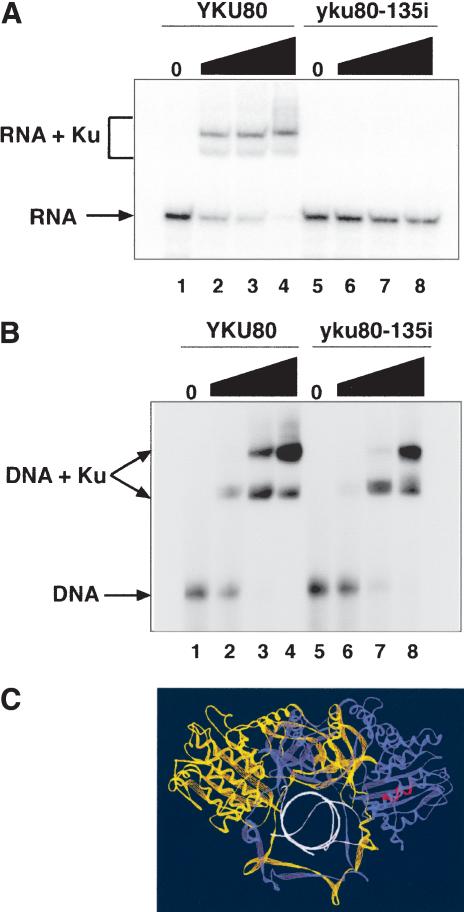
How does yku80-135i suppress the effects of TLC1 overexpression? The mutation might reduce Ku's ability to bind TLC1 RNA, or the mutation might enhance Ku's interactions with other silencing factors, leaving its ability to bind TLC1 RNA unchanged. To differentiate between these possibilities, we isolated mutant Ku protein from yeast containing the yku80-135i allele. Unlike the wild-type protein, the mutant Ku protein had no detectable TLC1 RNA-binding activity; no shifted species were seen, even in the presence of high levels of mutant Ku protein (Fig. 3A). However, the mutant Ku protein was not biochemically inert; it bound a radiolabeled DNA fragment with only a modest decrease in affinity compared with the wild-type protein (Fig. 3B).
Figure 3.
Mutant Ku protein binds DNA but not the TLC1 stem-loop RNA in vitro. (A) Gel-shift analysis of RNA-binding reactions. A total of 16 pM wild-type TLC1 stem-loop RNA was used as a substrate. Ku was present at 0 nM (lanes 1,5), 0.5 nM (lanes 2,6), 8 nM (lanes 3,7), and 40 nM (lanes 4,8). (B) Gel-shift analysis of DNA-binding reactions. A total of 190 pM ADE2 DNA was used as a substrate. Two shifted species are seen, probably reflecting Ku binding to one or both ends of the DNA fragment. Ku concentrations were the same as in A. (C) Structure of human Ku bound to DNA. Ku70 is in yellow, Ku80 is in blue, the DNA is in white, and the residues that correspond to the site of the 15-bp insertion are in red. Figure prepared using Swiss PBD Viewer and the coordinates from accession number 1JEY in the Protein Data Bank (Walker et al. 2001).
The simplest interpretation of these results is that the mutant Ku protein has lost the ability of wild-type Ku to bind TLC1 RNA. Alternatively, if TLC1 RNA binding is mediated by a bridging protein, then the mutant has lost the ability to interact with the bridge. Comparison of the wild-type and mutant Ku proteins on an overloaded silver-stained gel demonstrated that other proteins are present at low levels in these preparations (data not shown). However, the spectrum of copurifying species is the same in the wild-type and mutant samples. Thus, at this level of detection, there is no obvious candidate for a bridging protein present in the wild-type Ku sample but not the mutant Ku sample that would account for their different RNA binding activities.
Mapping the position of the yku80-135i insertion onto the crystal structure of human Ku (Walker et al. 2001) revealed that the 15-bp insertion lies in one of the peripheral globular domains of the protein, away from the conserved DNA binding cavity (Fig. 3C). Thus, residues important for DNA and TLC1 RNA binding appear to be distinct, raising the possibility that Ku can bind both molecules at the same time.
yku80-135i strains have normal chromosome end protection and DNA repair
Ku has multiple roles in the cell; in addition to its role in telomeric silencing, Ku also protects the chromosome end and participates in DNA repair via the NHEJ pathway (Porter et al. 1996; Boulton and Jackson 1998; Gravel et al. 1998; Nugent et al. 1998; Polotnianka et al. 1998; DuBois et al. 2002). We compared these activities in wild-type, yku80-135i and _yku80_Δ strains, to determine how extensively the 15-bp insertion in yku80-135i affected Ku's function.
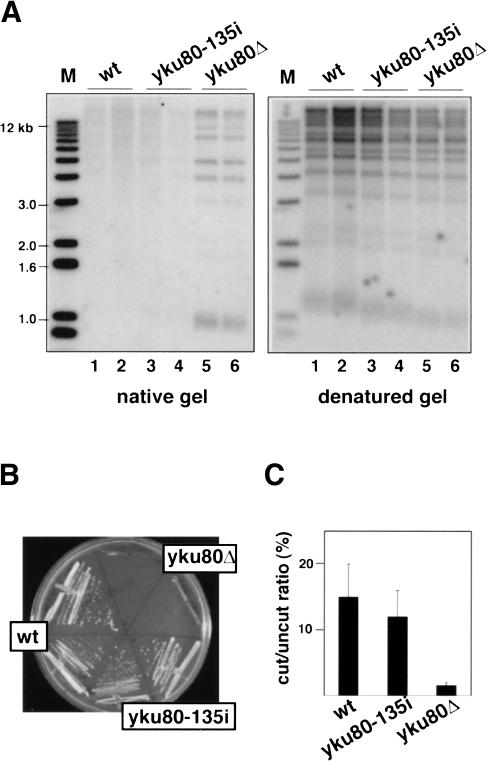
One method to evaluate the integrity of the chromosome end is to physically probe its structure. In wild-type cells, the chromosome end is not extensively single stranded throughout most of the cell cycle, although TG1-3 overhangs can be transiently detected in S phase (Wellinger et al. 1993; Dionne and Wellinger 1996). In contrast, chromosomes in _yku80_Δ strains have long, single-stranded TG1-3 overhangs that are stably maintained throughout the cell cycle (Gravel et al. 1998; Polotnianka et al. 1998). These overhangs can be detected by nondenaturing gel electrophoresis followed by in-gel hybridization with a C1-3A probe. Prominent bands were seen in the DNA samples harvested from _yku80_Δ strains (Fig. 4A), indicating that the telomeric DNA was extensively single stranded. However, yku80-135i strains did not have such a banding pattern; its telomeric DNA resembled that of wild-type yeast. Stripping the gel and reprobing it under denaturing conditions verified that the amount of DNA loaded in each lane was approximately equal.
Figure 4.
DNA end protection and NHEJ are not compromised in yku80-135i strains. (A) Analysis of telomere end structure. Two independent isolates of UCC6058 strains containing pRS315-YKU80, pRS315-yku80-135i, or pRS315 were analyzed. Genomic DNA was digested with _Xho_I and resolved by agarose gel electrophoresis, along with a DNA molecular weight ladder. The gel was hybridized with a radiolabeled C1-3A probe first under nondenaturing conditions and then under denaturing conditions. The terminal restriction fragments of chromosomes with subtelomeric Y′ elements are 1.0-1.3 kb in size (depending on strain background); chromosomes without Y′ elements give rise to terminal restriction fragments of diverse sizes. (B) Analysis of temperature sensitivity. Growth at 37°C of the strains in A was compared. (C) Transformation efficiencies of UCC5913, UCC3744, and UCC3745. Values represent the number of transformants recovered with linearized pBTM116 relative to the number recovered with supercoiled pBTM116. Experiments were done in triplicate.
A second measure of chromosome end protection is growth at high temperature. When grown at 37°C_, yku80_Δ mutants trigger the _RAD9_-dependent DNA damage checkpoint (Teo and Jackson 2001), apparently in response to telomere defects rather than NHEJ defects (Fellerhoff et al. 2000; Gravel and Wellinger 2002). In contrast to the _yku80_Δ strains, the yku80-135i mutants were capable of growth at 37°C (Fig. 4B), suggesting that the DNA damage checkpoint was not triggered.
Synthetic lethal experiments provide a third indication of the integrity of the chromosome end. Combining a _yku80_Δ mutation with the deletion of any of the components of telomerase results in cells that can neither protect their telomeric DNA repeats nor synthesize new ones; these cells die at all temperatures (Gravel et al. 1998; Nugent et al. 1998; Polotnianka et al. 1998). However, the yku80-135i mutation was not synthetically lethal with a _tlc1_Δ (data not shown). Thus, three different measures of chromosome end protection all suggest that this activity of wild-type Ku is retained in the yku80-135i mutant.
In contrast to its role at telomeres, Ku actively promotes DNA end-joining at the sites of double-strand breaks (Haber 2000). This activity can be evaluated using a plasmid rejoining assay, which compares the ability of a strain to be transformed with supercoiled versus linearized plasmids. Wild-type yeast use the NHEJ machinery to recircularize the linearized plasmid, and thus, can stably maintain it, whereas _yku80_Δ strains are defective in NHEJ, and consequently, are less capable of being transformed with a linearized plasmid (Boulton and Jackson 1996). We found that wild-type and yku80-135i strains were transformed with linearized plasmids with essentially the same efficiency (p = 0.6; Fig. 4C), suggesting that NHEJ was intact in the yku80-135i mutant.
Thus, the 15-bp insertion in yku80-135i has a very specific effect. It disrupts Ku's ability to bind the 48-nucleotide stem-loop of TLC1 RNA, but retains Ku's ability to bind DNA, as well as many of Ku's DNA-based activities, namely telomeric silencing, chromosome end protection, and DNA repair via NHEJ.
Telomere addition is compromised if Ku and TLC1 RNA cannotinteract
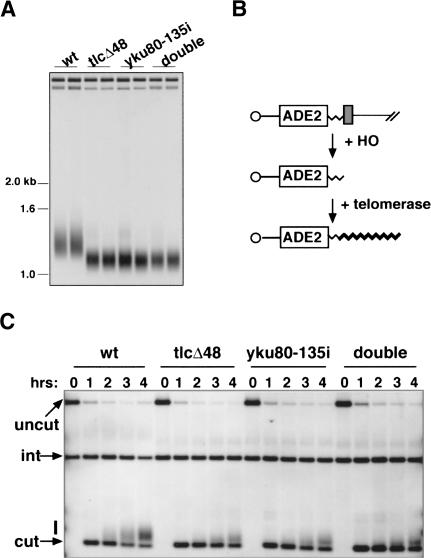
What cellular activities are affected if Ku cannot bind the TLC1 RNA stem-loop? Two lines of evidence suggest that telomere addition is less efficient under these conditions. First, we examined global telomere length in strains defective in the Ku-TLC1 RNA interaction. Consistent with our earlier findings that tlc1_Δ_48 strains had a shorter telomere length than wild-type strains (Peterson et al. 2001), we found that yku80-135i strains had the same short telomere length (Fig. 5A), and that telomeres in the double tlc1_Δ_48 yku80-135i mutant were no shorter than either single mutant. Serial passage of the yku80-135i and tlc1_Δ_48 yku80-135i strains demonstrated that, like tlc1_Δ_48 strains, the telomeres were stably maintained at this short length (data not shown). This contrasts with the phenotype of a complete deletion of TLC1, in which telomeres progressively shorten and the cells senesce (Singer and Gottschling 1994). The short telomere length of the yku80-135i and tlc1_Δ_48 yku80-135i mutants is unlikely to be due to catalytic defects in telomere synthesis, as telomerase has equivalent activity in vitro when purified from wild-type or tlc1_Δ_48 strains (Peterson et al. 2001). Rather, when Ku cannot bind the TLC1 RNA stem-loop, the efficiency of telomerase function in vivo appears to be compromised.
Figure 5.
Telomere length maintenance and de novo telomere addition are affected when Ku cannot interact with TLC1 RNA. (A) Southern blot analysis of telomere length in UCC5114, UCC5116, UCC5120, and UCC5118-2. The positions of relevant DNA size markers are indicated. (B) Schematic of the de novo telomere addition assay. The left arm of chromosome VII contains a short telomeric seed sequence (thin jagged line) and a recognition site for the HO endonuclease (gray box) next to an ADE2 marker. Upon induction of HO, the chromosome is cleaved to expose the telomeric seed sequence. Telomerase then adds new telomeric repeats onto it (thick jagged line). (C) Southern blot analysis of de novo telomere addition in UCC5913, UCC6073, UCC3744, and UCC3746. Blots were probed with an ADE2 fragment that also hybridizes to a 1.6-kb fragment from the genomic ade2-101 locus (labeled “int”), which serves as an internal loading control. The product of HO cleavage (labeled “cut”), and the smear of products reflecting new telomere addition (marked with a bar) are also indicated.
Results from de novo telomere addition assays support this view. For this experiment, the left arm of chromosome VII was engineered to contain a short telomeric seed sequence next to the recognition site for the HO endonuclease (Fig. 5B; Diede and Gottschling 1999). Upon induction of HO, the chromosome was cleaved and the telomeric seed exposed. Telomerase was then recruited to the newly revealed telomere, where it catalyzed the addition of new telomeric repeats. In wild-type cells, telomere addition was detectable after 2 h, and by 4 h, more than half of the broken ends had acquired new telomeric repeats (Fig. 5C). In contrast, the kinetics of telomere addition were slower in the mutants, and fewer ends acquired repeats. These results suggest that the recruitment of telomerase to the telomere, or the initiation of telomere synthesis once there, was less efficient in the mutants. In addition, the extent of telomere lengthening was reduced (i.e., those ends that did acquire telomeric repeats got fewer of them). This might reflect a role for Ku in telomerase processivity. Alternatively, if telomerase undergoes multiple rounds of binding and dissociating from the end during the course of telomere elongation, then the reduced telomere lengthening in the yku80-135i and tlc1_Δ_48 mutants may also be a consequence of a recruitment or initiation defect.
Does the Ku-TLC1 interaction enable telomerase to act at sites other than telomeres?
As discussed above, Ku binds not only to telomeres, but also to broken DNA ends and participates in their repair by NHEJ. However, given our findings, we asked whether Ku can also recruit telomerase to broken DNA ends, through its interaction with TLC1 RNA. That is, does telomerase provide Ku with a second option for dealing with a double-strand break?
Kolodner and colleagues developed a system in S. cerevisiae to examine how cells handle damaged DNA in ways that lead to gross chromosomal rearrangements (GCRs; Myung et al. 2001). In their experimental system,
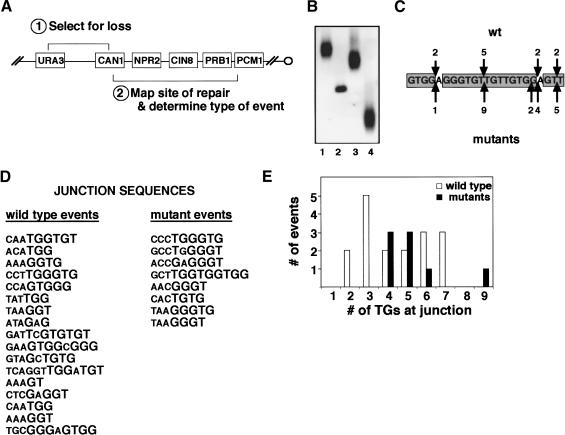
URA3 was placed 7.5 kb distal to CAN1 on the left end of chromosome V (Fig. 6A), and spontaneous events that resulted in the loss of both gene functions were isolated at very low frequency (-3 × 10-10 in wild-type cells). A total of 80% of the GCRs were the result of terminal deletions, in which new telomeres were added centromere-proximal to CAN1, presumably in response to DNA damage within that area.
Figure 6.
The Ku-TLC1 interaction affects telomere healing events. (A) Schematic of the relevant markers on chromosome VL. URA3 replaced the endogenous HXT13 gene, located -7.5 kb from CAN1 and -22 kb from the end of the chromosome. (B) Southern blot analysis of representative GCR events. Three telomere healing events (lanes 1,3,4) and one translocation event (lane 2) are shown. (C) The NPR2 hotspot. Nucleotides 451-473 are shown. Positions of telomere addition, and the number of independent events recovered, are indicated for wild-type strains (downward arrows) and all mutant strains (upward arrows). (D) Chromosome V sequences at the junctions of unique telomere addition events from wild-type and all mutant strains. Sequences run 5′-3′; telomeres were added proximal to the last residue indicated. (E) Histogram quantifying the number of TG residues at the junctions of the telomere addition events in D. Contiguous T1G1-3 residues were counted; one intervening non-T1G1-3 residue was allowed to be skipped. The skipped residue was not included in the count, and only one skip was made per sequence.
To explore whether the Ku-TLC1 interaction enables cells to use telomerase to heal broken DNA ends, we adopted the Kolodner system and modified it slightly. We treated actively growing cells with a pulse of methyl methanesulfonate (MMS), to induce a low level of DNA damage across the genome and thus increase the frequency of recovering GCR events. The cells were allowed a period of time to repair the damage, and then were plated on medium containing FOA and canavanine to select for GCR events. We used a combination of PCR and Southern analysis to map and characterize these events. The site of DNA repair must lie within the -12 kb between the end of CAN1 and the most distal essential gene on chromosome V, PCM1. PCR was used to identify which ORFs in this region were still present, and then Southern blots were used to examine a restriction fragment containing the repair site. If DNA repair occurred by telomere addition, the terminal restriction fragment was characteristically heterogeneous in length (Fig. 6B), due to the variability in the number of telomeric repeats added. In contrast, if DNA repair occurred by translocation of the broken DNA end to another chromosome, or by fusion to sequences distal to URA3, the restriction fragment containing the repair site was sharp in appearance. To confirm these identifications, blots were stripped and rehybridized with a telomeric DNA probe (data not shown).
In wild-type cells, the frequency of recovering FOARCANR colonies was -7 × 10-8, and the vast majority of these GCR events were the result of telomere healing (Table 1). This prevalence was seen in all four segments of the 12-kb region. Thus, treatment of the cells with MMS did increase the frequency of recovering GCR events, but did not change the bias in the mechanism used to repair the DNA damage. This conclusion is supported by recent work from Myung and Kolodner (2003).
Table 1.
Frequencies of recovering GCR events repaired by telomere addition or translocation
| # independent events | Frequency of healing (× 108)b | ||||||
|---|---|---|---|---|---|---|---|
| Repair locus (size) | Straina | GCR frequency (× 108) ± SD | w/telomere | w/translocation | w/telomere | (fold down) | w/translocation |
| CAN1 | wt | 1.06 ± 0.38 | 14 | 0 | 1.06 ± 0.31 | (1) | <0.23 |
| yku80-135i | 0.11 ± 0.04 | 1 | 10 | 0.01 ± 0.04 | (106) | 0.10 ± 0.04 | |
| (2.7 kb) | tlc1δ48 | 0.20 ± 0.06 | 1 | 4 | 0.04 ± 0.14 | (26) | 0.16 ± 0.10 |
| double | 0.10 ± 0.05 | 3 | 7 | 0.03 ± 0.04 | (35) | 0.07 ± 0.03 | |
| NPR2 | wt | 4.42 ± 1.61 | 18 | 1 | 4.19 ± 1.0 | (1) | 0.23 ± 0.82 |
| yku80-135i | 0.39 ± 0.13 | 5 | 3 | 0.24 ± 0.13 | (17) | 0.15 ± 0.19 | |
| (2.2 kb) | tlcδ48 | 1.08 ± 0.36 | 16 | 2 | 0.96 ± 0.26 | (4) | 0.12 ± 0.23 |
| double | 0.35 ± 0.18 | 9 | 4 | 0.24 ± 0.09 | (17) | 0.11 ± 0.11 | |
| CIN8 | wt | 1.12 ± 0.41 | 17 | 3 | 0.95 ± 0.25 | (1) | 0.17 ± 0.22 |
| + | yku80-135i | 0.14 ± 0.04 | 5 | 20 | 0.03 ± 0.02 | (32) | 0.10 ± 0.02 |
| PRB1 | tlcδ48 | 0.22 ± 0.08 | 4 | 7 | 0.08 ± 0.08 | (12) | 0.14 ± 0.06 |
| (6.7 kb) | double | 0.21 ± 0.11 | 3 | 18 | 0.03 ± 0.04 | (32) | 0.18 ± 0.04 |
The wild-type and mutant strains underwent chromosomal translocation at similar frequencies (Table 1). This result is consistent with our conclusion from the plasmid rejoining assay that yku80-135i strains retain the capacity to do NHEJ. In addition, the viability of the mutant strains upon exposure to this level of MMS was identical to that of the wild-type strain (data not shown).
However, when we examined the telomere addition events that arose in the mutant strains, an interesting dichotomy emerged. In the CAN1, CIN8, and PRB1 regions, the frequency of healing with a telomere was dramatically (12- to 106-fold) lower in the mutants (Table 1). In fact, DNA repair in these regions occurred more often by translocation than by telomere addition. In contrast, in the NPR2 region, telomere addition remained the predominant repair pathway used. The frequency of GCR events was also highest in this region in all strain backgrounds, and Southern blots indicated that the majority of these telomere additions occurred at approximately the same site. Sequencing confirmed that 11 telomere addition events in wild-type strains and 21 telomere addition events in the mutant strains occurred within a 23-bp TG-rich sequence in NPR2 (Fig. 6C). DNA damage distal to this site, followed by single-stranded 5′-3′ resection, would expose this TG-rich sequence, which could then serve as a high-affinity binding site for Cdc13p (Anderson et al. 2003), and correspondingly, telomerase could be recruited to this site without a requirement for Ku. In the CAN1, CIN8, and PRB1 regions, however, there are no extensive TG-rich sequences; telomere healing of DNA damage in these regions appears to be much more dependent on Ku.
A total of 45 wild-type and 43 mutant telomere healing events were sequenced. In some cases, both a wild-type and a mutant telomere healing event occurred at the same site, or within 20 bp of each other. We assume that at these sites, as in the NPR2 hotspot, factors other than the Ku-TLC1 interaction influence telomere addition.
However, another interesting dichotomy emerged when we examined the 17 wild-type and 8 mutant telomere healing events that occurred at unique positions. In the wild-type strains, telomeres were added at sequences containing a diverse number of TG1-3 residues (Fig. 6D,E), from as few as two to as many as seven residues. However, in the mutant strains, no telomere healing occurred at short TG tracts; telomere healing events were only recovered at junctions of four or more TG1-3 residues. We propose that the Ku-TLC1 interaction is required for telomerase to gain access to broken DNA ends with little TG sequence, whereas at sequences with longer TG tracts, other factors, such as Cdc13p binding or more extensive base-pairing with the TLC1 template region, can enable telomerase to act at a low frequency.
Discussion
Ku enhances telomerase activity at telomeres
Ku participates in many events at the ends of yeast chromosomes: Ku protects the end from undergoing degradation or fusion events, it is required to repress transcription of genes located near the ends, and it is involved in the clustering of telomeres at the nuclear periphery (Tuteja and Tuteja 2000). To this list of activities, we have now added a new role for Ku in modulating telomerase activity. Telomeres are shorter and the kinetics of telomere addition are slower in strains in which Ku is unable to interact with the 48-nucleotide stem-loop motif of TLC1 RNA. Our working hypothesis is that, by binding TLC1, Ku helps to recruit telomerase to the telomere, and thus increases the local concentration of the enzyme at its site of action. However, it is also possible that Ku influences telomerase activity at a step subsequent to its localization.
Thus, there are two interactions between telomere-binding proteins and telomerase components that impact telomerase function in vivo. In addition to the protein-RNA interaction between Ku and TLC1, there is also a protein-protein interaction between Cdc13p and Est1p that is essential for telomerase activity in vivo (Evans and Lundblad 1999; Pennock et al. 2001). The biochemical and structural properties of Ku and Cdc13p suggest that they bind telomeric DNA quite differently. Ku forms a ring that encircles DNA (Walker et al. 2001); it has a greater affinity for duplex DNA than single-stranded DNA (Falzon et al. 1993), and therefore, may sit at the single-strand/double-strand transition of the telomere. In contrast, Cdc13p lies along the single-stranded portion of the telomere and makes extensive base contacts with it (Mitton-Fry et al. 2002; Anderson et al. 2003). Thus, the two proteins could physically bind the telomere at the same time. Whether they also interact with telomerase in a concerted fashion, or whether they have distinct roles at (for example) different stages in the cell cycle, remains to be determined.
Ku facilitates the healing of broken DNA by the addition of a telomere
Telomerase does not function exclusively at telomeres. At low frequency, telomerase is also used to heal a broken chromosome by capping it with a telomere. In contrast to Cdc13p, Ku is a highly abundant protein whose ability to bind DNA ends is not limited by sequence or structure. Moreover, Ku has been shown to relocalize from telomeres to the sites of DNA damage (Martin et al. 1999). Therefore, Ku is a good candidate for recruiting telomerase to broken DNA ends.
Our analysis of GCR events provided three clues that the Ku-TLC1 interaction is important for telomere healing of DNA damage. First, the frequency of telomere healing was lower in the yku80-135i and tlc1_Δ_48 mutants. In the NPR2 hotspot, telomere healing was (on average) 13-fold lower in the mutants (Table 1), suggesting that here, as at native telomeres, the efficiency of telomerase recruitment is reduced when Ku cannot interact with TLC1 RNA. At all other loci examined, the frequency of telomere healing was even more severely reduced, down (on average) 40-fold in the mutants. Second, a larger fraction of the mutant telomere addition events (35 of 43) occurred in the NPR2 hotspot or at sequences that were used multiple times. Third, among the unique sites of telomere addition, almost half of the junctions in wild-type strains contained two to three TG residues, whereas the junctions in the mutants always contained four or more TG residues (Fig. 6). Together, these observations suggest that Ku enables telomerase to act at a wide variety of sites of DNA damage, but that when the Ku-TLC1 interaction is disrupted, telomere healing becomes more rare, occurring only if Cdc13p can get a good foothold on the chromosome and/or if the TLC1 template sequence can make extensive base-pair contacts with the broken DNA end.
Telomere addition at short TG sequences has been seen before. In an early study that enabled the S. cerevisiae telomerase template sequence to be predicted, Kramer and Haber (1993) found de novo telomere addition occurred at sequences with as few as a single G to up to 10 TG residues. Ku may have been contributing to the recruitment of telomerase to these sequences as well.
Ku mediates the fate of a double-strand break
Why would it be advantageous for the cell to have a mechanism for recruiting telomerase to double-strand breaks? Telomere healing is a drastic and inherently dangerous means of DNA repair, because all genetic information distal to the site of the break is lost. The low frequency at which telomere addition is observed in GCR experiments tells us that this means of DNA repair is, in fact, either rarely selected or rarely successful. DNA repair by homologous recombination and NHEJ are far more commonly observed, likely because these mechanisms are more effective at conserving the native genome. However, the cell may sometimes need to cap a broken chromosome with a telomere in order to survive. While in haploids, a terminal deletion may often be lethal, in diploids, the risk may be better tolerated. Therefore, having a multipotent protein like Ku—capable of recruiting either the NHEJ machinery or telomerase to a break—may be useful for cells in crisis.
If Ku controls access to a double-strand break, what regulates which repair machinery it selects? The stage of the cell cycle in which DNA damage is incurred, or the checkpoint response that the damage elicits, may favor Ku's interactions with one type of DNA repair machinery or another. There may also be molecular clocks at work: Ku might try first to engage the NHEJ machinery, and then, if unsuccessful, switch to trying to recruit telomerase. Alternatively, Ku might arrive at the double-strand break already in a complex with component(s) of one machine or the other, in which case the relative stabilities of those complexes versus their rates of accomplishing DNA repair would dictate which mechanism prevails.
Does Ku interact with telomerase in other species?
Comparison of the five Saccharomyces species whose genomes have been sequenced reveals that Ku80 has been highly conserved, including the N-terminal domain in which the yku80-135i insertion lies (Cliften et al. 2003). The telomerase RNAs have evolved more rapidly. However, in S. mikatae and S. kudriavzevii, we have identified sequences related to TLC1 that can fold into structures with great similarity to the 48-nucleotide stem-loop of S. cerevisiae (data not shown). Thus, we predict that the interaction between Ku and telomerase RNA will be conserved among these yeasts.
Ku has variable effects on telomere length in different species. Telomeres are shorter than wild type in Schizosaccharomyces pombe ku70 mutants (Baumann and Cech 2000), whereas telomeres are longer than wild type in Arabidopsis ku70 mutants (Riha and Shippen 2003), and telomeres of both short and long lengths have been reported for Ku80-/- mice (Samper et al. 2000; d'Adda di Fagagna et al. 2001). Because telomere length is set by a complex series of interactions between many telomere-binding proteins and positive and negative regulators of telomerase (McEachern et al. 2000), the effect of removing Ku from the mix could have different outcomes in different species or in different genetic backgrounds.
However, the potential for Ku to modulate telomerase activity in mammalian cells appears to exist. Yoo and Dynan (1998) reported that purified human Ku binds RNA. More recently, Chai et al. (2002) have shown that α-Ku70 and α-Ku80 antibodies can immunoprecipitate telomerase from human cell lines. Whether this is a protein-protein interaction or a protein-RNA interaction is not clear; these antibodies can immunoprecipitate hTERT, the catalytic subunit of human telomerase, when it is mixed with Ku70 and Ku80 in a rabbit reticulocyte lysate, but hTERT can functionally reconstitute with the rabbit telomerase RNA (Xiang et al. 2000), so it is possible that the interaction between Ku and TERT in these lysates is RNA mediated. On the other hand, the human telomerase RNA is much smaller than the yeast TLC1 RNA, and thus, has less potential to serve as a scaffold for protein binding; over time the Ku-telomerase interaction in humans may have evolved to rely on the protein rather than the RNA component of the enzyme. Regardless of the details, the biological advantage—increased efficiency of telomerase recruitment—may be the same in both organisms.
The putative interaction between Ku and human telomerase may be particularly important in tumor cells. Telomerase becomes reactivated in the vast majority of human tumors (McEachern et al. 2000); this has traditionally been thought to be necessary to overcome the telomere attrition that otherwise occurs upon DNA replication in telomerase-deficient cells. However, tumor cells must also cope with massive amounts of genomic instability (Lengauer et al. 1998). Therefore, a second reason for reactivating telomerase may be to provide tumor cells with an additional mechanism for healing DNA damage. In support of this idea, cell lines transformed with hTERT have been shown recently to exhibit less radiation sensitivity than their nontransformed counterparts, although the NHEJ activities of extracts made from these cell lines are identical (Sharma et al. 2003). Thus, Ku's role in bringing telomerase to multiple types of DNA ends may have been conserved across eukaryotes, signifying its importance in genome preservation.
Materials and methods
Yeast strains, medium, and methods
Yeast strains used in this work are listed in Table 2. Medium recipes and protocols for standard manipulations of yeast are described on the Gottschling laboratory web site (http://www.fhcrc.org/labs/gottschling).
Table 2.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| BJ2168 | MATaleu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407gal2 | Jones 1991 |
| UCC3505 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1::HIS3 adh4::URA3-TEL-VIIL ADE-TEL-VR | Singer and Gottschling 1994 |
| UCC6057 | UCC3505 yku70::KanMX | Peterson et al. 2001 |
| UCC6058 | UCC3505 yku80::KanMX | Peterson et al. 2001 |
| UCC5913 | MATa-inc ura3-52 lys2-801 ade2-101 trpΔ63 his3Δ200 leu2Δ1::GAL1-HO-LEU2 VII-L::ADE2-TG(1-3)-HO site-LYS2 | Diede and Gottschling 1999 |
| UCC6073 | MATa-inc ura3-52 lys 2-801 ade 2-101 trp 1Δ63 his 3Δ200 leu2Δ1::GAL1-HO-LEU2 VII-L::ADE2-TG(1-3)-HO site-LYS2 tlc 1Δ48 | This study |
| UCC3744 | UCC5913 yku80-135i | This study |
| UCC3745 | UCC5913 yku80::KanMX | This study |
| UCC3746 | UCC6073 yku80-135i | This study |
| BY4733 | MATahis3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | Brachmann et al. 1998 |
| UCC3755 | BY4733 yku80-135i | This study |
| UCC1540 | MATα his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 tlc1Δ48 | This study |
| UCC5114a | MATahis3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 | This study |
| UCC5116a | MATahis3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 tlc1Δ148 | This study |
| UCC5118-2a | MATahis3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 tlc1Δ48 yku80-135i | This study |
| UCC5119a | MATα his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 tlc1Δ48 yku80-135i | This study |
| UCC5120a | MATahis3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hxt13::URA3 yku80-135i | This study |
Plasmid construction and mutagenesis
pTCG, pTCG-3X stem, pTCG-3X mutant stem, pRS306-tlc1_Δ_48, pRS425TEF-YKU70, and pRS423TEF-YKU80 have been described previously (Peterson et al. 2001). Low-copy YKU70 and YKU80 plasmids were generated by amplifying YKU70 and YKU80 from the yeast genome, along with 1.5-1.7 kb of upstream sequence and 0.5 kb of downstream sequence, and cloning the PCR products into the _Bam_HI-_Sal_I sites (for YKU70) or the _Hind_III-_Pst_I sites (for YKU80) of pRS315 (Brachmann et al. 1998). pRS315-YKU70 complements _yku70_Δ; pRS315-YKU80 complements _yku80_Δ. pRS315-YKU70 and pRS315-YKU80 were mutagenized in vitro by Tn7 transposition, using the GPS-LS kit (NEB). One mutagenized pRS315-yku80 plasmid enabled cells to retain telomeric silencing despite TLC1 overexpression; this plasmid was subsequently named pRS315-yku80-135i. pRS423TEF-_YKU80_-TAP was constructed by amplifying the TAP-TRP cassette from pBS1479 (Rigaut et al. 1999) and co-transforming the resulting fragment with pRS423TEF-YKU80. The tagged Ku80 subunit retained the ability to complement _yku80_Δ. pRS423TEF-_yku80-135i_-TAP was created by replacing the _Spe_I-_Sph_I fragment of pRS423TEF-_YKU80_-TAP with a PCR fragment containing the N-terminal half of yku80-135i. Sequencing verified that the only mutation in the yku80-135i sequence was the 15-bp insertion. pRS306-yku80-135i was created by cloning a _Hind_III-_Not_I fragment from pRS315-yku80-135i into pRS306 (Brachmann et al. 1998); this plasmid was used to introduce yku80-135i into the genome by two-step gene replacement. pTCT7-3X stem and pTCT7-3X mutant stem were created by popping out the _Apa_I-_Bam_HI fragment containing the GAL1 promoter from pTCG-3X stem and pTCG-3X mutant stem, respectively.
Ku purification
Wild-type and mutant Ku were purified from BJ2168 cells containing pRS425TEF-YKU70 and either pRS423TEF-_YKU80_-TAP or pRS423TEF-_yku80-135i_-TAP. Cultures (1.5 L) were grown at 30°C in YC-trp-leu medium to a density of -3 × 107 cells/mL. Cells were harvested, washed once with water, once with TAP1 buffer (25 mM HEPES at pH 8, 200 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM DTT), and frozen in liquid nitrogen. Cells were broken by grinding the frozen cell pellet with an equal amount of dry ice in a coffee mill (Krups). The resulting powder was resuspended in one pellet volume of TAP1 buffer plus one tablet Mini-Complete protease inhibitors (Roche), and then sonicated with 4 × 15-sec bursts (Fisher 550 Sonic Dismembrator, setting #3). The extract was clarified by two rounds of centrifugation, first at 4000 rpm for 10 min in a JM-6 rotor, and then at 27,000 rpm for 90 min in an SW41 rotor. The cleared extract was adjusted to contain 10% glycerol, 10 mM Tris (pH 8) and 0.1% NP40, and Ku was purified by sequential passage of the extracts over IgG and calmodulin beads. The protocols and buffers of Rigaut et al. (1999) were followed, except that the first binding was done overnight in batch with 1 mL IgG-Sepharose fast flow beads (Amersham) per 10 mL of extract, and the final elution buffer contained 10% glycerol. Coomassie-stained protein gels were used to identify which fractions contained Ku; those fractions were pooled, dialyzed against Ku storage buffer (10 mM Tris at pH 8, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol), aliquoted, and stored at -80°C.
In vitro binding assays
RNA substrates were generated by in vitro transcription, using _Spe_I-linearized pTCT7-3X stem or pTCT7-3X mutant stem as a template. Reactions (20 μL) contained 40 mM Tris (pH 8), 10 mM MgCl2, 5 mM DTT, 0.15 mg/mL spermidine, 0.01% Triton X-100, 0.008% PEG, 12 mM ATP, GTP, and CTP, 0.01 mM UTP, 3 μM [α32P]UTP (800 Ci/mmole), 100 units T7 RNA polymerase (NEB), and 20 units SuperRNaseIN (Ambion). Reactions were incubated for 2 h at room temperature, supplemented with 1 mM CaCl2 and 10 units DNaseI (Roche), incubated for 15 min at 37°C, phenol-chloroform extracted, and run through a MiniQuick Spin column (Roche). A DNA substrate was generated by annealling two complementary ADE2 oligos (100 mers), end-labeling the resulting duplex DNA with T4 polynucleotide kinase (NEB) according to the manufacturer's instructions, and purifying it as described above.
Binding reactions (20 μL) contained 21 mM Tris (pH 7.5), 10 mM NaCl, 5 mM MgCl2, 0.05 mM EDTA,1 mM DTT, 11% glycerol, 100 μg/mL BSA, 12-16 pM RNA or 190 pM DNA, and the indicated amounts of Ku. Reactions were incubated on ice for 30 min and then loaded onto a 5% nondenaturing polyacrylamide gel containing 1 mM MgCl2 and 5% glycerol and run in a Tris-glycine buffer. The gel was dried on Whatmann paper and imaged with a Typhoon PhosphorImager (Molecular Dynamics).
In vivo telomere analysis
Telomere length was evaluated by Southern blot analysis, using _Xho_I-digested genomic DNA and a digoxigenin-labeled Y′ probe (Singer et al. 1998). Telomere end structure was evaluated by in-gel hybridization of _Xho_I-digested DNA as described (Gravel et al. 1998).
Plasmid rejoining assay
Strains were transformed with 200 ng of supercoiled or _Eco_RI-linearized pBTM116 as described (Boulton and Jackson 1996).
De novo telomere addition assay
The procedure of Diede and Gottschling (1999) was followed. In brief, log-phase cells were arrested with nocodazole (10 μg/mL) and then shifted into YEP + 3% galactose + nocodazole. Time points were collected hourly. DNA was harvested, digested with _Spe_I, and analyzed by Southern blot, using an ADE2 probe.
Gross chromosomal rearrangement assay
Cultures (5 mL) of each strain were grown in YEPD to log phase (1 × 107 cells/mL) in a rotating wheel at 30°C. A total of 0.015% MMS was added and the cultures were returned to the wheel for 1 h. The cells were spun down, washed once, resuspended in fresh YEPD, and returned to the wheel for -17 h. Cells were plated on YC + FOA + canavanine and on YEPD to determine the GCR frequency. FOARCANR colonies were analyzed by multiplex PCR to determine which ORFs were still present, and by Southern blot hybridization to determine whether a restriction fragment containing that last ORF was heterogeneous or sharp in appearance. The size of the restriction fragment also enabled the site of telomere addition to be estimated; those sites were then captured by two rounds of PCR using the strategy of Myung et al. (2001). The second PCR product was sequenced and compared with the chromosome V sequence available on SGD (http://www.yeastgenome.org). Oligo sequences used in this work are available upon request.
Acknowledgments
We thank members of the Gottschling lab for helpful discussions, and we thank Sue Biggins, Rich Gardner, Fred van Leeuwen, Michael McMurray, Gerry Smith, and Meng-Chao Yao for comments on the manuscript. This work was supported by National Institutes of Health grants F32 GM20104 (to A.E.S.) and R01 GM43893 (to D.E.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1125903.
References
- Anderson E.M., Halsey, W.A., and Wuttke, D.S. 2003. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry 42**:** 3751-3758. [DOI] [PubMed] [Google Scholar]
- Baumann P. and Cech, T.R. 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 11**:** 3265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biery M.C., Stewart, F.J., Stellwagen, A.E., Raleigh, E.A., and Craig, N.L. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28**:** 1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24**:** 4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17**:** 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:** 115-132. [DOI] [PubMed] [Google Scholar]
- Chai W., Ford, L.P., Lenertz, L., Wright, W.E. and Shay, J.W. 2002. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277**:** 47242-47247. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301**:** 71-76. [DOI] [PubMed] [Google Scholar]
- Cohn M. and Blackburn, E.H. 1995. Telomerase in yeast. Science 269**:** 396-400. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Hande, M.P., Tong, W.M., Roth, D., Lansdorp, P.M., Wang, Z.Q., and Jackson, S.P. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11**:** 1192-1196. [DOI] [PubMed] [Google Scholar]
- Diede S.J. and Gottschling, D.E. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 99**:** 723-733. [DOI] [PubMed] [Google Scholar]
- Dionne I. and Wellinger, R.J. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. 93**:** 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois M.L., Haimberger, Z.W., McIntosh, M.W., and Gottschling, D.E. 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161**:** 995-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad, V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286**:** 117-120. [DOI] [PubMed] [Google Scholar]
- Falzon M., Fewell, J.W., and Kuff, E.L. 1993. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268**:** 10546-10552. [PubMed] [Google Scholar]
- Fellerhoff B., Eckhardt-Schupp, F., and Friedl, A.A. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics 154**:** 1039-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S. and Wellinger, R.J. 2002. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22**:** 2182-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivee, M., Labrecque, P., and Wellinger, R.J. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280**:** 741-744. [DOI] [PubMed] [Google Scholar]
- Greider C.W. 1991. Telomeres. Curr. Opin. Cell Biol. 3**:** 444-451. [DOI] [PubMed] [Google Scholar]
- Haber J.E. 2000. Parters and pathways repairing a double-strand break. Trends Genet. 16**:** 259-264. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Evans, S.K., Weilbaecher, R.G., and Lundblad, V. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10**:** 809-812. [DOI] [PubMed] [Google Scholar]
- Jones E. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194**:** 428-453. [DOI] [PubMed] [Google Scholar]
- Kramer K.M. and Haber, J.E. 1993. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes & Dev. 7**:** 2345-2356. [DOI] [PubMed] [Google Scholar]
- Lendvay T.S., Morris, D.K., Sah, J., Balasubramanian, B., and Lundblad, V. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144**:** 1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler, K.W., and Vogelstein, B. 1998. Genetic instabilities in human cancers. Nature 396**:** 643-649. [DOI] [PubMed] [Google Scholar]
- Lin J.J. and Zakian, V.A. 1995. An in vitro assay for Saccharomyces telomerase requires EST1. Cell 81**:** 1127-1135. [DOI] [PubMed] [Google Scholar]
- Lingner J., Hughes, T.R., Shevchenko, A., Mann, M., Lundblad, V., and Cech, T.R. 1997a. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276**:** 561-567. [DOI] [PubMed] [Google Scholar]
- Lingner J., Cech, T.R., Hughes, T.R., and Lundblad, V. 1997b. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. 94**:** 11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. and Szostak, J.W. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57**:** 633-643. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche, T., Suka, N., Grunstein, M., and Gasser, S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97**:** 621-633. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Krauskopf, A., and Blackburn, E.H. 2000. Telomeres and their control. Annu. Rev. Genet. 34**:** 331-358. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R.M., Anderson, E.M., Hughes, T.R., Lundblad, V., and Wuttke, D.S. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296**:** 145-147. [DOI] [PubMed] [Google Scholar]
- Myung K. and Kolodner, R.D. 2003. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair 2**:** 243-258. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta, A., and Kolodner, R.D. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104**:** 397-408. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes, T.R., Lue, N.F., and Lundblad, V. 1996. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274**:** 249-252. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E., and Lundblad, V. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8**:** 657-660. [DOI] [PubMed] [Google Scholar]
- Pennock E., Buckley, K., and Lundblad, V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104**:** 387-396. [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Stellwagen, A.E., Diede, S.J., Singer, M.S., Haimberger, Z.W., Johnson, C.O., Tzoneva, M., and Gottschling, D.E. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27**:** 64-67. [DOI] [PubMed] [Google Scholar]
- Polotnianka R.M., Li, J., and Lustig, A.J. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8**:** 831-834. [DOI] [PubMed] [Google Scholar]
- Porter S.E., Greenwell, P.W., Ritchie, K.B., and Petes, T.D. 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24**:** 582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio, O.M., Zierath, P.D., Billington, B.L., Chhablani, S.K., and Gottschling, D.E. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes & Dev. 7**:** 1133-1145. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17**:** 1030-1032. [DOI] [PubMed] [Google Scholar]
- Riha K. and Shippen, D.E. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. 100**:** 611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Goytisolo, F.A., Slijepcevic, P., van Buul, P.P., and Blasco, M.A. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1**:** 244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G.G., Gupta, A., Wang, H., Scherthan, H., Dhar, S., Gandhi, V., Iliakis, G., Shay, J.W., Young, C.S.H., and Pandita, T.K. 2003. hTERT associates with human telomeres and enhances genomic stability and DNA repair. Oncogene 22**:** 131-146. [DOI] [PubMed] [Google Scholar]
- Singer M.S. and Gottschling, D.E. 1994. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266**:** 404-409. [DOI] [PubMed] [Google Scholar]
- Singer M.S., Kahana, A., Wolf, A.J., Meisinger, L.L., Peterson, S.E., Goggin, C., Mahowald, M., and Gottschling, D.E. 1998. Identification of high copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150**:** 613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner B.R., Hidaka, K., and Futcher, B. 1996. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl Acad. Sci. 93**:** 2817-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.H. and Jackson, S.P. 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2**:** 197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R. and Tuteja, N. 2000. Ku autoantigen: A multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35**:** 1-33. [DOI] [PubMed] [Google Scholar]
- Walker J.R., Corpina, R.A., and Goldberg, J. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412**:** 607-614. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Wolf, A.J., and Zakian, V.A. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72**:** 51-60. [DOI] [PubMed] [Google Scholar]
- Xiang H., Wang, J., Mao, Y.W., and Li, D.W. 2000. hTERT can function with rabbit telomerase RNA: Regulation of gene expression and attenuation of apoptosis. Biochem. Biophys. Res. Commun. 278**:** 503-510. [DOI] [PubMed] [Google Scholar]
- Yoo S. and Dynan, W.S. 1998. Characterization of the RNA binding properties of Ku protein. Biochemistry 37**:** 1336-1343. [DOI] [PubMed] [Google Scholar]