Protective immunity against Helicobacter is characterized by a unique transcriptional signature (original) (raw)
Abstract
Immunization with a whole-cell sonicate vaccine of Helicobacter felis in conjunction with cholera toxin as a mucosal adjuvant induces long-term protective immunity in a majority of laboratory mice. We have combined gene expression profiling and immunohistochemical analysis on a set of immunized animals to better understand the mechanism of protection. The stomachs of protected animals exhibited a strikingly different transcriptional profile compared with those of nonprotected or control mice, indicating that vaccination targets the appropriate site and leaves a molecular signature. Among the genes whose up-regulation is significantly correlated with protection are a number of adipocyte-specific factors. These include the fat-cell-specific cytokines adipsin, resistin, and adiponectin and the adipocyte surface marker CD36. Interestingly, potentially protective T and B lymphocytes can be found embedded in the adipose tissue surrounding protected stomachs but never in control or unprotected stomachs. Adipsin-specific immunohistochemical staining of protected stomach sections further revealed molecular cross-talk between adjacent lymphoid and adipose cell populations. We propose a mechanism of protection that involves the effector responses of either or both lymphocyte subclasses as well as the previously unappreciated paracrine functions of adipose tissue surrounding the resident lymphocytes.
Keywords: rodent, lymphocytes, bacterial infections, vaccination, mucosa
Gastric infection with the bacterium Helicobacter pylori is associated with the development of acute chronic gastritis, peptic ulcer disease, and two gastric malignancies, adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma (1–4). Therefore, successful eradication of H. pylori from infected populations is considered an important public health goal. The currently used eradication strategy focuses on treatment of those individuals that display overt disease symptoms with a combination of two to three antibiotics and a proton pump inhibitor (5). However, the increasing prevalence of antibiotic resistance (6), the lack of protection against reinfection, and the high cost limit the usefulness of this strategy for control of _H. pylori_-related diseases. The development of a prophylactic vaccine with possible therapeutic applications is widely viewed as a promising complementary approach.
The first animal model for Helicobacter infection became available in 1990 with the demonstration that Helicobacter felis could colonize the murine stomach and induce chronic active gastritis (7). Using this model, Chen et al. (8) were able to demonstrate that immunization with a whole-cell H. felis sonicate oral vaccine in combination with cholera toxin as a mucosal adjuvant can protect the host from later challenge with large viable doses of the same strain. Moreover, the organism load of an existing H. felis infection could also be significantly reduced with the same vaccine regimen, thereby demonstrating the effect of therapeutic immunization (9). Observations made by using the H. felis model system were subsequently confirmed by using a mouse-adapted strain of H. pylori (10, 11).
The protective mechanism of this and other vaccination strategies against H. pylori is currently being actively investigated. Due to the localization of H. pylori in the mucus layer of the gastric epithelium, it was initially assumed that protective immunity was mediated by IgA secreted into the stomach lumen. Evidence from challenge experiments using B lymphocyte-deficient mouse strains has ruled out this possibility, because these animals retain the same level of protection as wild-type mice (12–14). Additional experiments have shown that MHC I-deficient mice, which lack the CD8+ population of T cells, are fully protected, whereas MHC II deficiency results in lack of protection (12, 15). Hence, successful immunization against H. pylori seems to depend on the CD4+ population of T cells. Adoptive transfer experiments of _Helicobacter-_specific Th1 and Th2 cells further indicate that only a Th2-polarized response can mediate protective immunity (16). In contrast, colonization of the human stomach with H. pylori induces a proinflammatory Th1 response characterized by high levels of IFNγ and IL-12, which is clearly ineffective in combating infection (17, 18) and does not prevent reinfection after antibiotic therapy. Several Helicobacter vaccine protocols effective in small animal models have been tested in human trials (19–23); however, none of them was successful in triggering a strong systemic response in the majority of vaccinees, suggesting that large discrepancies exist between murine and human hosts.
By combining the well characterized whole-cell sonicate/cholera toxin vaccine (8, 14, 24–26) with gene expression profiling of protected vs. unprotected stomachs, we attempt to shed light on the issue of protective immunity from a very different experimental angle. As shown earlier, this vaccine protocol stimulates long-term, probably life-long, immunity (25) and prevents the pathological changes normally associated with chronic Helicobacter infection of the stomach (26). Here we present data indicating that the protective response occurs locally in the gastric mucosal environment, is clearly reflected at the gene expression level, and involves a number of previously recognized as well as novel cell types.
Materials and Methods
Animals. Specific pathogen-free female BALB/c mice were obtained from the Animal Resource Centre, Canning Vale, Australia, at 7 wk of age. All protocols involving animal experimentation were approved by the Animal Care and Ethics Committee at the University of New South Wales.
Immunization and Infection Experiments. The protocol for prophylactic immunization with 1 mg of H. felis sonicate and 10 μg of cholera toxin (Sigma) was described previously (26). For challenge at 2 mo postimmunization, mice were infected intragastrically with 0.1 ml of viable H. felis bacterial culture (≈108 bacteria) twice during a 3-d period.
Sampling and Histological Assessment. Mice were killed 22 mo after bacterial challenge, and the stomachs were removed. Half of each stomach was fixed in 10% buffered formalin before embedding in paraffin, and the rest was snap-frozen in liquid nitrogen for RNA preparation. Sections were cut and stained with May–Grünwald–Giemsa to assess bacterial colonization and with hematoxylin/eosin for histological analysis.
Microarray Analysis. Gene expression profiling with 38,000 element-spotted murine cDNA microarrays was performed as described (27). Microarray data were stored in the Stanford Microarray Database (28). Data were filtered with respect to spot quality (spots with regression correlations <0.6 were omitted) and data distribution (genes whose log2 of red/green normalized ratio is more than 2 standard deviations away from the mean in at least five arrays were selected) before clustering. Only genes for which information was available for >70% of arrays were included. Data were log2 transformed and analyzed by using cluster and treeview (29). Statistical analysis was done by using the significance analysis of microarrays algorithm (30). Data from all of the arrays used in this paper are available at http://genome-www4.stanford.edu/MicroArray/SMD.
Immunohistochemistry. Sequential 4-μm paraffin sections were stained with antibodies specific for the following murine antigens: CD45R/B220 on B cells (PharMingen/BD Biosciences), CD3 on T cells (Biomeda, Foster City, CA), adipsin on adipocytes (Santa Cruz Biotechnology) and F4/80 on macrophages (Abcam Limited, Cambridge, U.K.). The detection was performed by using biotinylated secondary antibodies in combination with horseradish-peroxidase-coupled streptavidin (Jackson ImmunoResearch) and the substrate DAB (Research Genetics/Invitrogen, Carlsbad, CA). All sections were counterstained with hematoxylin, dehydrated, and mounted by using synthetic non-aqueous mounting medium. A Zeiss Axiophot microscope equipped with an AxioCam digital camera and axiovision 3.1. software (Zeiss) were used for documentation.
Results
Gene Expression Profiling of Control, Infected, and Immunized Mice. We analyzed 32 mice in the present study: of these, 5 belonged to an untreated control group (group A), 8 were infected with H. felis (group B), and 19 were immunized with a whole-cell sonicate of H. felis and cholera toxin before challenge with H. felis (group C) 2 mo later. The mice were then maintained for 22 mo to allow sufficient time for development of _Helicobacter_-specific pathology before histological and gene expression analysis. Bacterial colonization was assessed by culture, histology, and ureB-specific RT-PCR (data not shown). Of the 19 immunized animals, only 5 were colonized, suggesting that the vaccine was protective in 74% of the cases. In contrast, all _H. pylori_-challenged unvaccinated animals were colonized successfully. Colonization in both groups was most dense in the antral region and in the antral/body transitional zone.
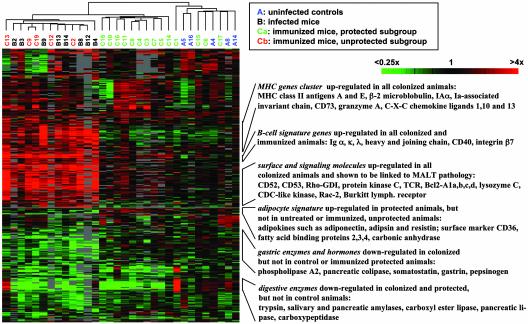
RNA was isolated from whole stomachs of all 32 animals and hybridized to a 38,000-element murine cDNA microarray. A hierarchical clustering algorithm was used to group genes and samples in an “unsupervised” fashion on the basis of similarities of gene expression (29). The relationships among the samples are summarized in a dendrogram (Fig. 1). All 13 colonized animals cluster together tightly and form one arm of the dendrogram. Interestingly, this branch contains both the eight challenged animals that did not receive the vaccine (black) and the five animals that were immunized unsuccessfully (the “unprotected” subgroup, depicted in red). The control animals (in blue) form a separate branch that also contains four mice of the “protected” subgroup of immunized animals. All other protected animals (green) cluster together in a third branch. Infection status is therefore revealed as a strong force in driving the clustering. Among the genes up-regulated in the infected group were lymphocyte-specific surface markers and signaling molecules previously linked to the development of MALT pathology such as CD52 and calgranulin A (27). Similarly, a number of gastric enzymes and hormones are specifically down-regulated in this group, most notably somatostatin, gastrin, phospholipase A2, and progastricsin. This trend most likely reflects the massive loss of certain epithelial and endocrine cell types (atrophy) that is a common feature of _Helicobacter_-induced pathology. In addition, the fact that a majority of immunized protected animals forms a distinct branch suggests that the immunization procedure alone also acts as a strong driving force, indicating that the used vaccine regimen elicits an immune response locally at the site of potential infection.
Fig. 1.
Molecular signature of immunized, _Helicobacter-_infected, and control murine stomachs. Data are a measure of relative gene expression and represent the quotient of the hybridization of the fluorescent cDNA probe prepared from each stomach sample compared with a reference pool. Red and green represent high and low experimental sample/reference ratios, respectively (see scale bar). Gray signifies technically inadequate or missing data. The color coding of array names is indicated in the box. Selected gene clusters are designated and described briefly.
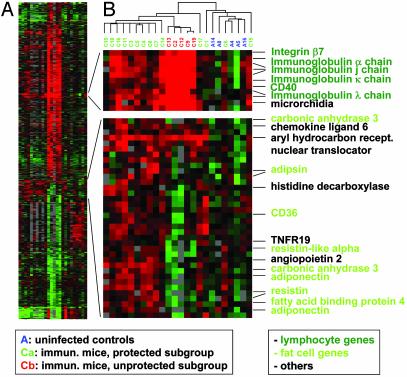
Statistical Comparison of the Gene Expression Profiles of Protected and Unprotected Stomachs. The lack of protective immunity in roughly one-fourth of the vaccinated animals turned out to be an asset in the comparison of protected with unprotected stomachs. In a statistical analysis using the multiclass function of the significance analysis of microarrays algorithm (30), three groups (control, immunized/protected, and immunized/unprotected) were subjected to a three-way comparison. The eight animals of the infected unvaccinated group were excluded from this analysis, because they did not contribute additional information (data not shown). Seven hundred and seventy-six genes (Fig. 2_A_) differed significantly between at least two of the three groups when a 5% false positive discovery rate cutoff was used. The majority of these genes distinguish the infected from all other samples and have been discussed (27). All significant genes were converted to a cluster-readable format by the program samster (downloadable from http://falkow.stanford.edu/whatwedo/software).
Fig. 2.
Three-way significance analysis of microarrays comparison of protected, unprotected, and control stomachs. (A) Overview of all 776 genes that differ significantly between at least two of the three groups when a 5% false positive discovery rate cutoff was used. (B) Two enlarged sections are shown. (Upper) Genes that are up-regulated in both the unprotected and protected subgroups of animals compared with the controls, albeit much stronger in the former group. (Lower) Genes that are up-regulated in the protected vs. unprotected or control groups. Selected genes are designated. The color coding of arrays and genes is indicated in the boxes.
Two up-regulated gene clusters were identified that contain genes whose expression correlates very tightly with protection (Fig. 2_B_). One of them represents a lymphocyte signature with mostly B cell-specific genes such as the Ig genes encoding the α, κ, λ, and joining chains. The same cluster also includes the B cell surface marker CD40 as well as integrin β7, a component of the homing receptor of both B and T cells for mucosal surfaces of the gastrointestinal tract. Whereas this cluster correlates well with protection, earlier experiments suggested that its up-regulation is not sufficient for protection: the same lymphocyte signature is found in stomachs from which H. pylori has been eradicated by antibiotic therapy, but these cured mice can be reinfected readily even with the same Helicobacter strain (A.M., J.O., A.L., and S.F., unpublished data).
The other cluster correlating tightly with protection contains a number of genes specifically expressed in fat cells/adipocytes. Among these are three adipocyte-specific cytokines, the so-called “adipokines,” adipsin, resistin, and adiponectin (formerly known as adipocyte complement-related protein of 30 kDa, or Acrp30). Other genes in this cluster include the fat cell-specific fatty acid-binding protein 4, carbonic anhydrase 3, and the surface marker CD36. In general, fat cell-specific genes are up-regulated in protected, but down-regulated in unprotected, immunized stomachs relative to the controls. The angiogenesis promoter angiopoietin 2 is also contained in this cluster, and fat tissue has previously been shown to be a major source of this secreted protein. Indeed, induction of angiopoietin 2 by the adipokine leptin has been shown to trigger the remodeling of fat tissue microvasculature (31). Another interesting gene expressed strongly in protected stomachs encodes histidine decarboxylase, a signature protein of enterochromaffin-like cells, an important endocrine cell type of the stomach. Histidine decarboxylase is a key enzyme in the histamine synthesis pathway, and secreted histamine in turn modulates gastric acid secretion.
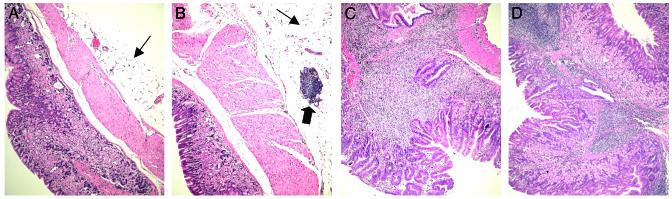
Histological Comparison of Murine Stomach Sections. The described differences in overall gastric gene expression suggest dramatic differences in cellular composition among the four groups of the study. In order to discern which additional cell types are present due to the immunization procedure and where they are localized in the stomach, we assessed the overall stomach histology on hematoxylin/eosin-stained sections for representatives of every group (Fig. 3). As reported (32), life-long infection of BALB/c mice with either H. pylori or H. felis results in a MALT-type pathology (Fig. 3 C and D) characterized by massive lymphocytic infiltration, destruction of the gastric epithelium, and, in a minority of cases, progression to MALT lymphoma. As suggested by the gene expression results, no morphological differences were observed between colonized mice that had received the vaccine (immunized/unprotected) and those that had been infected without prior vaccination.
Fig. 3.
Histological comparison of stomachs representative of each group of animals [control (A), immunized/protected (B), infected (C), and immunized/unprotected (D)]. The sections were stained with hematoxylin/eosin and examined at ×20. The thin arrows point to fat tissue in control and protected stomachs, and the thick arrow points to a cluster of lymphocytes embedded in the fat tissue.
Uninfected stomachs are usually embedded in a layer of fat tissue (Fig. 3_A_, indicated by thin arrow), the thickness of which varies slightly from individual to individual and from section to section. In contrast, infected stomachs are usually surrounded by a strongly diminished or even entirely disrupted fat layer, correlating with the loss of adipocyte-specific gene expression observed in infected stomachs relative to controls (Fig. 2). In contrast, protected stomachs exhibit a thicker fat layer than controls (Fig. 3 A and B), again confirming the gene expression results (Fig. 2).
Furthermore, the fat tissue of protected stomachs contains clusters of lymphocytes that cannot be found in controls (thick arrow). The striking presence of these clusters correlates with protection and is very likely reflected by the lymphocyte-specific gene cluster shown in Fig. 2.
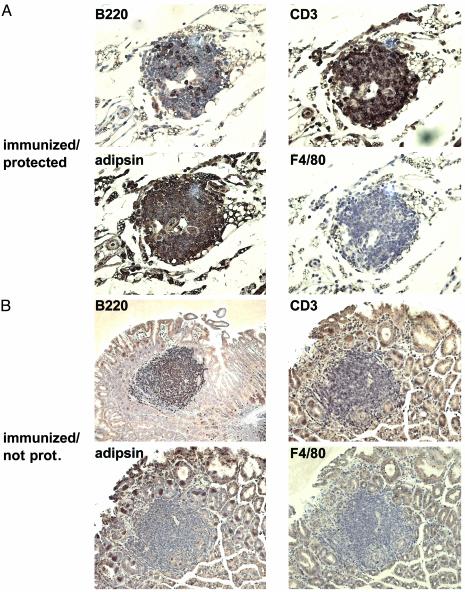
Immunohistochemical Analysis of Protected and Unprotected Stomachs. On the basis of the assumption that adipose tissue functions are crucial in protective immunity against Helicobacter and the observation that lymphocytic clusters reside in fat depots in protected stomachs, we hypothesized that there should be crosstalk between the two tissue types. The secreted adipokines are likely mediators of cross-talk with lymphocytes and have previously been shown to play this role (33); therefore, adipsin was chosen as a representative member of this group. Sequential 4-μm sections of paraffin-embedded stomach tissue were stained with an adipsin-specific antibody as well as antibodies against known marker proteins for T cells (CD3), B cells (B220/CD45R), and macrophages (F4/80) to determine the cellular composition of lymphocytic clusters in protected stomachs vs. MALT-type lymphoid aggregates in unprotected stomachs (Fig. 4).
Fig. 4.
Immunohistochemical staining for antigens specific for cell types potentially involved in a protective immune response. Four sequential sections were cut from paraffin-embedded tissue, stained with the indicated antibodies, and examined at ×40. A fat-embedded lymphocytic cluster typical of protected stomachs is shown in A, and a lymphoid aggregate typical of MALT tissue in infected mice is shown in B.
A representative cluster embedded in adipose tissue of a protected stomach is shown in Fig. 4_A_ Top: whereas very few cells stain strongly with the B cell-specific anti-B220 antibody, the vast majority stain positive for CD3, revealing a B/T cell ratio of ≈1 in 4. As expected, both markers show a characteristic membrane-staining pattern consistent with the surface localization of both antigens. No macrophages are present in the cluster. The surrounding fat tissue stains strongly for adipsin; interestingly, the adipsin signal further extends into the lymphocytic cluster, where it displays a membrane pattern similar to the two surface markers B220 and CD3, suggesting that adipsin binds to receptors present on lymphocytes of both subgroups.
In contrast, a typical lymphoid aggregate residing in the gastric epithelium of an unprotected colonized stomach (Fig. 4_B_) shows an almost inverse staining pattern: clearly, the vast majority (≈90%) of lymphocytes belong to the B subtype, as would be expected from a B cell lymphoproliferative disorder. A small but significant number of T cells can also be detected that have previously been shown to be _H. pylori_-specific and essential for the antigen-dependent proliferation of B cells in MALT lymphoma (34). In contrast to the clusters typical of protected stomachs, MALT aggregates do not stain with an adipsin-specific antibody, indicating that the proposed cross-talk between lymphoid and adipose tissue is restricted to protected stomachs and correlates with protection.
Discussion
Understanding the protective mechanism of any vaccination protocol is an essential prerequisite in developing a rational approach to future vaccine optimization and design. The conventional method of elucidating the mechanistic action of a vaccine is to analyze the effector functions of a small number of well studied cell types at the systemic level, i.e., in body fluids and ex vivo. This approach may not be suitable when mucosal immunization strategies are investigated, because a localized response at the site of potential pathogen entry or colonization may be more efficient and crucial at clearing such an infection than a systemic response.
By generating hypotheses from gene expression profiles of stomachs of vaccinated animals and testing them by immunohistochemistry, we have put special emphasis on studying the target site of a Helicobacter vaccine. Whereas H. pylori can persistently colonize the mucus lining of human stomachs for decades and is perfectly adapted to this niche, it has no genetic attributes to invade deeper tissues or to enter the bloodstream. Therefore, unlike other mucosal surface colonizers (Neisseria meningitidis, Streptococcus pneumoniae, or Haemophilus influenzae), H. pylori never becomes systemic. The segregation of animals into the three distinct branches representing controls, colonized, and protected animals, respectively (Fig. 1), demonstrates a profound effect of both Helicobacter infection and whole-cell vaccination on gastric gene expression profiles and indicates a strong localized response to the vaccine. Only one previous study has provided evidence for a localized gastric B cell response by measuring antigen-specific IgA and IgG in biopsy specimens of vaccinated individuals who had received a combination of killed H. pylori whole-cell vaccine and mutant LTR192G (35).
Our comparison of protected and unprotected or control mice establishes a link between the production and secretion of adipokines and a protective response. One of these factors, adipsin, could furthermore be shown to bind to the surfaces of lymphocytes that are organized in clusters and embedded in adipose tissue of protected stomachs (Fig. 4_A_) implying that cross-talk occurs between the two cell types. Indeed, the paracrine relationships between adipose and lymphoid tissues, both of which can be found in close proximity, i.e., in the perinodal depots and the omentum, have recently been a matter of intense research (33, 36–40). The omentum is a bilayered sheet of mesothelial cells that consists in large part of adipocytes and connects the stomach, spleen, pancreas, and transverse colon. It contains aggregates of immune cells (“milky spots”) that are believed to play a role in natural immunity (39). A special population of B cells (termed B1 cells) homes to the omentum through the interaction of the chemokine CXCL13 with its receptor on the B1 cell surface, CXCR5, and is believed to play a prominent role in natural antibody production (40). Indeed, CXCL13–/– mice are deficient in mounting an antiphosphorylcholine response against an i.p. injected streptococcal vaccine (40). Whether the aggregates we find surrounding the stomachs protected from Helicobacter infection are part of the omentum structure remains to be established.
Adipocytes themselves are increasingly no longer viewed only as the primary site for whole body energy storage but may need to be redefined as a major endocrine cell type that releases hormones in response to extracellular stimuli or changes in metabolic status.
The best-studied adipokine to date is leptin, which has complex functions in the regulation of immunity and inflammation (36). In experimental animals, leptin levels are acutely increased by inflammatory stimuli such as lipopolysaccharide and proinflammatory cytokines (41, 42), which in turn up-regulate the production of Th-1-specific cytokines (43). The functions of other members of the adipokine family are less well understood. Adiponectin and resistin are antagonists mediating insulin sensitivity and resistence, respectively (44, 45). Adipsin, also known as adipocyte complement factor D precursor, is a component of the alternative complement pathway, has serine protease activity, and is secreted into the bloodstream (46).
In our study, all three adipokines are positively coregulated in stomachs protected against Helicobacter (Fig. 2). Several protective mechanisms based on the sum of the above-mentioned findings are conceivable. (i) Soluble factors secreted by adipocytes spread through the epithelium into the stomach lumen and directly mediate a protective effect by neutralizing Helicobacter, adipsin being the most likely candidate in conjunction with antigen-specific mucosal IgA, the secretion of which has been shown to be triggered by a very similar vaccine protocol in humans (35). (ii) Secreted adipocyte factors play an indirect role by stimulating the effector functions of resident T cell populations. This proposition is supported by numerous studies demonstrating an essential role of CD4+ T cells in the protection against Helicobacter (12, 15, 47). Molecular cross-talk is likely to occur in both directions, because activated lymphocytes secrete cytokines that in turn activate perinodal adipose tissue by stimulating increased lipolysis both in vitro (48) and in vivo (49). (iii) Antigen-specific T and B cells residing in fat-embedded clusters act in concert to neutralize Helicobacter, and the observed increase in fat tissue characteristic of protected stomachs is a consequence rather than a cause of protection.
Acknowledgments
We thank D. Scotty Merrell and Yueh-Hsiu Chien for helpful discussions and suggestions. We are indebted to the staff of the Stanford Functional Genomics Facility for providing high-quality mouse arrays and to John Wilson for help with the animal experiments. This work was supported by National Institutes of Health Grants AI38459 and CA92229 (to S.F.) and Deutsche Forschungsgemeinschaft Grant MU1675/1-1 (to A.M.). J.O. is funded by National Health and Medical Research Council Grant 9936694.
Abbreviation: MALT, mucosa-associated lymphoid tissue.
References
- 1.Wyatt, J. I. & Rathbone, B. J. (1988) Scand. J. Gastroenterol. 142**,** 44–49. [PubMed] [Google Scholar]
- 2.Stolte, M. & Eidt, S. (1989) J. Clin. Pathol. 42**,** 1269–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genta, R. M., Hamner, H. W. & Graham, D. Y. (1993) Hum. Pathol. 24**,** 577–583. [DOI] [PubMed] [Google Scholar]
- 4.Isaacson, P. G. (1999) Ann. Oncol. 10**,** 637–645. [DOI] [PubMed] [Google Scholar]
- 5.Unge, P. (1999) Curr. Top. Microbiol. Immunol. 241**,** 261–300. [DOI] [PubMed] [Google Scholar]
- 6.Glupczynski, Y. (1998) Acta Gastroenterol. Belg. 61**,** 357–366. [PubMed] [Google Scholar]
- 7.Lee, A., Fox, J. G., Otto, G. & Murphy, J. (1990) Gastroenterology 99**,** 1315–1323. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M., Lee, A. & Hazell, S. (1992) Lancet 339**,** 1120–1121. [DOI] [PubMed] [Google Scholar]
- 9.Doidge, C., Crust, I., Lee, A., Buck, F., Hazell, S. & Manne, U. (1994) Lancet 343**,** 914–915. [DOI] [PubMed] [Google Scholar]
- 10.Marchetti, M., Arico, B., Burroni, D., Figura, N., Rappuoli, R. & Ghiara, P. (1995) Science 267**,** 1655–1658. [DOI] [PubMed] [Google Scholar]
- 11.Lee, A, O'Rourke, J., De Ungria, M. C., Robertson, B., Daskalopoulos, G. & Dixon, M. F. (1997) Gastroenterology 112**,** 1386–1397. [DOI] [PubMed] [Google Scholar]
- 12.Ermak, T. H., Giannasca, P. J., Nichols, R., Myers, G. A., Nedrud, J., Weltzin, R., Lee, C. K., Kleanthous, H. & Monath, T. P. (1998) J. Exp. Med. 188**,** 2277–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard, T. G., Czinn, S. J., Redline, R. W., Sigmund, N., Harriman, G. & Nedrud, J. G. (1999) Cell. Immunol. 191**,** 74–80. [DOI] [PubMed] [Google Scholar]
- 14.Sutton, P., Wilson, J., Kosaka, T., Wolowczuk, I. & Lee, A. (2000) Immunol. Cell. Biol. 78**,** 28–30. [DOI] [PubMed] [Google Scholar]
- 15.Pappo, J., Torrey, D., Castriotta, L., Savinainen, A., Kabok, Z. & Ibraghimov, A. (1999) Infect. Immun. 67**,** 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammadi, M., Nedrud, J., Redline, R., Lycke, N. & Czinn, S. J. (1997) Gastroenterology 113**,** 1848–1857. [DOI] [PubMed] [Google Scholar]
- 17.Bamford, K. B., Fan, X., Crowe, S. E., Leary, J. F., Gourley, W. K., Luthra, G. K., Brooks, E. G., Graham, D. Y., Reyes, V. E. & Ernst, P. B. (1998) Gastroenterology 114**,** 482–492. [DOI] [PubMed] [Google Scholar]
- 18.Smythies, L. E., Waites, K. B., Lindsey, J. R., Harris, P. R., Ghiara, P. & Smith, P. D. (2000) J. Immunol. 165**,** 1022–1029. [DOI] [PubMed] [Google Scholar]
- 19.DiPetrillo, M. D., Tibbetts, T., Kleanthous, H., Killeen, K. P. & Hohmann, E. L. (1999) Vaccine 18**,** 449–459. [DOI] [PubMed] [Google Scholar]
- 20.Angelakopoulos, H. & Hohmann, E. L. (2000) Infect. Immun. 68**,** 2135–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bumann, D., Metzger, W. G., Mansouri, E., Palme, O., Wendland, M., Hurwitz, R., Haas, G., Aebischer, T., von Specht, B. U. & Meyer, T. F. (2001) Vaccine 20**,** 845–852. [DOI] [PubMed] [Google Scholar]
- 22.Kotloff, K. L., Sztein, M. B., Wasserman, S. S., Losonsky, G. A., DiLorenzo, S. C. & Walker, R. I. (2001) Infect. Immun. 69**,** 3581–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michetti, P., Kreiss, C., Kotloff, K. L., Porta, N., Blanco, J. L., Bachmann, D., Herranz, M., Saldinger, P. F., Corthesy-Theulaz, I., Losonsky, G., et al. (1999) Gastroenterology 116**,** 804–812. [DOI] [PubMed] [Google Scholar]
- 24.Lee, A. & Chen, M. (1994) Infect. Immun. 62**,** 3594–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radcliff, F. J., Chen, M. & Lee, A. (1996) Vaccine 14**,** 780–784. [DOI] [PubMed] [Google Scholar]
- 26.Sutton, P., Danon, S. J., Walker, M., Thompson, L. J., Wilson, J., Kosaka, T. & Lee, A. (2001) Gut 49**,** 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, A., O'Rourke, J., Grimm, J., Guillemin, K., Dixon, M. F., Lee, A. & Falkow, S. (2003) Proc. Natl. Acad. Sci. USA 100**,** 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherlock, G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29**,** 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95**,** 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98**,** 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen, B., Barkan, D., Levy, Y., Goldberg, I., Fridman, E., Kopolovic, J. & Rubinstein, M. (2001) J. Biol. Chem. 276**,** 7697–7700. [DOI] [PubMed] [Google Scholar]
- 32.Enno, A., O'Rourke, J., Rolfe Howlett, C., Jack, A., Dixon, M. F. & Lee, A. (1995) Am. J. Pathol. 147**,** 217–222. [PMC free article] [PubMed] [Google Scholar]
- 33.Pond, C. M. (2003) Trends Immunol. 24**,** 13–18. [DOI] [PubMed] [Google Scholar]
- 34.Du, M. Q. & Isaacson, P. G. (2002) Lancet Oncol. 3**,** 97–104. [DOI] [PubMed] [Google Scholar]
- 35.Losonsky, G. A., Kotloff, K. L. & Walker, R. I. (2003) Vaccine 21**,** 562–565. [DOI] [PubMed] [Google Scholar]
- 36.Fantuzzi, G. & Faggioni, R. (2000) J. Leukocyte Biol. 68**,** 437–446. [PubMed] [Google Scholar]
- 37.Saltiel, A. R. (2001) Nat. Med. 7**,** 887–888. [DOI] [PubMed] [Google Scholar]
- 38.Mora, S. & Pessin, J. E. (2002) Diabetes Metab. Res. Rev. 18**,** 345–356. [DOI] [PubMed] [Google Scholar]
- 39.Williams, R. & White, H. (1986) Curr. Probl. Surg. 23**,** 789–865. [DOI] [PubMed] [Google Scholar]
- 40.Ansel, K. M., Harris, R. B. S. & Cyster, J. G. (2002) Immunity 16**,** 67–76. [DOI] [PubMed] [Google Scholar]
- 41.Grunfeld, C., Zhao, C., Fuller, J., Pollack, A., Moser, A., Friedman, J. & Feingold, K. R. (1996) J. Clin. Invest. 97**,** 2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarraf, P., Frederich, R. C., Turner, E. M., Ma, G., Jaskowiak, N. T., Rivet, D. J., 3rd, Flier, J. S., Lowell, B. B., Fraker, D. L. & Alexander, H. R. (1997) J. Exp. Med. 185**,** 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lord, G. M., Matarese, G., Howard, J. K., Baker, R. J., Bloom, S. R. & Lechler, R. I. (1998) Nature 394**,** 897–901. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi, T., Kamon, J., Waki, H., Terauchi, Y., Kubota, N., Hara, K., Mori, Y., Ide, T., Murakami, K., Tsuboyama-Kasaoka, N., et al. (2001) Nat. Med. 7**,** 941–946. [DOI] [PubMed] [Google Scholar]
- 45.Steppan, C. M., Bailey, S. T., Bhat, S., Brown, E. J., Banerjee, R. R., Wright, C. M., Patel, H. R., Ahima, R. S. & Lazar, M. A. (2001) Nature 409**,** 307–312. [DOI] [PubMed] [Google Scholar]
- 46.White, R. T., Damm, D., Hancock, N., Rosen, B. S., Lowell, B. B., Usher, P., Flier, J. S. & Spiegelman, B. M. (1992) J. Biol. Chem. 267**,** 9210–9213. [PubMed] [Google Scholar]
- 47.Lucas, B., Bumann, D., Walduck, A., Koesling, J., Develioglu, L., Meyer, T. F. & Aebischer, T. (2001) Infect. Immun. 69**,** 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pond, C. M. & Mattacks, C. A. (1995) J. Lipid Res. 36**,** 2219–2231. [PubMed] [Google Scholar]
- 49.Pond, C. M. & Mattacks, C. A. (1998) Immunol. Lett. 63**,** 159–167. [DOI] [PubMed] [Google Scholar]