Sorting of Yeast Membrane Proteins into an Endosome-to-Golgi Pathway Involves Direct Interaction of Their Cytosolic Domains with Vps35p (original) (raw)
Abstract
Resident late-Golgi membrane proteins in Saccharomyces cerevisiae are selectively retrieved from a prevacuolar–endosomal compartment, a process dependent on aromatic amino acid–based sorting determinants on their cytosolic domains. The formation of retrograde vesicles from the prevacuolar compartment and the selective recruitment of vesicular cargo are thought to be mediated by a peripheral membrane retromer protein complex. We previously described mutations in one of the retromer subunit proteins, Vps35p, which caused cargo-specific defects in retrieval. By genetic and biochemical means we now show that Vps35p directly associates with the cytosolic domains of cargo proteins. Chemical cross-linking, followed by coimmunoprecipitation, demonstrated that Vps35p interacts with the cytosolic domain of A-ALP, a model late-Golgi membrane protein, in a retrieval signal–dependent manner. Furthermore, mutations in the cytosolic domains of A-ALP and another cargo protein, Vps10p, were identified that suppressed cargo-specific mutations in Vps35p but did not suppress the retrieval defects of a vps35 null mutation. Suppression was shown to be due to an improvement in protein sorting at the prevacuolar compartment. These data strongly support a model in which Vps35p acts as a “receptor” protein for recognition of the retrieval signal domains of cargo proteins during their recruitment into retrograde vesicles.
Keywords: Saccharomyces cerevisiae, vps, protein sorting, endosome, Golgi
Introduction
Membrane and protein traffic in the secretory and endocytic pathways is mediated by vesicular intermediates that carry cargo molecules between membrane enclosed compartments. Genetic and biochemical approaches are revealing the roles of the complex protein machinery involved in formation of a vesicle from a donor compartment and its subsequent fusion with the appropriate acceptor compartment (Rothman 1994; Schekman and Orci 1996; Robinson 1997). The mechanisms that govern the active exclusion or sequestering of cargo proteins in vesicles have been the subject of much interest because they are important determinants in maintaining the appropriate structural and functional organization of the organelles of the secretory and endocytic pathways. However, the molecular mechanisms responsible for selective transport remain incompletely defined.
Coat proteins associate with the cytosolic face of organelles and perform two distinct functions in vesicular transport: (a) cargo recognition and concentration; and (b) vesicle formation. Vesicle formation is initiated by the recruitment of a small GTPase in its GTP-bound state (Palmer et al. 1993; Matsuoka et al. 1998; Spang et al. 1998). The subsequent association of coat protein subunits to the membrane induces membrane deformation and appearance of the coated bud. Cargo selection occurs via the recognition of cytosolic sorting signals on the cargo molecules by components of the coat (Schekman and Orci 1996; Robinson 1997).
The study of trafficking and sorting of TGN resident proteins has enhanced our understanding of the process of cargo selection by vesicle coats. The TGN is a major intersection point and sorting station for the secretory and endocytic pathways, and plays a critical role in the proteolytic maturation of secreted proteins. Membrane proteins with functional roles in the TGN tend to have complex cellular itineraries. For example, in animal cells the membrane proteins furin, TGN38, and the mannose 6–phosphate (M6P) receptor can cycle between the TGN, endosomes, and the plasma membrane (Duncan and Kornfeld 1988; Stanely and Howell 1993; Molloy et al. 1999). In the case of the M6P receptor, the receptor bound to its lysosomal hydrolase ligand is sorted into vesicles at the TGN by the clathrin/adaptor protein (AP)-1 coat complex. The heterotetrameric AP-1 complex binds to a tyrosine-based sorting signal in the cytosolic domain of the M6P receptor (Glickman et al. 1989; Le Borgne et al. 1993; Ohno et al. 1995). Clathrin subsequently assembles onto the membrane associated AP-1 complex and triggers formation of the budded vesicle (Schmid 1997). The M6P receptor–ligand complex is then delivered to a late endosome where the ligand dissociates. The free receptor can be retrieved back to the TGN for another round of sorting. A requirement for the rab9 (Lombardi et al. 1993; Riederer et al. 1994) and dynamin (Nicoziani et al. 2000) GTPases in the retrieval pathway has been demonstrated although a vesicle coat complex mediating retrieval has not yet been identified. However, dynamin has a well-characterized role in vesicle formation (McNiven et al. 2000) thus it is likely that this step is vesicle mediated. The sorting of the M6P receptor into the retrieval pathway involves binding of a phenylalanine–tryptophan signal in its cytosolic domain to the endosomal protein TIP47 (Diaz and Pfeffer 1998).
The yeast late-Golgi, as defined by the presence of proteolytic enzymes dipeptidyl aminopeptidase (DPAP) A, Kex2p, and Kex1p, is considered to be the functional equivalent of the mammalian TGN (Brigance et al. 2000). The amenability of the yeast TGN–endosomal system to analysis using both genetic and biochemical approaches has also contributed substantially to our understanding of selective vesicular transport. Kex2p, Kex1p, and DPAP A as well as the sorting receptor, Vps10p, cycle between the TGN and endosomes but in contrast to mammalian TGN proteins they do not appear to visit the plasma membrane (Cooper and Bussey 1992; Roberts et al. 1992; Wilcox et al. 1992; Bryant and Stevens 1997). These proteins have large lumenal domains, a single transmembrane spanning domain, and cytosolic domains of ∼90–160 amino acids. Similar to the M6P receptor, Vps10p binds to the soluble vacuolar hydrolase carboxypeptidase Y (CPY) in the TGN and receptor–ligand complexes are transported to the prevacuolar–endosomal compartment (PVC) (Vida et al. 1993; Marcusson et al. 1994; Cooper and Stevens 1996). CPY then dissociates and is transported to the lysosome-like vacuole.
Whereas clathrin appears to play a role in the vesicle-mediated transport of TGN membrane proteins from the TGN to the PVC (Seeger and Payne 1992a), this transport step does not appear to require cytosolic tail sorting signals (Roberts et al. 1992; Redding et al. 1996b). In contrast, retrieval of DPAP A, Kex2p, and Vps10p from the PVC is dependent on aromatic amino acid–based signals in their cytosolic domains (Wilcox et al. 1992; Nothwehr et al. 1993; Cereghino et al. 1995; Cooper and Stevens 1996; Bryant and Stevens 1997). Mutation of these signals causes the proteins to be mislocalized to the vacuole, reflecting default transport to the vacuole in absence of packaging into retrograde vesicles. In animal cells, aromatic sorting signals consisting of YXXØ (Y is tyrosine, X is any amino acid, and Ø is a residue with a bulky hydrophobic side chain) have been shown to interact with the μ subunit of AP complexes present in clathrin–AP vesicle coats (Bonifacino and Dell'Angelica 1999; Heilker et al. 1999). The aromatic retrieval signals in yeast TGN proteins only loosely conform to the YXXØ consensus and F is preferred over Y in the case of DPAP A (Nothwehr et al. 1993). Clathrin is required for proper localization of Kex2p and DPAP A (Payne and Schekman 1989; Seeger and Payne 1992b). However, clathrin most likely exerts this affect at the TGN itself since the affects of clathrin inactivation on Kex2p localization were shown to be independent of its retrieval signal (Redding et al. 1996b). AP gene disruptions do not appear to cause a defect in Kex2p localization, although in some cases such mutations exhibit synthetic defects with clathrin heavy chain mutations (Phan et al. 1994; Rad et al. 1995; Stepp et al. 1995). Nevertheless, at present it seems unlikely that clathrin–AP complexes are involved in sorting of yeast TGN proteins at the PVC.
Genetic screens have led to identification of several proteins that function in PVC-to-Golgi retrieval of yeast TGN proteins. The vps and pep mutants were obtained based on the missorting of CPY (Jones 1977; Bankaitis et al. 1986; Rothman and Stevens 1986; Robinson et al. 1988) whereas the grd and soi mutants were obtained based on mislocalization of DPAP A and Kex2p to the vacuole (Nothwehr et al. 1996; Redding et al. 1996a). An initial study focusing on the genes required for retrieval showed that loss of function of the VPS29, VPS30, or VPS35 genes caused Vps10p to be mislocalized to the vacuolar membrane in a manner dependent on the PVC target membrane SNAP receptor Pep12p but independent of late secretory functions (Seaman et al. 1997). An assay for PVC-to-Golgi transport subsequently demonstrated that retrieval of both Vps10p and the model TGN protein A-ALP was dependent on the function of Vps35p (Nothwehr et al. 1999). In accordance with these results, Vps35p and Vps29p are known to associate with the cytoplasmic face of the PVC as is also the case for two other proteins involved in the retrieval step, Vps5p and Vps17p (Horazdovsky et al. 1997; Nothwehr and Hindes 1997; Seaman et al. 1997, Seaman et al. 1998). Furthermore, biochemical experiments demonstrated that Vps35p and Vps29p interact and form a multimeric complex with Vps5p, Vps17p, and Vps26p. These proteins, collectively called the retromer complex, were also shown to associate with vesicles and thus have been proposed to comprise a vesicle coat structure (Seaman et al. 1998). Interestingly, the retromer proteins are strongly conserved from yeast to mammals suggesting that a retromer complex may also be used in mammals for retrieval of proteins from late endosomes.
The retromer model predicts that one or more of these subunits must associate directly with the retrieval signals contained within the cytosolic domains of cargo proteins. Vps35p is a good candidate for such a receptor protein because several vps35 mutant alleles have been identified that exhibit cargo-specific defects in retrieval (Nothwehr et al. 1999). One interpretation of these results is that Vps35p contains a pocket for binding to the retrieval signals from multiple cargo proteins but the structural features important for binding to each cargo protein are distinct. In this study we set out to directly test whether Vps35p interacts with the cargo proteins A-ALP and Vps10p. Using cross-linking and coimmunoprecipitation we find that a pool of A-ALP associates with Vps35p in a manner dependent upon the A-ALP retrieval signal. Furthermore, mutations in the cytosolic domains of A-ALP and Vps10p were obtained that suppressed the corresponding cargo-specific mutations in VPS35 but did not suppress a complete deletion of VPS35. Suppression was shown to occur at the level of sorting at the PVC. These results indicate that Vps35p plays a role in cargo selection at the PVC via direct interaction with the cytosolic domains of cargo proteins.
Materials and Methods
Antibodies
The anti-ALP polyclonal antibody was described previously (Nothwehr et al. 1996). The anti-CPY and anti-Vps35p polyclonal antibodies were gifts from S. Emr (University of California, San Diego, CA); the anti-Vma2p and anti-Vps10p polyclonal antibodies were gifts from T. Stevens (University of Oregon, Eugene, OR). Mouse monoclonal antibodies against CPY (mAb 10A5-B5) and Vma2p (mAb 13D11-B2) were obtained from Molecular Probes. Mouse anti–hemagglutinin (HA) monoclonal HA.11 ascites fluid was obtained from Covance Research Products. All secondary antibodies used for immunofluorescence experiments were obtained from Jackson ImmunoResearch Laboratories with the exception of the Alexa 488–conjugated goat anti–mouse antibody that was obtained from Molecular Probes.
Genetic Screens for Suppressors of Mutant vps35 Alleles
Random PCR mutagenesis and screening for suppressor mutations within the cytosolic domain of A-ALP that suppress the vps35-101 allele were performed as follows. A 0.44-kbp PCR fragment corresponding to the region encoding the cytosolic domain of DPAP A and short flanking regions was amplified under mutagenic conditions (Cadwell and Joyce 1992) using plasmid pAH16 (Nothwehr et al. 1999) as a template. Next, a unique XbaI site was introduced immediately after the initiator methionine of the STE13-PH08 insert in pSN55 (Table ) resulting in plasmid pSH3. A gapped vector was generated by digesting pSH3 with XbaI–BglII to release a 0.35-kbp fragment precisely corresponding to the region encoding the cytosolic domain of DPAP A. The gapped vector was recombined with the mutagenized PCR fragment in vivo by cotransforming the two DNA samples into yeast strain SNY105 (Table ) and selecting for transformants on media lacking uracil. A total of 20,000 transformants were screened using an ALP activity assay (Chapman and Munro 1994; Nothwehr et al. 1996). Transformants that exhibited reduced ALP activity were then further screened for A-ALP processing by immunoprecipitation of A-ALP (see below) and SDS-PAGE analysis. Plasmids were rescued from yeast clones that exhibited reduced processing over the control (SNY105 carrying pSN55) and were transformed back into SNY105 to confirm that suppression was linked to the plasmid. The DPAP A cytosolic domain-encoding region was then subjected to DNA sequence analysis using a dRhodamine Terminator Sequencing kit and a model 310 automatic sequencer (both from Applied Biosystems). Plasmid pSN55-23.2 contained only one mutation that altered the amino acid sequence whereas pSN55-23.1 and pSN55-8.1 contained multiple mutations. The single mutation responsible for suppression was identified by swapping restriction fragments into the wild-type parental plasmid pSN345 (subcloning described below) and then testing for suppression.
Table 1.
Plasmids Used in This Study
| Plasmid | Description | Reference or source |
|---|---|---|
| pSN55 | CEN-URA3 plasmid encoding A-ALP | Nothwehr et al. 1993 |
| pSN345 | CEN-URA3 plasmid encoding A-ALP | This study |
| pSN346 | CEN-URA3 plasmid encoding A-ALP with a N88I mutation | This study |
| pSN347 | CEN-URA3 plasmid encoding A-ALP with a N92I mutation | This study |
| pSN55-23.2 | CEN-URA3 plasmid encoding A-ALP with a N88K mutation | This study |
| pSN100 | CEN-URA3 plasmid encoding A-ALP with F85A and F87A mutations | Nothwehr et al. 1993 |
| pAH98 | CEN-URA3 plasmid encoding A-ALP under control of the GAL1 promoter | This study |
| pSN343 | CEN-URA3 plasmid encoding GAL1 promoter-driven mutant A-ALP with a deletion of codons 2 through 100 | This study |
| pSN350 | CEN-TRP1 plasmid containing the vps35-105 allele | This study |
| pHY5 | CEN-URA3 containing the VPS27 gene under GAL1 promoter control | T.H. Stevens |
| pSH11 | Integrating plasmid for introducing vps10-Q 1499 L allele (pRS306-based) | This study |
| pLC115 | Integrating plasmid for introducing VPS10::HA allele (pRS306-based) | Conibear and Stevens 2000 |
| pSN348 | Integrating plasmid for introducing vps10-Q 1499 L::HA allele (pRS306-based) | This study |
Table 2.
Saccharomyces cerevisiae Strains Used in This Study
| Strain | Description | Reference |
|---|---|---|
| SNY36-9A | MAT a _his3-Δ_200 leu2-3,112 pho8_Δ::ADE2 suc2-Δ_9 trp1-901 ura3-52 | Nothwehr et al. 1995 |
| SNY80 | SNY36-9A vps35_Δ::HIS3_ | This study |
| SNY110 | SNY36-9A vps35-101 | This study |
| SNY114 | SNY36-9A vps35-105 | This study |
| AHY81 | SNY36-9A (converted to diploid) vps35-105/vps35-105 vps10/vps10 | This study |
| SNY71 | SNY36-9A pep4_Δ::LEU2_ | This study |
| SHY28 | SNY36-9A vps10-Q 1499 L | This study |
| SHY29 | SNY36-9A vps10-Q 1499 Lvps35_Δ::HIS3_ | This study |
| SHY30 | SNY36-9A vps10-Q 1499 Lvps35-105 | This study |
| SNY132 | SNY36-9A pep4_Δ::LEU2 VPS10::HA vps35_Δ_::HIS3_ | This study |
| SNY133 | SNY36-9A pep4_Δ::TRP1 VPS10::HA vps35-105_ | This study |
| SNY135 | SNY36-9A pep4_Δ::LEU2 vps10-Q_ 1499 L::HA | This study |
| SNY136 | SNY36-9A pep4_Δ::LEU2 vps10-Q_ 1499 L::HA vps35_Δ::HIS3_ | This study |
| SNY137 | SNY36-9A pep4_Δ::TRP1 vps10-Q_ 1499 L::HA vps35-105 | This study |
| W303-1A | MAT a ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| AHY63 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 | This study |
| AHY66 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 vps35_Δ::HIS3_ | This study |
| SNY102 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 vps35-101 | This study |
| SNY126 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 vps29::HIS3 | This study |
| SNY128 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 VPS10::HA vps27_Δ::LEU2_ | This study |
| SNY129 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 VPS10::HA vps27_Δ::LEU2 vps35_Δ_::HIS3_ | This study |
| SNY134 | W303-1A pep4-Δ_H3 pho8_Δ::ADE2 vps10-Q 1499 L::HA vps27_Δ::LEU2 vps35_Δ_::HIS3_ | This study |
| SEY6210 | _MAT_α _his3-_Δ_200 leu2-3,112 lys2-801 suc2-_Δ_9 trp1-Δ_901 ura3-52 | Robinson et al. 1988 |
| SNY132 | SEY6210 pep4_Δ::TRP1 pho8_Δ_::LEU2 VPS10::HA vps35_Δ_::HIS3_ | This study |
| SNY136 | SEY6210 pep4_Δ::TRP1 pho8_Δ_::LEU2 vps10-Q_ 1499 L::HA vps35_Δ::HIS3_ | This study |
| PBY1 | SEY6210 pep4_Δ::TRP1 pho8_Δ_::LEU2 vps35_Δ_::HIS3_ | This study |
| PBY2 | SEY6210 pep4_Δ::TRP1 pho8_Δ_::LEU2 vps35-101_ | This study |
| SHY26 | SEY6210 pho8-Δ_X vps27_Δ::LEU2 | This study |
| SNY105 | SEY6210 (converted to diploid) pho8_Δ::LEU2/pho8_Δ_::LEU2 vps35-101/vps35-101_ | This study |
Suppressor mutations in the Vps10p cytosolic domain were generated by amplifying a 1.2-kbp PCR fragment containing the coding region for the Vps10p cytosolic domain under mutagenic conditions (Cadwell and Joyce 1992). pAAC201 (a gift from A. Cooper, University of Missouri, Kansas City, MO) consists of the VPS10 gene in a yeast centromere (CEN)–LEU2 vector. To generate the gapped vector, pAAC201 was digested with PstI–StuI thereby removing a 0.69-kbp fragment containing the region encoding nearly the entire Vps10p cytosolic domain. The gapped vector and PCR fragment were cotransformed into strain AHY81 (Table ) and ∼10,000 transformants were screened for the absence of aberrant CPY secretion. Plasmids were rescued from clones that secreted less CPY than the control (AHY81 carrying pAAC201) and were retransformed back into AHY81 to verify that suppression was due to mutation of the plasmid. Two suppressor plasmids were obtained using this approach (p81.257 and p81.262). DNA sequencing revealed that each plasmid contained a single mutation that altered the amino acid sequence of the cytosolic domain.
Plasmids and Yeast Strain Construction
Most of the plasmids and yeast strains used in this study are indicated in Table and Table , respectively. Plasmid pSN345 is a derivative of pSN55 in which a region of the polylinker between the KpnI and EcoRI sites has been removed by digesting with KpnI–EcoRI, filling in the sticky ends, and religating. Plasmids pSN55-23.1 and pSN55-8.1 were generated by the procedure for random mutagenesis of the DPAP A cytosolic domain described above. These plasmids consist of pSN55 with multiple mutations in the cytosolic domain region. To generate plasmids that contain just the suppressor mutation, the 0.94-kbp HindIII fragments from pSN55-23.1 and pSN55-8.1 were swapped for the corresponding HindIII fragment in pSN345, resulting in plasmids pSN346 and pSN347, respectively.
The VPS10::HA allele (consisting of three copies of the influenza HA epitope fused to the 3′ end of the VPS10 open reading frame) was introduced into yeast using pLC115 as previously described (Conibear and Stevens 2000). To make a construct for integration of the vps10-Q1499L into the VPS10 locus, the 2.3-kbp HindIII fragment from p81.257 (described above) was inserted into the HindIII site of pRS306. The resulting plasmid, pSH11, was digested with BclI and transformed into yeast to Ura+ prototrophy. Ura+ transformants were then grown on 5-fluoroorotic acid to select for Ura− loop-out strains. The Q1499L mutation removes a BclI site, thus screening for the presence of this mutation was carried out by PCR amplification of a region containing the relevant portion of VPS10 followed by incubation of the PCR product with BclI to determine if the site was present. The vps10-Q1499L::HA allele was constructed by digesting pSH11 with MscI–BclI to release a 322-bp fragment. The vector DNA was then ligated to the corresponding 0.44-kbp MscI–BclI fragment from pLC115 resulting in pSN348. The vps10-Q1499L::HA allele in pSN348 was introduced into yeast using the same approach used to introduce the vps10-Q1499L allele.
Radiolabeling, Immunoprecipitation, and Subcellular Fractionation
Yeast strains were propagated at 30°C for all pulse-chase experiments. The procedures for immunoprecipitation of CPY (Vater et al. 1992) and A-ALP (Nothwehr et al. 1996) have been previously described. Radioactively labeled proteins were quantified from gels using a Phosphorimager system (Fuji Photo Film Co.). Half-times of processing were determined by plotting the log of the percentage of unprocessed A-ALP at each time point followed by linear regression analysis. Subcellular fractionation of labeled yeast spheroplasts was carried out as previously described (Nothwehr et al. 1999) except that rabbit anti–Vps10p antibody was used for immunoprecipitation from each fraction.
Cross-linking and Coimmunoprecipitation
Yeast strains were grown overnight at 30°C in minimal selective media lacking methionine and cysteine and containing either glucose or galactose as a carbon source depending on whether GAL1 promoter induction was desired. 1 OD600 unit of cells in 1 ml of media was pulse-labeled with [35S]Express label (NEN Life Science Products) for 30 min and chased with 50 μg/ml unlabeled methionine and cysteine for 30 min. The cells were spheroplasted and osmotically lysed by resuspending cell pellets with 250 μl phosphate-buffered saline containing 0.5 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A. To initiate the cross-linking reaction, 5 μl of dithio_bis_(succinimidylpropionate) (DSP; Pierce Chemical Co.) stock (20 mg/ml in DMSO) was added. Control samples received DMSO alone. The reactions were incubated for 30 min at 23°C, quenched by addition of 25 μl 1 M Tris, pH 8.0, and held on ice for 5 min. 40 μl of 10 mg/ml bovine serum albumin and 50 μl of 8 M urea/1% SDS were then added and the extracts were denatured at 100°C for 5 min. After diluting with buffer to 1 ml the extracts contained the following: 35 mM Tris, pH 8.0, 0.1% Triton X-100, 0.05% SDS, 0.4 M urea, and 2 mM EDTA. Polyclonal antibodies against either Vps35p or ALP were then added at levels sufficient for quantitative immunoprecipitation. After incubation for 2 h at 4°C immune complexes were precipitated using IgG Sorb (The Enzyme Center). The precipitates were washed three times with 1 ml volumes of a solution containing 10 mM Tris, pH 8.0, 0.1% SDS, 0.1% Triton X-100, and 2 mM EDTA. The immunoprecipitated proteins were released by resuspending the IgG Sorb pellet in 50 μl 8M urea/1% SDS/5% 2-mercaptoethanol and incubating at 100°C for 5 min. The eluted proteins were subjected to immunoprecipitation as before and analyzed by SDS-PAGE and fluorography.
To determine the percent cross-linking, radioactively labeled proteins were quantified from gels as described above. The percent cross-linking was calculated by comparing the amount of product (e.g., Vps35p) obtained where two different antibodies were used in each precipitation (e.g., anti-ALP then anti-Vps35p) to the amount of product obtained where both precipitations were carried out using the same antibody (e.g,. anti-Vps35p). Samples compared in such a manner (e.g., Fig. 1 A, lanes 1–4) were derived from the same labeled culture to avoid any differences due to labeling efficiency between cultures.
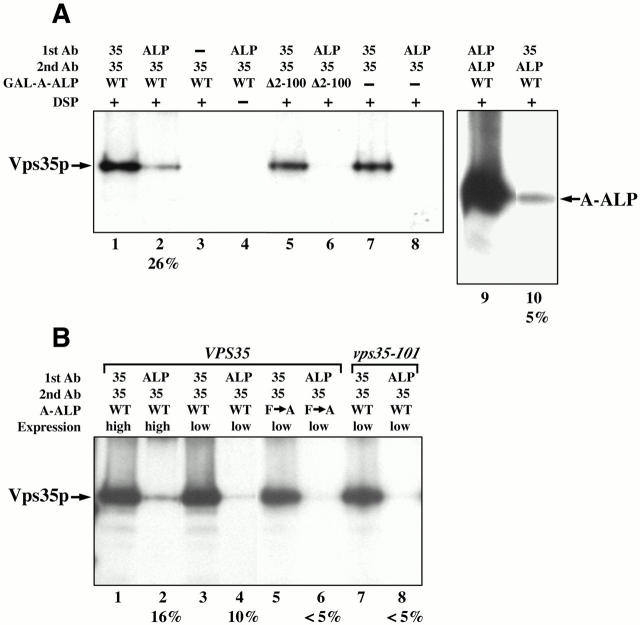
Figure 1.
Vps35p and A-ALP are members of a protein complex. (A) Extracts from 35S-labeled yeast strains were cross-linked with DSP (lanes 1–3 and 5–10) and immunoprecipitated using the indicated antibodies (1st Ab) where 35 and ALP refer to anti-Vps35p and anti-ALP, respectively. Addition of no antibody or no DSP is indicated (−) for samples in lanes 3 and 4, respectively. The immunoprecipitated proteins were denatured, reduced, and then precipitated again (2nd Ab) before being analyzed by SDS-PAGE and fluorography. In lanes 1–4, 5 and 6, 7 and 8, and 9 and 10 the following yeast strain–plasmid combinations were analyzed, respectively: AHY63–pAH98, AHY63–pSN343, AHY63 (no plasmid), and AHY63–pAH98. (B) The same cross-linking approach as described in (A) was used. The following yeast strain–plasmid combinations were used in lanes 1 and 2, 3 and 4, 5 and 6, and 7 and 8, respectively: AHY63–pAH98, AHY63–pSN55, AHY63–pSN100, and SNY102–pSN55. Wild-type or mutant A-ALP was expressed from the GAL1 promoter (high) or from the endogenous STE13 promoter (low), as indicated. In both A and B the percent cross-linking (see Materials and Methods) obtained for some samples is indicated under the corresponding row numbers for each panel.
Fluorescence Microscopy
The procedures for preparation of fixed spheroplasted yeast cells, attachment to microscope slides, and costaining of the A-ALP fusion protein and Vma2p were previously described (Roberts et al. 1991; Nothwehr et al. 1995). All secondary antibodies were diluted 1:500 before used unless otherwise noted.
Simultaneous detection of Vps10-HA and Vma2p was carried out by incubating with the following reagents followed by extensive washing: (a) mouse anti–HA and rabbit anti–Vma2p; (b) biotin conjugated goat anti–rabbit immunoglobulin G (IgG; heavy and light chain [H+L]); and (c) goat anti–mouse IgG (H+L) conjugated to Alexa 488 (1:100 dilution) and streptavidin conjugated to Texas red.
Yeast cells were photographed using an Olympus BX-60 epifluorescence microscope (Olympus). Film negatives were digitized using a Kodak Professional RFS 2035 Plus film scanner and adjusted using Adobe Photoshop 5.5 (Adobe Systems).
Results
Vps35p Forms a Complex with the TGN Resident Protein A-ALP
To test whether Vps35p directly interacts with the cytosolic domains of cargo proteins during retrieval we initially used a coimmunoprecipitation approach focusing on interaction with the model TGN protein A-ALP. A-ALP consists of the cytosolic domain of DPAP A (product of the STE13 gene) fused to the transmembrane and lumenal domains of alkaline phosphatase. Trafficking of this fusion protein appears identical to that of DPAP A (Nothwehr et al. 1993). A-ALP appears to cycle to and from the PVC relatively infrequently (Bryant and Stevens 1997) thus only a small percentage of A-ALP would be expected to be associated with the retrieval apparatus at any given time. Overexpression of TGN resident proteins has been shown to reduce their efficiency of retention (Wilcox et al. 1992; Nothwehr et al. 1993) suggesting saturation of the retrieval apparatus. Therefore, we tested for interaction between A-ALP and Vps35p in strains overexpressing A-ALP in order to maximize the amount of A-ALP engaging the retrieval apparatus. In addition, coimmunoprecipitation was carried out from cell extracts treated with the bifunctional cross-linking reagent DSP since the retrieval apparatus–cargo interaction is transient and thus likely to be relatively weak.
Initial experiments were carried out using a yeast strain carrying a low copy number CEN-plasmid containing a GAL1 promoter-driven A-ALP construct. A-ALP was overproduced ∼15-fold when the strain was grown in media containing galactose as the sole carbon source (data not shown). Strains were pulsed with [35S]methionine/cysteine for 30 min and chased with unlabeled amino acids for 30 min. The cells were spheroplasted, osmotically lysed under nondenaturing conditions, and treated with DSP. After cross-linking, the reactions were quenched, the proteins were denatured, and then incubated with antibody against either Vps35p or A-ALP followed by addition of fixed Staphylococcus aureus cells (IgG Sorb). The immunoprecipitated and washed proteins were then eluted from the antibody/IgG Sorb by denaturing under reducing conditions in order to reduce the disulfide bond in DSP and thus break the cross-links. The proteins were then subjected to a second immunoprecipitation and analyzed by SDS-PAGE and fluorography.
A typical experiment is shown in Fig. 1 A. Consistent with the presence of an A-ALP–Vps35p complex, we observed that Vps35p was immunoprecipitated when anti-ALP was used in the first precipitation followed by anti-Vps35p (lane 2). Quantification of this amount of Vps35p (lane 2), and comparison with that obtained using Vps35p for both immunoprecipitations (lane 1), indicates that 26% of the total Vps35p was cross-linked to A-ALP. Cross-linking was consistently observed using this approach although the percentage obtained varied somewhat from experiment to experiment (Fig. 1, compare lanes 1 and 2 of A and B). A number of control experiments indicated that the Vps35p in lane 2 was actually cross-linked to A-ALP. No Vps35p was immunoprecipitated if the anti-ALP antibody was not added in the first immunoprecipitation (lane 3) or if DSP was not added (lane 4). In the absence of A-ALP expression, no Vps35p was immunoprecipitated (lane 8). Furthermore, cross-linking of Vps35p and A-ALP was dependent on the DPAP A cytosolic domain since no cross-linking was detected to an A-ALP deletion mutant lacking residues 2–100 of the 118–amino acid cytosolic domain (lanes 5 and 6). As a further test of cross-linking between the two proteins, the order of the antibody incubations was reversed to see if anti-Vps35p could precipitate A-ALP. A small fraction (5%) of the A-ALP was immunoprecipitated (compare lanes 9 and 10) consistent with precipitation of a cross-linked product in the first immunoprecipitation. Similar controls as those described above demonstrated that the immunoprecipitation of A-ALP by the Vps35p antibody was specific (data not shown).
To address the possibility that overexpression of A-ALP might lead to nonspecific cross-linking between Vps35p and A-ALP, we applied this approach to strains expressing A-ALP at a normal level under control of the endogenous STE13 promoter. At low expression levels cross-linking was observed (Fig. 1 B, lanes 3 and 4) albeit at a somewhat lower level: 10% of total Vps35p compared with 16% cross-linking observed for a strain overproducing A-ALP (lanes 1 and 2). If the cross-linking represents the bona fide physical association of A-ALP with Vps35p that occurs during the process of protein sorting, this interaction should be reduced by mutations that specifically affect retrieval. We analyzed A(F →A)-ALP in which the two critical phenylalanine residues of the retrieval signal (F85 and F87) have been mutated to alanines (Nothwehr et al. 1993). In addition, a vps35-101 strain expressing a mutant form of Vps35p that is defective for retrieval of A-ALP but normal for retrieval of Vps10p (Nothwehr et al. 1999) was analyzed. In both control reactions there was a marked reduction in cross-linking (<5%) compared with that observed using wild-type alleles of both proteins (10%). Thus at normal expression levels cross-linking is clearly sensitive to subtle mutations in A-ALP and Vps35p that are known to affect retrieval. We conclude from these data that Vps35p and A-ALP associate in vivo during protein sorting at the PVC.
Identification of Mutations in the A-ALP Cytosolic Domain That Can Specifically Suppress the vps35-101 Allele
Vps35p is a subunit of the large peripheral membrane retromer complex proposed to be involved in protein sorting and vesicle formation at the PVC (Seaman et al. 1998). Whereas the cross-linking data described above were consistent with a direct interaction between Vps35p and the A-ALP cytosolic domain, it was possible that one or more retromer subunits might be sandwiched between A-ALP and Vps35p in the complex. To address this issue, and to test for interactions using an independent method, we used a genetic suppression approach.
The vps35-101 and vps35-105 alleles were previously shown to be cargo-specific alleles that were defective for A-ALP and Vps10p retrieval, respectively (Nothwehr et al. 1999). Because Vps35p expressed from these alleles exhibits function toward a subset of cargo proteins it is likely that the point mutation responsible for the phenotype of each mutant allele has a subtle effect on the tertiary structure of the protein. If these mutations interfere with binding of Vps35p to the cytosolic domain of A-ALP or Vps10p, it may be possible to restore binding, and thus sorting, by introducing compensating suppressor mutations into their cytosolic domains. The identification of such mutations would indicate direct interaction between Vps35p and the cytosolic domain of cargo proteins.
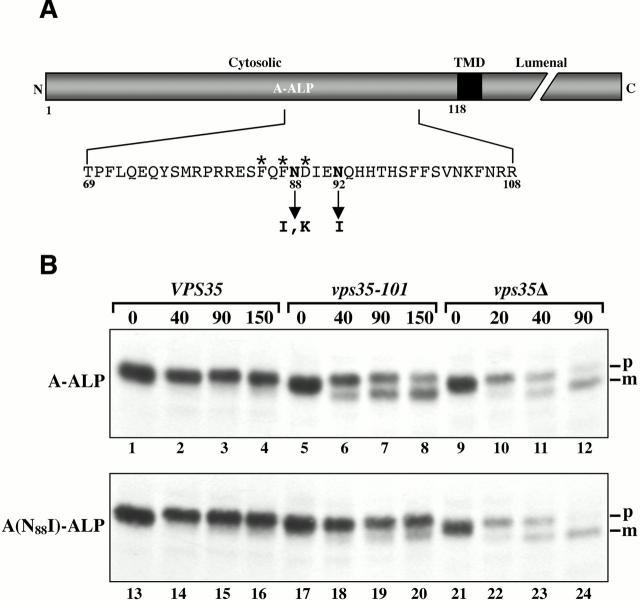
We first attempted to generate suppressor mutations in the cytosolic domain of A-ALP that would restore its retrieval in a vps35-101 strain. Vacuolar mislocalization of A-ALP caused by poor PVC retrieval results in processing of its COOH-terminal ALP propeptide by a vacuolar protease (Nothwehr et al. 1993). Upon processing, A-ALP becomes enzymatically active thus vacuolar processing can be detected by both an increase in gel mobility and by an increase in activity. Using PCR mutagenesis, a plasmid construct encoding A-ALP was randomly mutagenized in the region encoding the cytosolic domain. Other regions of the plasmid were unmutagenized. A library of vps35-101 yeast clones carrying the mutagenized plasmids was initially screened using an ALP activity assay (Chapman and Munro 1994; Nothwehr et al. 1996). Plasmids rescued from clones exhibiting reduced ALP activity and a slower rate of A-ALP processing were transformed back into the vps35-101 strain and into a _vps35_Δ strain to assess whether suppression was linked to the plasmid and was allele specific. Three plasmids were obtained that exhibited much slower processing in the vps35-101 strain but did not suppress the _vps35_Δ allele. In all three cases, the suppression phenotype was due to mutation of a single base pair as determined by DNA sequencing and restriction fragment swapping experiments. Interestingly, two of the mutations were at codon 88 (N88I and N88K) and the third was nearby at codon 92 (N92I) (Fig. 2 A).
Figure 2.
(A) The positions of three independent mutations in the A-ALP cytoplasmic domain that specifically suppress the vps35-101 allele are shown. A schematic diagram of A-ALP is shown with the lumenal, transmembrane (TMD), and cytosolic domains indicated. The cytosolic domain residues previously shown to comprise a PVC-to-TGN retrieval signal (Nothwehr et al. 1993) are labeled with asterisks and the suppressor mutations (N88I, N88K, and N92I) are indicated with arrows. (B) The N88I mutation in the cytosolic domain of A-ALP reduces its rate of vacuolar processing in a vps35-101 strain. Yeast strains were pulse labeled with [35S]methionine/cysteine for 10 min and chased for the times indicated in minutes above the upper panel. The cells were lysed and wild-type or mutant A-ALP was immunoprecipitated and analyzed by SDS-PAGE and fluorography. The following strain–plasmid combinations were analyzed in lanes 1–4, 5–8, 9–12, 13–16, 17–20, and 21–24, respectively: SNY36-9A–pSN345, SNY110–pSN345, SNY80–pSN345, SNY36–9A–pSN346, SNY110–pSN346, and SNY80–pSN346. The positions of precursor (p) and processed mature (m) A-ALP are indicated.
Fig. 2 B shows a representative experiment in which wild-type and A(N88I)-ALP were immunoprecipitated from strains that were radioactively labeled for 10 min and chased for the indicated times. A-ALP processing in the vps35-101 strain was markedly slower for the A(N88I)-ALP mutant compared with wild-type A-ALP (compare lanes 5–8 to 17–20). Quantification of the half-time of processing in VPS35, vps35-101, and _vps35_Δ strains has been carried out for wild-type A-ALP and each of the three mutants (Table ). All three mutations exhibit dramatic suppression of vps35-101 although N88I was more effective than the other two. None of the three mutations exhibited any significant change in the rate of processing in the VPS35 or _vps35_Δ strains (Fig. 2 B and Table ) and are therefore allele specific.
Table 3.
Phenotypes of A-ALP Mutants That Suppress vps35-101 and Nature of the Responsible Mutations
| A-ALP construct | Cytosolic domain mutation | A-ALP processing half-time in strains: | |||
|---|---|---|---|---|---|
| Vps+ | vps35-101 | _vps35_Δ | _vps27_Δ | ||
| pSN345 | N.A. | >180 | 96 | 34 | 47 |
| pSN346 | N88I | >180 | >180 | 29 | 42 |
| pSN55-23.2 | N88K | >180 | 143 | 29 | 47 |
| pSN347 | N92I | >180 | 155 | 39 | 42 |
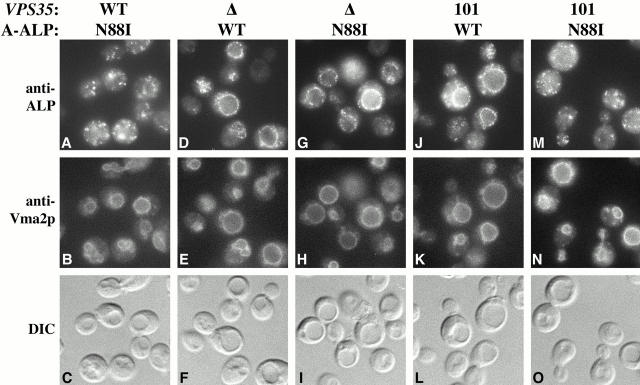
The fact that the A-ALP mutants are processed with similar kinetics to that of wild-type A-ALP in the _vps35_Δ strain suggested that the mutants are transported through ER and Golgi compartments with kinetics similar to that of wild-type A-ALP. Two additional experiments were performed to determine whether suppression was indeed due to an improvement of PVC-to-Golgi retrieval as opposed to slower than normal transport through the early secretory pathway or slower exit from the TGN. To see whether A(N88I)-ALP was localized to the TGN in vps35-101 cells, as would be expected for enhancement of PVC-to-Golgi retrieval, its localization was analyzed by indirect immunofluorescence microscopy. The data in Fig. 3 (A–C) indicate that in cells containing wild-type VPS35 the A(N88I)-ALP mutant exhibited a punctate staining pattern typical of yeast TGN membrane proteins (Redding et al. 1991; Roberts et al. 1992). In the _vps35_Δ strain both A-ALP and A(N88I)-ALP were localized predominantly to the vacuolar membrane as demonstrated by comparing their staining pattern to that of the vacuolar membrane marker Vma2p and to the crater-like outline of the vacuole in cells viewed by differential interference contrast (DIC) optics (Fig. 3, D–F and G–I). These results indicate a lack of suppression of the _vps35_Δ allele by the N88I mutation. Similarly, A-ALP was localized to the vacuolar membrane in the vps35-101 mutant (Fig. 3, J–L) consistent with our previous observations (Nothwehr et al. 1999). However, A(N88I)-ALP exhibited a predominantly punctate TGN-like staining pattern in the vps35-101 strain (Fig. 3, M–O) consistent with its very slow rate of processing (Fig. 2 B).
Figure 3.
An N88I mutation in the cytosolic domain of A-ALP restores Golgi localization to A-ALP expressed in a vps35-101 strain. The following strains (shown respectively from left to right) were analyzed: SNY71–pSN346, PBY1–pSN345, PBY1–pSN346, PBY2–pSN345 and PBY2–pSN346. Strains propagated at 30°C were fixed, spheroplasted, and costained with antibodies against ALP and Vma2p. After subsequent detection with fluorochromes (see Materials and Methods) the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas red.
As another approach to assess whether the decrease in suppressor A-ALP processing in the vps35-101 mutant was due to a decreased rate of transport between the ER and the PVC, the rate of processing was analyzed in a _vps27_Δ yeast strain. In class E vps mutants such as vps27 both PVC-to-TGN and PVC-to-vacuole trafficking is blocked (Piper et al. 1995). The blockage leads to an enlarged PVC containing active vacuolar proteases as well as trapped TGN proteins that are unable to be retrieved. Therefore, the rate of processing in vps27 cells should reflect the rate of transport from the ER to the PVC. The rate of processing of the three suppressor A-ALP mutants in a _vps27_Δ strain was essentially the same as that of wild-type A-ALP (Table ) again indicating that the anterograde trafficking of the mutant proteins was normal. Taken together, these results indicate that the mutant A-ALP proteins suppress vps35-101 by improving sorting at the PVC.
A Point Mutation in the Vps10p Cytosolic Domain Specifically Suppresses the vps35-105 Allele
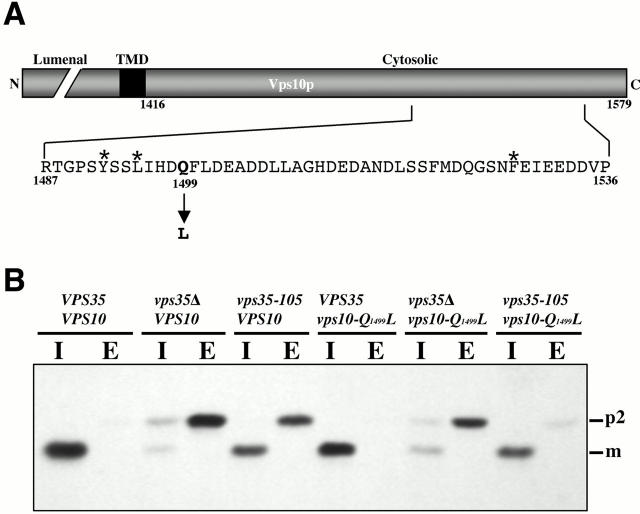
The discovery of mutant VPS35 alleles specifically defective for either A-ALP or Vps10p retrieval (Nothwehr et al. 1999), coupled with the data above indicating a direct interaction between Vps35p and A-ALP, motivated us to next address whether Vps35p and Vps10p interact. As a test of interaction between the two proteins we used a genetic suppressor approach similar to that described above for A-ALP. A region of the VPS10 open reading frame containing the cytosolic domain-encoding region was randomly mutagenized and the mutant plasmids were introduced into a vps10 vps35-105 strain. The vps35-105 allele is defective for retrieval of Vps10p but nearly normal for A-ALP retrieval (Nothwehr et al. 1999). A major effect of the perturbation of Vps10p trafficking is aberrant secretion of CPY (Marcusson et al. 1994; Cooper and Stevens 1996). Thus vps10 vps35-105 transformants carrying the mutagenized plasmids were screened using a colony-blotting assay that detects secretion of CPY with the objective of finding clones that secrete less CPY than the starting strain. Two mutant plasmids (p81.257 and p81.262) were obtained that suppressed the vps35-105 mutation but did not suppress the _vps35_Δ mutation. Interestingly, DNA sequencing of the mutagenized region revealed that both plasmids had exactly the same mutation in the cytosolic domain that changed the Q1499 codon to a L codon (Fig. 4 A). Plasmid p81.262, but not p81.257, had an additional mutation that did not change the encoded amino acid sequence. The fact that the Q1499L mutation was independently obtained in two different plasmids emphasizes the importance of this amino acid replacement in suppression of the vps35-105 allele.
Figure 4.
A Q1499L mutation in Vps10p suppresses the CPY sorting defect of a vps35-105 strain. (A) A schematic diagram of Vps10p is shown with the lumenal, transmembrane (TMD), and cytosolic domains indicated. The cytosolic domain residues previously shown to comprise a PVC-to-TGN retrieval signal (Cooper and Stevens 1996) are indicated with asterisks and the suppressor mutation is indicated with an arrow. (B) SNY36-9A, SNY80, SNY114, SHY28, SHY29, and SHY30 cells (left to right) were pulsed with [35S]methionine/cysteine for 10 min and chased for 45 min with unlabeled amino acids. CPY was immunoprecipitated from intracellular (I) and extracellular (E) fractions and analyzed by SDS-PAGE and fluorography. The positions of the Golgi (p2) and vacuolar (m) forms of CPY are indicated.
A number of experiments were used to evaluate and quantify suppression by the vps10-Q1499L allele. A pulse-chase immunoprecipitation approach was used to quantify the extent of aberrant CPY secretion in vps35-105 vps10-Q1499L and control strains (Fig. 4 B). Strains were radioactively pulsed for 10 min, chased for 45 min, and CPY was immunoprecipitated from intracellular and extracellular fractions. Experiments identical to that shown in Fig. 4 B have been repeated several times and the average of the percent CPY secretion obtained for each strain is shown in Table . As expected for a vacuolar protein, practically all of the CPY was intracellular in the VPS35 VPS10 strain. In vps mutants CPY tends to be secreted in the “p2” form which has been modified by ER and Golgi glycosyltransferases but has not underwent vacuolar proteolytic processing to the mature form. Accordingly, we observed marked CPY secretion in the _vps35_Δ VPS10 and vps35-105 VPS10 strains: 78 and 45%, respectively. The vps10-Q1499L allele appears to encode a fully functional CPY receptor since little or no CPY secretion is observed in the VPS35 background. Similarly, in the _vps35_Δ background comparable amounts of CPY secretion are observed for the wild-type and Q1499L mutant (78 and 75%, respectively). Thus the mutation does not suppress the _vps35_Δ allele. However, comparison of CPY secretion from the vps35-105 VPS10 and vps35-105 vps10-Q1499L strains (45 and 14%, respectively) indicates that the Q1499L mutation strongly suppresses the mutant form of Vps35p expressed from the vps35-105 allele.
Table 4.
Quantification of CPY Secretion from Strains Expressing Wild-type or Suppressor Mutant Forms of VPS10
| Genotype | Percent CPY secretion ± SD | n |
|---|---|---|
| VPS35 VPS10 | <5 | 1 |
| vps35_Δ_ VPS10 | 78 ± 0.4 | 4 |
| vps35-105 VPS10 | 45 ± 2.5 | 4 |
| VPS35 vps10-Q1499L | <5 | 1 |
| vps35_Δ_ vps10-Q1499L | 75 ± 1.1 | 4 |
| vps35-105 vps10-Q1499L | 14 ± 1.8 | 5 |
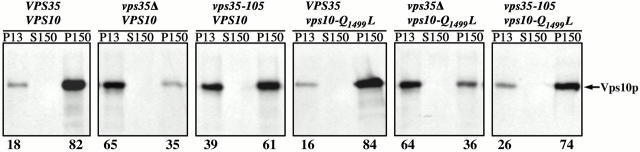
We next evaluated the trafficking and localization of the Vps10-Q1499L protein in vps35-105 cells directly. Vacuole-localized Vps10p in vps35 cells is unprocessed by vacuolar proteases (Seaman et al. 1997), precluding the analysis of processing kinetics. We instead used subcellular fractionation and immunofluorescence microscopy to localize Vps10-Q1499L. Cells were converted to spheroplasts, pulse labeled for 25 min at 30°C with [35S]methionine/cysteine, and then chased for 40 min with excess unlabeled amino acids to allow Vps10p to reach its steady-state cellular location. The spheroplasts were gently lysed and centrifuged at 450 g to pellet any unlysed cells. The supernatant was then centrifuged at 13,000 g to generate a pellet fraction (P13) and the supernatant centrifuged at 150,000 g to generate pellet (P150) and supernatant (S150) fractions. Vps10p was then immunoprecipitated from the three fractions and analyzed by SDS-PAGE and fluorography. Under similar experimental conditions the majority of Vps10p in wild-type cells was previously found in the high speed pellet fraction (P150) that contains vesicular, endosomal, and Golgi membranes (Marcusson et al. 1994; Seaman et al. 1997). A minor pool was found in the P13 fraction that contains ER, plasma membrane, and vacuoles. Consistent with these previous studies, we observed 82% of Vps10p in the P150 fraction with the remaining 18% found in the P13 fraction in the wild-type VPS35 VPS10 strain (Fig. 5). As expected, given the role of Vps35p in Vps10p PVC-to-Golgi retrieval, much of the Vps10p was found in the P13 fraction in the _vps35_Δ VPS10 strain (65%) and the vps35-105 VPS10 strain (39%). The Vps10-Q1499L mutant behaved essentially the same as wild-type Vps10p in the VPS35 and _vps35_Δ strains. However, in the vps35-105 background much less of the Vps10-Q1499L protein was found in the P13 fraction (26%) as compared with wild-type Vps10p (39%). Thus the Q1499L mutation allows Vps10p to maintain a more wild-type distribution in vps35-105 cells but has no affect in _vps35_Δ cells.
Figure 5.
A Q1499L mutation in Vps10p suppresses the vacuolar mislocalization defect in vps35-105 cells. Spheroplasted cells were pulsed for 25 min with [35S]methionine/cysteine and chased for 45 min with unlabeled amino acids. The spheroplasts were lysed and lysates were centrifuged at 13,000 g to generate a pellet fraction (P13). The supernatant was then centrifuged at 150,000 g to generate pellet (P150) and supernatant (S150) fractions. Wild-type or mutant Vps10p was then immunoprecipitated from each fraction and analyzed by SDS-PAGE and fluorography. The relative percentage of Vps10p present in the P13 and P150 fractions as determined by Phosphorimager analysis is indicated below each panel. The strains are described in the legend to Fig. 4 B.
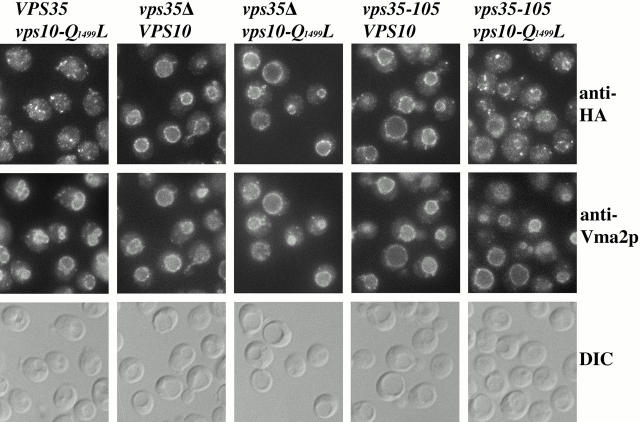
Localization of wild-type and mutant Vps10p was also analyzed by immunofluorescence microscopy. To facilitate detection, three copies of the HA epitope tag were fused to the COOH terminus of wild-type and mutant Vps10p. COOH-terminal addition of the HA tag does not appear to affect the function or trafficking of Vps10p since VPS10::HA strains exhibit normal CPY sorting (data not shown). The Vps10-Q1499L protein in the VPS35 vps10-Q1499L::HA strain exhibits a punctate staining pattern (Fig. 6) typical of TGN resident proteins. In contrast, both wild-type and mutant Vps10 proteins exhibit striking vacuolar localization in _vps35_Δ cells. Wild-type Vps10p is also clearly mislocalized to the vacuole in the vps35-105 background. However, the suppressor Vps10-Q1499L mutant in the vps35-105 background exhibits a mostly punctate Golgi-like staining pattern along with a minor amount of vacuolar membrane staining (compare the anti-HA and anti-Vma2p panels).
Figure 6.
The Vps10-Q1499L mutant protein exhibits a predominantly Golgi localization pattern in vps35-105 cells. The following strains (shown respectively from left to right) were analyzed: SNY135, SNY132, SNY136, SNY133, and SNY137. Strains propagated at 30°C were fixed, spheroplasted, and costained with antibodies against the HA epitope and Vma2p. After subsequent detection with fluorochromes (see Materials and Methods) the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas red.
In summary, assessment of Vps10p trafficking by CPY immunoprecipitation, subcellular fractionation, and immunofluorescence together indicate that the Q1499L mutation in Vps10p suppresses vps35-105 in an allele-specific fashion.
The Q1499L Mutation Improves Vps10p Sorting at the PVC in vps35-105 Cells
We next addressed whether suppression of the Vps10p localization defect in vps35-105 cells was due to improved protein sorting at the PVC as would be expected if the Q1499L mutation improved protein–protein interaction with mutant Vps35p. A previously developed assay for PVC-to-Golgi retrieval (Bryant and Stevens 1997) was used to analyze retrieval of Vps10-Q1499L by immunofluorescence microscopy. In this assay, _vps27_Δ strains carry a GAL1 promoter-driven VPS27 construct. When grown on raffinose as a carbon source Vps27p is not expressed and the cells exhibit a phenotype in which TGN proteins are trapped within an exaggerated “class E” PVC. The class E compartment also contains certain vacuolar proteins such as the vacuolar proton-translocating ATPase (Raymond et al. 1992), of which Vma2p is a subunit. When Vps27p synthesis is induced by addition of galactose, both the PVC-to-TGN and PVC-to-vacuole transport pathways resume. Using such an assay Vps35p was previously shown to be required for PVC retrieval since a _vps35_Δ mutation resulted in TGN proteins being transported from the PVC to the vacuole rather than back to the Golgi (Nothwehr et al. 1999).
A PVC-to-Golgi retrieval assay analyzing trafficking of wild-type and mutant Vps10p in VPS35 VPS10::HA, vps35-105 VPS10::HA, and vps35-105 vps10-Q1499L::HA strains is shown in Fig. 7. When the three strains are grown on raffinose (0 min time point) both Vps10-HA and Vma2p are localized to one to two discrete structures per cell that are usually located adjacent to the vacuolar membrane (compare to DIC image). Cells were occasionally observed in which Vma2p localized to the vacuolar membrane as well. After addition of galactose for 90 min Vps10-HA in the VPS35 VPS10::HA strain exhibited a punctate staining pattern indicating retrieval back to the TGN whereas Vma2p was transported to the vacuolar membrane. As expected, in the vps35-105 VPS10::HA strain after 90 min induction, retrieval was defective and Vps10-HA was transported to the vacuolar membrane (compare to Vma2p). However, in the 90 min induced vps35-105 vps10-Q1499L::HA strain Vps10-Q1499L-HA exhibited a predominantly punctate staining pattern with a minor amount of vacuolar membrane staining. This pattern was similar to that observed for vps35-105 vps10-Q1499L::HA cells in the steady-state (Fig. 6). These results indicate that the majority of the Vps10-Q1499L-HA was retrieved back to the TGN. Therefore, the basis of the _vps10-Q1499L_–mediated suppression of vps35-105 is due to improved sorting at the PVC.
Figure 7.
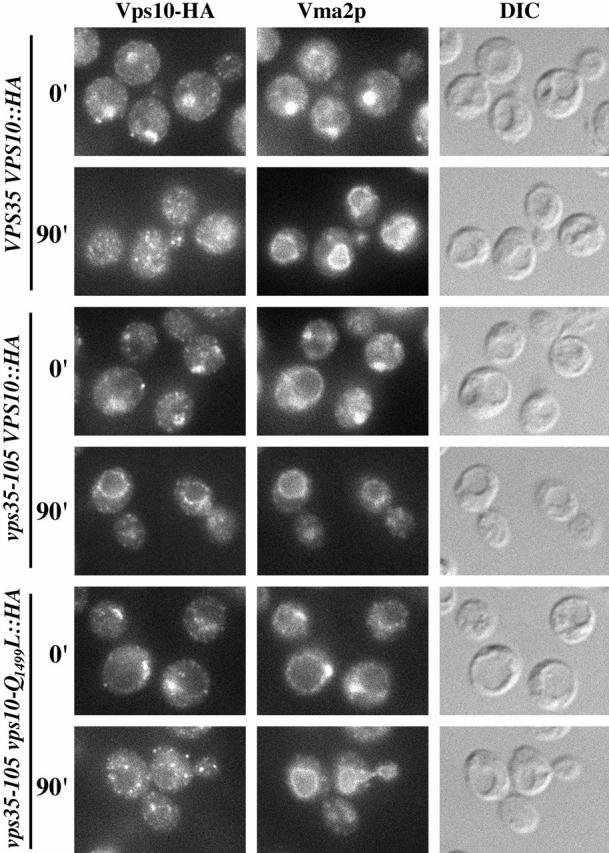
Suppression of the vps35-105 allele by the Q1499L mutation in Vps10p is due to improved sorting at the PVC. Strain SNY128 carrying pHY5 and strains SNY129 and SNY134, each carrying pHY5 and pSN350 were analyzed (shown respectively from top to bottom). All strains carry a _vps27_Δ mutation (Table ) and pHY5 expresses VPS27 under the GAL1 promoter (Table ). Strains were propagated in media containing raffinose. At the 0 min time point the cultures were adjusted to 2% galactose to induce expression of VPS27. After 0 and 90 min of galactose addition cells were fixed, spheroplasted, costained with antibodies against the HA epitope and Vma2p, and analyzed as described in the legend to Fig. 6.
Discussion
Retrieval of cargo proteins from endosomes to the Golgi apparatus is a complex process mediated by several peripheral membrane proteins. Subunits of the retromer complex involved in yeast endosomal retrieval are highly similar to proteins found in several higher eukaryotic organisms including humans. Thus retromer-mediated retrieval likely exists in these organisms as well. In this study we used two independent approaches to demonstrate that sorting of two cargo proteins, Vps10p and A-ALP, into the retrieval pathway involves physical interaction with Vps35p.
Our results provide definitive evidence supporting previous studies casting Vps35p as a prime candidate for being a “receptor” protein that recognizes the retrieval signals present on cargo proteins. For example, in yeast strains lacking function of another retromer subunit, Vps29p, Vps10p fails to be retrieved and is mislocalized to the vacuole. In wild-type cells most of Vps35p was found in a membrane fraction that was pelleted only after a high speed (100,000 g) spin, however, in _vps29_Δ cells Vps35p instead pelleted in a 13,000 g spin (Seaman et al. 1997). One interpretation of these results is that Vps35p associates with cargo such as Vps10p at the PVC and remains associated as cargo proteins are mislocalized to the vacuole, an organelle that fractionates in the 13,000 g pellet. In addition, mutant alleles of VPS35 have been described that fail to retrieve one type of cargo protein but do retrieve a different type of cargo (Nothwehr et al. 1999).
A chemical cross-linking approach was used to demonstrate that the cytosolic domain of A-ALP forms a complex with Vps35p. Vps35p has been shown on density gradients to fractionate in two peaks: one that corresponds to endosomal membranes and another denser peak that may correspond to vesicular membranes (Seaman et al. 1998). Given that A-ALP is predominantly localized to the TGN in the steady state (Nothwehr et al. 1993; Bryant and Stevens 1997) a transient interaction of A-ALP with the sorting machinery at the PVC would be expected. Because Vps35p also interacts with Vps10p and is likely to interact with other cargo proteins such as Kex2p and Kex1p, the percentage of total Vps35p that interacts with A-ALP would most likely be small. Accordingly, we detected that around 15–25% of Vps35p cross-linked to A-ALP when A-ALP was overproduced using the GAL1 promoter and about 10% of Vps35p cross-linked when A-ALP was expressed at normal levels. Of course the actual percentage of Vps35p that associates at any given time with A-ALP is probably somewhat higher since it is unlikely that every A-ALP–Vps35p binding pair was covalently attached during the cross-linking reaction. Thus far we have been unsuccessful at reproducibly demonstrating cross-linking between Vps10p and Vps35p. To cross-link two proteins the bifunctional cross-linker DSP must react with a primary amine group on both proteins and it is possible that the interface of the Vps10p–Vps35p complex does not contain such groups in an appropriate spatial arrangement.
Characteristics of the association of A-ALP and Vps35p indicated that it reflected a sorting process at the PVC. Little or no cross-linking was observed in a strain containing the vps35-101 allele. vps35-101 encodes Vps35p with a D123N mutation previously shown to prevent retrieval of A-ALP but not to affect retrieval of Vps10p. The Vps35-D123N mutant protein exhibited normal association to membranes and sorted Vps10p normally at the PVC (Nothwehr et al. 1999) thus it is most likely localized to the periphery of the PVC in a wild-type manner. If our cross-linking results were simply reflecting nonspecific association of PVC-localized A-ALP with Vps35p then A-ALP should cross-link to wild-type Vps35p and the Vps35-D123N mutant to a similar extent. Furthermore, cross-linking was sensitive to mutation of the two F residues that comprise the most important elements of the retrieval signal (Nothwehr et al. 1993).
A genetic suppression approach was used to further address the nature of Vps35p interaction with A-ALP and Vps10p. Mutations in the cytosolic domains of each protein were found that suppressed the cargo-specific vps35-101 and vps35-105 alleles in an allele-specific manner. Suppression appeared to be due to improved sorting at the PVC rather than prolonged retention in some organelle in the secretory pathway. For example, the A(N88I)-ALP mutant protein reached the exaggerated PVC compartment in vps27 cells with similar kinetics as wild-type A-ALP. In addition, the Vps10-Q1499L suppressor protein apparently exhibits normal cycling between the TGN and PVC as judged by its ability to sort CPY with wild-type efficiency. Finally, determination of the fate of wild-type and mutant Vps10p in _vps27_Δ strains after induction of Vps27p synthesis demonstrated that the Q1499L mutation in Vps10p improved its sorting at the PVC in vps35-105 cells.
Allele-specific suppression has been extensively used to detect protein–protein interactions and in some cases interactions detected using this approach have also been characterized biochemically (Adams et al. 1989; Mortin 1990; Phizicky and Fields 1995). In one study, suppressor mutations were isolated in SAC6 that restored binding to a mutant form of actin incapable of binding to wild-type SAC6. The mutations in SAC6 restored function by increasing binding affinity for the mutant form of actin (Sandrock et al. 1997). It is likely that the mechanism of suppression by mutations in VPS35 occurs by a similar mechanism.
Both the original mutations in VPS35 and the suppressor mutations in the cargo proteins have been identified (Fig. 2 A and 4 A), allowing consideration of the possible effects of these mutations on protein–protein interaction. The mutations responsible for the cargo-specific phenotypes in vps35-101 and vps35-105 are D123N and D528G suggesting that acidic residues may be important for cytosolic domain binding. Although far apart in the primary sequence, it is possible that the D123 and D528 mutations lie close to each other in the tertiary structure and may contribute to retrieval sequence recognition within the context of a single binding site. Alternatively, the D123 and D528 residues may play analogous roles in two distinct binding sites. Both the Y1492 residue of Vps10p and the FXFXD89 residues of DPAP A have been shown to be important determinants for retrieval (Nothwehr et al. 1993; Cooper and Stevens 1996). Residues 81–91 of the DPAP A cytosolic domain can function independently as a retrieval signal (Nothwehr et al. 1993; Bryant and Stevens 1997) suggesting that a relatively small region of the cytosolic domain is recognized by Vps35p. Interestingly, the suppressor mutations in both DPAP A and Vps10p are located very close to the aromatic amino acid signals (Fig. 2 A and 4 A). These results are consistent with a model in which the D123 and D528 residues of Vps35p are important for retrieval signal–specific binding to a subregion of the Vps10p and DPAP A cytosolic domains and that mutation of these residues reduced binding affinity. The suppressor mutations, that occurred just a few residues from the retrieval signals, would then restore binding affinity. Apparently the amino acid changes leading to suppression of the D123G and D528N mutations in Vps35p are also compatible with binding to wild-type Vps35p since no defect in retrieval due to these mutations was observed in VPS35 cells. An exceedingly small number of amino acid changes appear to be capable of suppression as suggested by the fact that both Vps10p suppressor mutants contained mutations in the same codon and the three A-ALP suppressor mutations were clustered between codons 88 and 92. Analysis of the physical–chemical characteristics of the residues involved does not suggest a straightforward explanation of how binding was restored. Resolving this issue will require structural analysis of a cytosolic domain–Vps35p complex.
This current study along with previous reports on the function of the retromer subunits supports a model in which Vps35p interacts with the cytosolic domains of cargo proteins such as Vps10p and A-ALP (Fig. 8). This interaction would then cause those proteins to enter budding vesicles for delivery of proteins to the TGN. The allele-specific suppression data argue that the interaction between the cytosolic domains of A-ALP and Vps10p with Vps35p is direct rather than being mediated by another member of the complex. However, the possibility remains that Vps35p might interact with cargo proteins in collaboration with another protein, i.e., both proteins would physically contact the cytosolic domain. The retromer complex has been shown to consist of two subcomplexes: a Vps35p–Vps29p–Vps26p subcomplex and a Vps5p–Vps17p subcomplex (Seaman et al. 1998). Mutations in VPS5 or VPS17 result in more global trafficking defects than mutations in VPS35, VPS29, or VPS26 (Horazdovsky et al. 1997; Nothwehr and Hindes 1997; Seaman et al. 1997). These observations have led to the proposal that the Vps35p-containing subcomplex is responsible for selective recruitment of cargo into vesicles at the PVC whereas the Vps5p and Vps17p are structural components of a vesicle coat (Seaman et al. 1998). Thus Vps35p may collaborate with Vps29p or Vps26p in binding to cargo molecules. Interestingly, Vps35p membrane association is not perturbed in vps29 mutants whereas Vps29p membrane association is fully dependent on Vps35p. These data strongly suggest that Vps29p may bind only to Vps35p (and possibly other coat proteins) rather than to the cargo protein.
Figure 8.
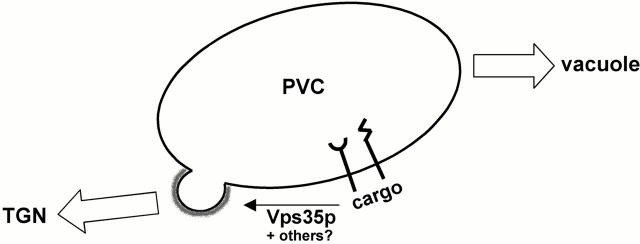
A schematic model of the sorting of cargo proteins from the PVC to TGN in yeast is shown. At the PVC ,Vps35p associates with the cytosolic domains of cargo proteins such as Vps10p and DPAP A via their retrieval signals. This interaction leads to concentration of cargo within forming vesicles that then bud off and are delivered to the TGN. Other proteins that may participate in this process include Vps29p, Vps26p, and Grd19p. A vesicle coat containing Vps5p and Vps17p is represented by the gray halo. For additional details see the text.
Another protein that might collaborate with Vps35p in binding to cargo proteins is Grd19p, an 18.7-kD NADPH oxidase p40 (PX) domain containing protein (Voos and Stevens 1998). PX domains have been found in a number of proteins involved in membrane traffic including the sorting nexin family of which VPS5 is a member (Horazdovsky et al. 1997; Nothwehr and Hindes 1997). Interestingly, grd19 mutants are defective for retrieval of A-ALP but nearly normal for retrieval of Vps10p. Grd19p was shown to bind to the DPAP A cytosolic domain in an assay involving incubation of yeast cytosol with a DPAP A cytosolic domain–6xHis fusion protein purified from Escherichia coli. However, it is possible that Grd19p might not bind directly to DPAP A but rather may bind via some other protein such as Vps35p. Moreover, the in vitro binding of Grd19p to DPAP A was only slightly affected by a deletion that removed the FXFXD retrieval signal indicating that Grd19p can bind to features outside of the retrieval signal region. Thus it is possible that Grd19p is a previously unidentified component of the retromer complex that is necessary for recruitment of DPAP A into PVC-to-TGN vesicles. The fact that in grd19 mutants the retromer can still function in Vps10p retrieval indicates that Vps35p does not need Grd19p to bind to the Vps10p retrieval signal. Vps35p and Grd19p appear to play fundamentally different roles since the interaction of Vps35p with the cytosolic domain of A-ALP (DPAP A) is dependent on the FXFXD signal and appears to be direct. These data, taken together with our identification of allele-specific suppressor mutations in Vps10p and A-ALP that occurred very close to the retrieval signals, strongly suggests that Vps35p binds to cargo proteins via their retrieval signals.
Acknowledgments
We acknowledge the excellent technical assistance of Anna Hindes during the early phases of this work. Scott Emr and Tom Stevens generously provided antibodies. David Eide, Liz Conibear, and Steve Alexander provided valuable critical evaluation of the manuscript.
This work was supported by a grant awarded to S.F. Nothwehr from the National Institutes of Health (GM-53449).
Footnotes
Abbreviations used in this paper: AP, adaptor protein; CEN, yeast centromere; CPY, carboxypeptidase Y; DIC, differential interference contrast; DPAP, dipeptidyl aminopeptidase; DSP, dithio_bis_(succinimidylpropionate); M6P, mannose 6–phosphate; PVC, prevacuolar–endosomal compartment.
References
- Adams A.E.M., Botstein D., Drubin D.G. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science. 1989;243:231–233. doi: 10.1126/science.2643162. [DOI] [PubMed] [Google Scholar]
- Bankaitis V.A., Johnson L.M., Emr S.D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Dell'Angelica E.C. Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigance W.T., Barlowe C., Graham T.R. Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol. Biol. Cell. 2000;11:171–182. doi: 10.1091/mbc.11.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N.J., Stevens T.H. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J. Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell R.C., Joyce G.G. Randomization of genes by PCR mutagenesis. PCR Meth. Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cereghino J.L., Marcusson E.G., Emr S.D. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R., Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E., Stevens T.H. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A., Bussey H. Yeast Kex1p is a Golgi-associated membrane proteindeletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J. Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.A., Stevens T.H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E., Pfeffer S.R. TIP47 - a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Duncan J.R., Kornfeld S. Intracellular movement of two mannose 6-phosphate receptorsreturn to the Golgi apparatus. J. Cell Biol. 1988;106:617–628. doi: 10.1083/jcb.106.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman J.N., Conibear E., Pearse B.M.F. Specificity of binding of clathrin adaptors to signals on the mannose-6-phosphate/insulin-like growth factor II receptor. EMBO J. 1989;8:1041–1047. doi: 10.1002/j.1460-2075.1989.tb03471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilker R., Spiess M., Crottet P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999;21:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Horazdovsky B.F., Davies B.A., Seaman M.N.J., McLaughlin S.A., Yoon S., Emr S.D. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.W. Proteinase mutants of Saccharomyces cerevisiae . Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R., Schmidt A., Mauxion F., Griffiths G., Hoflack B. Binding of AP-1 Golgi adaptors to membranes requires phosphorylated cytoplasmic domains of the mannose-6-phosphate/insulin-like growth factor II receptor. J. Biol. Chem. 1993;268:22552–22556. [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M.A., Goda Y., Zerial M., Pfeffer S.R. Rab9 functions in transport between late endosomes and the trans-Golgi network in vitro . EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson E.G., Horazdovsky B.F., Cereghino J.L., Gharakhanian E., Emr S.D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- McNiven M.A., Cao H., Pitts K.R., Yoon Y. The dynamin family of mechanoenzymespinching in new places. Trends Biochem. Sci. 2000;25:115–120. doi: 10.1016/s0968-0004(99)01538-8. [DOI] [PubMed] [Google Scholar]
- Molloy S.S., Anderson E.D., Jean F., Thomas G. Bi-cycling the furin pathwayfrom TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/s0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- Mortin M.A. Use of second-site suppressor mutations in Drosophila to identify components of the transcriptional machinery. Proc. Natl. Acad. Sci. USA. 1990;87:4864–4868. doi: 10.1073/pnas.87.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoziani P., Vilhardt F., Llorente A., Hilout L., Courtoy P.J., Sandvig K., van Deurs B. Role for dynamin in late endosome dynamics and trafficking of the cation-independent mannose 6-phosphate receptor. Mol. Biol. Cell. 2000;11:481–495. doi: 10.1091/mbc.11.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Roberts C.J., Stevens T.H. Membrane protein retention in the yeast Golgi apparatusdipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J. Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Conibear E., Stevens T.H. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J. Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Bryant N.J., Stevens T.H. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol. Cell. Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Hindes A.E. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J. Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Nothwehr S.F., Bruinsma P., Strawn L.S. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol. Biol. Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno J., Stewart J., Fournier M., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., Bonifacino J.S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Palmer D.J., Helms J.B., Beckers C.J., Orci L., Rothman J.E. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- Payne G.S., Schekman R. Clathrina role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Phan H.L., Finlay J.A., Chu D.S., Tan P.K., Kirchhausen T., Payne G.S. The Saccharomyces cerevisiae APS1 gene encodes a homolog of the small subunit of the mammalian clathrin AP-1 complexevidence for functional interaction with clathrin at the Golgi complex. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M., Fields S. Protein-protein interactionsmethods for detection and analysis. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R.C., Cooper A.A., Yang H., Stevens T.H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae . J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad M.R., Phan H.L., Kirchrath L., Tan P.K., Kirchhausen T., Hollenberg C.P., Payne G.S. Saccharomyces cerevisiae Apl2p, a homologue of the mammalian clathrin AP beta subunit, plays a role in clathrin-dependent Golgi functions. J. Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson I., Vater C.A., Stevens T.H. Morphological classification of the yeast vacuolar protein sorting mutantsevidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R.S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae . J. Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Brickner J.H., Marschall L.G., Nichols J.W., Fuller R.S. Allele-specific suppression of a defective _trans_-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins Mol. Cell. Biol. 16 1996. 6208 6217a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Seeger M., Payne G.S., Fuller R.S. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p Mol. Biol. Cell 7 1996. 1667 1677b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M.A., Soldati T., Shapiro A.D., Lin J., Pfeffer S.R. Lysosome biogenesis requires rab9 function and receptor recycling from endosomes to the trans-Golgi network. J. Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.J., Raymond C.K., Yamashiro C.T., Stevens T.H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roberts C.J., Nothwehr S.F., Stevens T.H. Membrane protein sorting in the yeast secretory pathwayevidence that the vacuole may be the default compartment. J. Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.S., Klionsky D.J., Banta L.M., Emr S.D. Protein sorting in Saccharomyces cerevisiaeisolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.S. Coats and vesicle budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- Rothman J. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman J.H., Stevens T.H. Protein sorting in yeastmutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Sandrock T.M., O'Dell J.L., Adams A.E.M. Allele-specific suppression by formation of new protein-protein interactions in yeast. Genetics. 1997;147:1635–1642. doi: 10.1093/genetics/147.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schmid S.L. Clathrin-coated vesicle formation and protein sorting - an integrated process. Ann. Rev. Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Seaman M.N.J., Marcusson E.G., Cereghino J.L., Emr S.D. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the Vps29, Vps30, and Vps35 gene products. J. Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., McCaffery J.M., Emr S.D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G.S. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast EMBO J. 11 1992. 2811 2818a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G.S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in _Saccharomyces cerevisiae_J. Cell Biol. 118 1992. 531 540b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Matsuoka K., Hamamoto S., Schekman R., Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc. Natl. Acad. Sci. USA. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanely K.K., Howell K.E. TGN38/41a molecule on the move. Trends Cell Biol. 1993;3:252–255. doi: 10.1016/0962-8924(93)90046-4. [DOI] [PubMed] [Google Scholar]
- Stepp J.D., Pellicena Palle A., Hamilton S., Kirchhausen T., Lemmon S.K. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol. Biol. Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater C.A., Raymond C.K., Ekena K., Howald-Stevenson I., Stevens T.H. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J. Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., Huyer G., Emr S.D. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J. Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W., Stevens T.H. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J. Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C.A., Redding K., Wright R., Fuller R.S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol. Biol. Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]