Identification of MAGE-3 Epitopes Presented by HLA-DR Molecules to CD4+ T Lymphocytes (original) (raw)
Abstract
_MAGE_-type genes are expressed by many tumors of different histological types and not by normal cells, except for male germline cells, which do not express major histocompatibility complex (MHC) molecules. Therefore, the antigens encoded by _MAGE_-type genes are strictly tumor specific and common to many tumors. We describe here the identification of the first _MAGE_-encoded epitopes presented by histocompatibility leukocyte antigen (HLA) class II molecules to CD4+ T lymphocytes. Monocyte-derived dendritic cells were loaded with a MAGE-3 recombinant protein and used to stimulate autologous CD4+ T cells. We isolated CD4+ T cell clones that recognized two different MAGE-3 epitopes, MAGE-3114–127 and MAGE-3121–134, both presented by the HLA-DR13 molecule, which is expressed in 20% of Caucasians. The second epitope is also encoded by MAGE-1, -2, and -6. Our procedure should be applicable to other proteins for the identification of new tumor-specific antigens presented by HLA class II molecules. The knowledge of such antigens will be useful for evaluation of the immune response of cancer patients immunized with proteins or with recombinant viruses carrying entire genes coding for tumor antigens. The use of antigenic peptides presented by class II in addition to peptides presented by class I may also improve the efficacy of therapeutic antitumor vaccination.
Keywords: human, invariant chain, peptide, tumor, histocompatibility leukocyte antigen class II
From cultures of blood lymphocytes or tumor-infiltrating lymphocytes of cancer patients stimulated with autologous tumor cells, it is possible to isolate CTLs that show specificity for tumor cells (1, 2). These CTLs have been used as tools to isolate genes that code for tumor antigens, such as those of the MAGE gene family (3, 4). The MAGE antigens are of particular interest for cancer immunotherapy because of their strict tumoral specificity and because they are shared by many tumors. MAGE genes are activated in tumors of many different histological types. They are silent in normal cells, except in testicular germ cells, which do not express MHC class I molecules and are therefore incapable of presenting antigens to CTLs (5, 6). Other families of genes with the same pattern of expression have been identified (7, 8). In addition to the genes identified with the approach involving antitumor CTLs, several other _MAGE_-type genes, i.e., genes expressed only in tumors and in male germline cells, have been identified by purely genetic approaches or by the use of antibodies present in the sera of cancer patients (9–11). As a result, there is now a large supply of sequences potentially coding for tumor-specific shared antigens.
Among the MAGE genes, MAGE-3 is one of the most frequently expressed in tumors. For instance, it is expressed in 76% of metastatic melanomas (12). Five antigenic peptides presented by class I molecules have been identified in the MAGE-3 protein. They are presented to CTLs by HLA-A1, -A2 (two epitopes), -A24, and -B44 (12–17). Some of these epitopes were identified as targets of CTL clones obtained by stimulation of T lymphocytes with autologous tumor cells. Others were identified by “reverse immunology” approaches, based on the knowledge of gene sequences and in vitro stimulation of lymphocytes of noncancerous individuals with candidate peptides considered likely to bind to a given HLA. MAGE-3 codes for a protein of 315 amino acids, and it is very likely that many other peptides derived from MAGE-3 bind to various HLA molecules and hence could be additional target antigens for an antitumor CTL response.
So far, the identification of tumor antigens has focused mainly on antigens recognized by CTLs, almost all of which are CD8+, even though the importance of CD4+ T cells in antitumor response has been demonstrated in numerous animals models (18–21). Only a few examples of antigens recognized by CD4+ T cells on human tumors have been described. CD4+ T cells that were raised in vitro against a peptide centered on the fusion region of bcr-abl were found to recognize HLA-DR4 leukemic blasts expressing bcr-abl (22). The same approach had been previously used to identify an epitope presented by HLA-DQ7 molecules in a mutated region of K-ras (23). Antigenic peptides encoded by the melanocyte differentiation proteins tyrosinase and gp100 have also been identified (24–26).
No antigen presented by a class II molecule has yet been reported for a _MAGE-_type gene. In an attempt to characterize such an antigen for MAGE-3, we used monocyte- derived dendritic cells loaded with a MAGE-3 recombinant protein to stimulate autologous CD4+ T cells. We report here the identification of MAGE-3 epitopes presented by HLA-DR molecules.
Materials and Methods
Cell Lines, Media, and Reagents.
The EBV-transformed B (EBV-B) cell lines and tumor cell line MZ2-MEL.43 were cultured in IMDM (GIBCO BRL) supplemented with 10% FCS (GIBCO BRL), 0.24 mM l-asparagine, 0.55 mM l-arginine, 1.5 mM l-glutamine (AAG), 100 U/ml penicillin, and 100 μg/ml streptomycin. The PhoenixAMPHO cell line (provided by Dr. Nolan, Stanford University, Stanford, CA) is a high-titer amphotropic retrovirus–producing cell line that was generated by stable transfection of 293T cells with a Moloney GagPol-IRES-Lyt 2 construct with a Rous sarcoma virus (RSV) promoter and a pPGK hygro selectable marker. These cells were then stably transfected with the Moloney amphotropic envelope gene driven by a CMV promoter and coselected with the diphtheria toxin resistance gene (pHED-7). This producer cell line is helper virus–free. PhoenixAMPHO cells were cultured in DMEM (Life Technologies) supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, and antibiotics.
Human recombinant IL-2 was purchased from Eurocetus, IL-7 from Genzyme, GM-CSF from Schering Plough, and TNF-α from R&D Systems. Human recombinant IL-4, IL-6, and IL-12 were produced in our laboratory.
MAGE-3 Protein.
The recombinant MAGE-3 protein was produced by SmithKline Beecham Pharmaceuticals. The full-length MAGE-3 sequence, preceded by the amino acid (aa)1 sequence MHHHHHHHGG, was engineered into a vector bearing the pMB1 replicon and the PL short promoter. This vector was used to transform Escherichia coli strain AR58 (27). Bacteria were grown in LB medium containing 50 μg/ml kanamycin at 30°C. After heat induction, the MAGE-3 expression product became detectable as a 46-kD band when assayed by Western blot (SDS-PAGE 12.5%; revealed by a rabbit polyclonal antiserum to MAGE-3). Protein purification was carried out at room temperature, and involved the following steps: cell lysis and centrifugation, repeated washing of the centrifugation pellet followed by solubilization of the pellet containing MAGE-3, immobilized metal ion affinity chromatography, anion exchange chromatography, concentration, and dialysis. The resulting MAGE-3 protein was >95% pure, as assessed by Coomassie blue staining.
Construction of pMFG-Ii.MAGE-3.
The plasmid IipSV51L, containing a cDNA encoding the human invariant chain (Ii), was provided by Dr. J. Pieters (Basel Institute for Immunology, Basel, Switzerland). The MFG plasmid was provided by Dr. O. Danos (Somatix Therapy Corp., Alameda, CA). The MFG retroviral vector is derived from the Moloney murine leukemia virus, which lacks a drug resistance marker and does not express any other potential antigenic protein except for the inserted cDNA (28). The cDNA encoding the NH2-terminal end (i.e., the cytoplasmic tail and the transmembrane region) of the human (hu)-Ii polypeptide (residues 1–80) was amplified by PCR using IipSV51L as the template. The following primers were used: hu-Ii sense, 5′-TTT_CCATGG_ATGACCAGCGCGAC-3′, and hu-Ii antisense, 5′-TTT_GGATCC_GGAAGCTTCATGCGCAGGTTC-3′ (the recognition sites for NcoI and BamHI are in italics). The PCR product was cloned into pCR2.1 and sequenced. The NcoI-BamHI amplification product was cloned into pMFG and opened with the enzymes NcoI and BamHI, resulting in pMFG-Ii. A BglII recognition site, replacing the ATG codon and in frame with the BamHI site at the 3′ end of the truncated Ii-cDNA, was introduced at the 5′ end of the MAGE-3 cDNA by PCR using the following primers: BglII sense, 5′-TTT_AGATCT_TGAGCAGAGGAGTCAGC-3′, and BglII antisense: 5′-CCC_AGATCT_TCACTCTTCCCCCTCTCTC-3′ (the recognition sites for BglII are in italics). The PCR product (BglII. MAGE-3.BglII) was cloned into pCR2.1 and sequenced. The recombinant plasmid, pMFG-Ii, was reopened with BamHI, and the BglII.MAGE-3.BglII amplification product was ligated to the compatible ends. Recombinant plasmids containing the MAGE-3 cDNA in frame and in the right orientation were identified by restriction fragment analysis.
Production of High-titer Ii.MAGE-3–encoding Recombinant Retrovirus.
The MAGE-3–encoding retroviral vector plasmid MFG-Ii.MAGE-3 was introduced into the PhoenixAMPHO packaging cells by transfection. The transfection procedure is a modification of the calcium phosphate–mediated transfection protocol of Graham and Van der Eb (29). 24 h before transfection, PhoenixAMPHO cells were plated in cell growth medium in a 75-cm2 tissue culture flask (Falcon; Becton Dickinson Labware). After adding the cells, the flask was gently shaken forward and backward to distribute cells evenly in the flask bottom. The cells were incubated at 37°C and 5% CO2. At the time of transfection, when the cells should have reached a confluence of 70–80%, the medium was removed and replaced by 14 ml fresh PhoenixAMPHO cell growth medium containing 25 mM chloroquine (Sigma). A transfection cocktail was prepared in a 50-ml tube by adding 40 μg retroviral vector plasmid DNA to water and diluting to 1,575 μl final vol. To this DNA solution 225 μl of 2 M CaCl2 was added. Then, 1,800 μl of 2× HeBS (50 mM Hepes, 10 mM KCl, 12 mM dextrose, 280 mM NaCl, and 1.5 mM Na2HPO4 dissolved in distilled water, filtered through a 0.2-μm filter, and stored at −20°C) was added in drops to the DNA/CaCl2 solution by bubbling vigorously for 15 s with an automatic pipette. The DNA/ CaCl2/HeBS mix was added immediately in drops onto the cells, and the flask was gently swirled to ensure uniform mixing of DNA/CaPO4 particles. The cells were incubated at 37°C/5% CO2 for 7–9 h, and the chloroquine-containing medium was changed for fresh PhoenixAMPHO cell growth medium. Approximately 24 h before the harvest of the retroviral supernatant, the PhoenixAMPHO medium was removed and gently replaced by 9 ml of IMDM containing only 2.5% FCS. The retroviral supernatant was harvested 48 h after transfection by removing the medium from the cells and filtering through a 0.45-μm filter to remove cell debris. After harvest and filtration, the virus-containing medium was kept on ice, aliquoted in appropriate volumes in 15-ml polypropylene tubes, and stored at −80°C.
Retroviral Transduction of Cell Lines.
Target cells were resuspended in 60-mm tissue culture plates (Falcon) at a density of 106 cells in 4 ml of infection cocktail containing 50% viral supernatant in growth medium and 6 μg/ml of protamine sulfate. The plates were centrifuged for 2 h at 32°C and 1,200 rpm, followed by another 2 h of incubation in a humidified incubator at 37°C. Cells were then transferred to 4 ml of growth medium. This transduction cycle was carried out immediately after plating the cells and was repeated at 24 and 48 h.
Dendritic Cells.
Blood cells were collected as buffy coat preparations from hemochromatosis patients. PBMCs were isolated by Lymphoprep (Nycomed Pharma) density gradient centrifugation. To generate autologous dendritic cells, PBMCs were depleted from T lymphocytes by rosetting with sheep erythrocytes (Bio Mérieux) treated with 2-aminoethylisothiouronium (Sigma). The lymphocyte-depleted PBMCs were left to adhere for 2 h at 37°C in culture flasks (Falcon) at a density of 2 × 106 cells/ml in RPMI 1640 medium supplemented with AAG and 1% autologous plasma (hereafter referred to as complete RPMI medium). Nonadherent cells were discarded, and adherent cells were cultured in the presence of IL-4 (100 U/ml) and GM-CSF (100 ng/ ml) in complete RPMI medium. Cultures were fed on days 2 and 4 by replacing half of the medium with fresh medium plus IL-4 (100 U/ml) and GM-CSF (100 ng/ml). On day 5, the nonadherent cell population was used as a source of enriched dendritic cells.
CD4+ Responder T Cells.
Rosetted T cells were treated with NH4Cl (160 mM) to lyse the sheep erythrocytes and washed. CD4+ T lymphocytes were isolated from rosetted T cells by negative selection using an anti-CD8 mAb coupled to magnetic microbeads (Miltenyi Biotech) and by sorting through a MACS®, as recommended by the manufacturer. The lymphocytes were frozen and then thawed the day before the coculture with dendritic cells.
Mixed Lymphocyte/Dendritic Cell Culture.
Autologous dendritic cells (5 × 105/ml) were incubated at 37°C, 5% CO2 for 18–20 h in complete medium supplemented with IL-4 (100 U/ ml), GM-CSF (100 ng/ml), and TNF-α (1 ng/ml) in the presence of the recombinant MAGE-3 protein (20 μg/ml). Cells were washed and added at 104 per round-bottomed microwell to 105 CD4+ lymphocytes in 200 μl IMDM supplemented with AAG and 10% human serum (hereafter referred to as complete IMDM) in the presence of IL-6 (1,000 U/ml) and IL-12 (10 ng/ ml). The CD4+ lymphocytes were restimulated on days 7, 14, and 21 with autologous dendritic cells freshly loaded with the MAGE-3 protein and were grown in complete IMDM supplemented with IL-2 (10 U/ml) and IL-7 (5 ng/ml). Due to the limited supply of dendritic cells for melanoma patient 7002, we used only 6 × 103 dendritic cells instead of 104 and restimulations were performed on days 10 and 20 instead of 7, 14, and 21. The microcultures containing proliferating CD4+ T cells were assessed on days 35–37 for their capacity to produce TNF and/or IFN-γ when stimulated with autologous EBV-B cells loaded with protein MAGE-3. Autologous EBV-B cells were incubated for 18–20 h in the presence of 20 μg/ml of protein MAGE-3, or OVA (Sigma) as a negative control. Protein-pulsed EBV-B cells were washed and distributed at 5,000 cells per round-bottomed microwell together with 2,500 CD4+ T lymphocytes in 150 μl of complete IMDM supplemented with IL-2 (25 U/ml). After 20 h, the supernatant was collected and its TNF content was determined by testing its cytotoxic effect on WEHI-164 clone 13 cells (30) in a MTT colorimetric assay (31, 32). IFN-γ released in the supernatant was measured by ELISA using reagents from Medgenix Diagnostics-Biosource. Inhibition with mAbs W6/32 (anti–HLA class I) or 2B6 (anti–HLA-DR) was performed by addition of a 1:20 dilution of ascites during the experiment.
CD4+ T Cell Clones.
The microcultures that recognized cells loaded with the MAGE-3 protein were cloned by limiting dilution, using as stimulating cells either the autologous EBV-B cell line loaded with the MAGE-3 protein or the autologous EBV-B cell line transduced with a retrovirus encoding Ii.MAGE-3. Allogeneic EBV-B cells (LG2-EBV) were added as feeder cells. CD4+ T cell clones were grown in complete IMDM supplemented with IL-2 (50 U/ml), IL-7 (5 ng/ml), and 0.5 μg/ml purified PHA (HA 16; Murex Diagnostics). The clones were supplemented with fresh culture medium once a week and passaged with feeder cells (1.5 × 106 allogeneic PBLs plus 5 × 105 LG2-EBV per well in a 24-well plate) at 1–2-wk intervals. Occasionally, clones were restimulated with 2 × 105 autologous EBV-B cells transduced with Ii.MAGE-3 as stimulating cells and 106 LG2-EBV. Established CD4+ T cell clones were then tested for TNF and/or IFN-γ production after stimulation with autologous EBV-B cells pulsed with protein MAGE-3 and MAGE-3– derived peptides.
Recognition Assays with Peptides.
Peptides were synthesized on solid phase using F-moc for transient NH2-terminal protection and were characterized using mass spectrometry. All peptides were >80% pure, as indicated by analytical HPLC. Lyophilized peptides were dissolved in DMSO and used at a concentration of 5 μg/ml. EBV-B cells were incubated for 2 h at 37°C in the presence of the different peptides, the indicated concentrations representing their concentrations during the incubation step. They were distributed at 5,000 cells per round-bottomed microwell together with 2,500 CD4+ T lymphocytes in 150 μl of complete IMDM supplemented with IL-2 (25 U/ml). Supernatants were harvested after 20 h and assessed for TNF and/or IFN-γ production. The results represent the average of duplicate or triplicate cultures.
Recognition Assays with Cell Lysates.
The MAGE-3 cDNA sequence cloned into expression vector pCEP-4 was transiently transfected into the 293-EBNA cell line by Lipofectamine® (GIBCO BRL). In brief, 5 × 104 293-EBNA cells per flat-bottomed microwell were transfected with pCEP4-MAGE-3 and 1 μl of Lipofectamine® in Optimem medium (GIBCO BRL). After 24 h, transfected 293-EBNA cells were lysed in 50 μl of complete RPMI medium by three cycles of rapid freeze–thawing. Monocyte-derived dendritic cells expressing HLA-DR13 molecules were then added (1.5 × 104 cells per well) to the lysates of transfected 293-EBNA cells and kept at 37°C for 24 h. The dendritic cells were then washed, and 2,500 CD4+ lymphocytes were added in 150 μl of complete IMDM supplemented with IL-2 (25 U/ml). Supernatants were harvested after 20 h and assessed for IFN-γ production.
Results
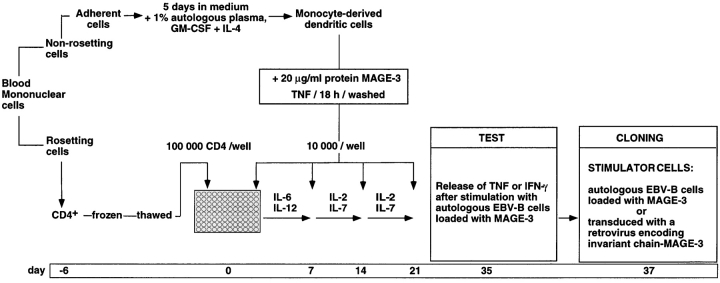
Blood monocytes were cultured in medium supplemented with GM-CSF and IL-4 to favor their differentiation into dendritic cells (Fig. 1). Autologous plasma was used to avoid loading the dendritic cells with bovine or allogeneic proteins. After 5 d, the dendritic cells were incubated overnight with 20 μg/ml of a MAGE-3 protein produced in E. coli and with TNF-α to induce their maturation. 96 microcultures were set up with 105 autologous responder CD4+ T cells and 104 dendritic cells loaded with protein MAGE-3 as stimulator cells. IL-6 and IL-12 were added during the first week to activate the T cells. The responder T cells received three additional weekly restimulations with dendritic cells pulsed with protein MAGE-3 in the presence of IL-2 and IL-7. After a resting period of 2 wk, the responder cells of each microculture were tested on day 35 for TNF or, more often, for IFN-γ production after stimulation for 20 h with autologous EBV-B cells loaded with protein MAGE-3.
Figure 1.
Overview of the procedure used to obtain anti–MAGE-3 CD4+ T cell clones.
CD4+ T Cell Clones Directed against a MAGE-3 Antigen.
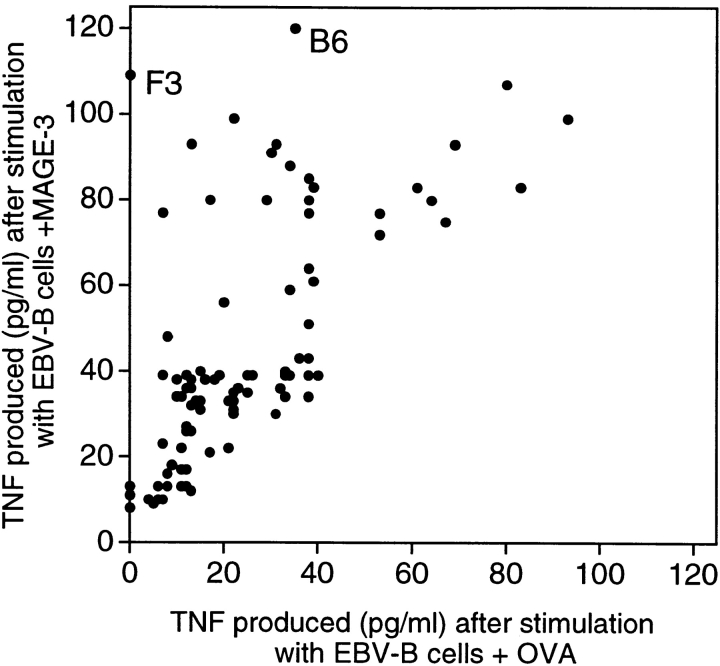
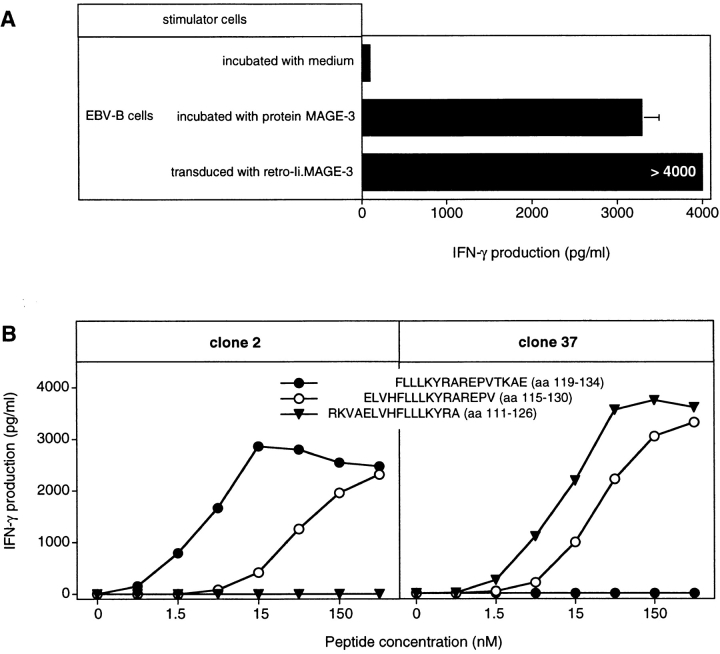
CD4+ T cells of hemochromatosis patient LB 1555, stimulated with dendritic cells loaded with protein MAGE-3, were tested for their ability to produce TNF upon stimulation with EBV-B cells loaded with either protein MAGE-3 or OVA. It was possible to measure the production of TNF by the T cells because, contrary to most EBV-B cell lines, that of patient LB 1555 did not produce TNF. The microculture (B6 in Fig. 2) that produced the highest level of TNF after stimulation with protein MAGE-3 was cloned by limiting dilution using autologous EBV-B cells loaded with protein MAGE-3 as stimulator cells. These stimulator cells were used for the cloning step because of the limited supply of autologous dendritic cells. A positive CD4+ clone was obtained, referred to hereafter as clone 37. It recognized autologous EBV-B cells loaded with protein MAGE-3. Even though the protein, which had been produced in bacteria, was >95% pure, we could not at this stage exclude the possibility that clone 37 recognized a bacterial contaminant. Therefore, we prepared a set of peptides of 16 aa, which overlapped by 12 and covered the entire MAGE-3 protein sequence. Each of these peptides was incubated with autologous EBV-B cells and tested for recognition by clone 37. We identified two overlapping peptides, MAGE-3111–126 and MAGE-3115–130 (see below), that stimulated production of both TNF (data not shown) and IFN-γ (Fig. 3 A). This demonstrated unambiguously that clone 37 recognized a MAGE-3 antigen and not a contaminant.
Figure 2.
Production of TNF by CD4+ T cells stimulated with autologous EBV-B cells incubated with protein MAGE-3. EBV-B cells from hemochromatosis patient LB 1555 were incubated with 20 μg/ml of OVA or MAGE-3 protein for 20 h. 5,000 cells were distributed in microwells to which an aliquot of each of the 96 CD4 microcultures was added (∼3,000 effector cells). TNF production was estimated after overnight culture, by the toxicity of the supernatants for the TNF-sensitive WEHI 164-13 cells. The dots represent the average result obtained with two aliquots of each CD4 microculture.
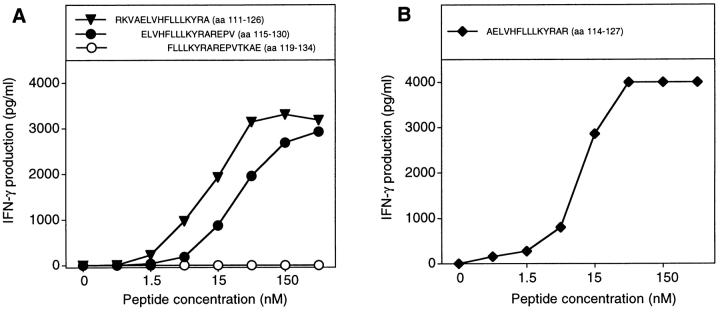
Figure 3.
MAGE-3 peptides recognized by clone 37. (A) Stimulation of clone 37 by two overlapping MAGE-3 peptides. Autologous EBV-B cells were incubated for 2 h with different concentrations of the peptides. Clone 37 (2,500 cells) was then cocultured with 5,000 peptide-pulsed cells for 20 h. IFN-γ production in the supernatant was measured by ELISA. (B) Stimulation by the shortest peptide recognized by clone 37. Conditions were as in A.
A CD4+ clone was obtained from another microculture which was also very well stimulated by cells loaded with protein MAGE-3 (F3 in Fig. 2). However, here the screening with the set of overlapping MAGE-3 peptides was negative. We found that the antigen recognized by this CD4+ clone was present in an HPLC fraction other than the one containing the MAGE-3 protein (data not shown). Our tentative conclusion is that this clone recognizes a bacterial contaminant.
Additional CD4+ clones were obtained from five independent microcultures, set up with cells from another hemochromatosis patient. Their ability to recognize cells loaded with protein MAGE-3 appeared to decrease upon further purification of the protein by HPLC. Two of these clones were also tested for their response to the set of MAGE-3 peptides, with negative results, suggesting further that they recognized a contaminant in the MAGE-3 batch. Therefore, it appears that bacterial contaminants in this MAGE-3 protein batch were more likely to activate CD4+ T cells than was MAGE-3 itself, so that CD4+ clones specific for MAGE-3 constituted only a minor fraction of the clones obtained with this procedure.
Clone 37 Recognizes Peptide AELVHFLLLKYRAR on HLA-DR13.
CD4+ clone 37 recognized two peptides which overlapped by 12 aa, namely RKVAELVHFLLLKYRA (aa 111–126) and ELVHFLLLKYRAREPV (aa 115–130) (Fig. 3 A). The recognition by clone 37 of cells loaded with protein MAGE-3 was abolished by an anti– HLA-DR antibody (data not shown). Patient LB 1555 was serologically typed DR3, DR13, and DR52. To identify the presenting HLA-DR molecule, we tested additional EBV-B cell lines expressing DR3, DR13, or DR52. All and only those expressing DR13 were able to present the MAGE-3111–126 and MAGE-3115–130 peptides to clone 37 (Table I). 31 different HLA-DR13 alleles have been described to date (33). LB 1555 expresses the DRB1*1302 allele, and the other DR13 cell lines tested express DRB1* 1301 or DRB1*1302, the latter two representing 80% of the HLA-DR13 alleles.
Table I.
HLA-DR13 Cells Present the Two Peptides MAGE-3111–126 and MAGE-3115–130 to CD4+ Clone 37
| EBV-B cell line | Serological specificity | IFN-γ production by clone 37 stimulated with peptide | |
|---|---|---|---|
| MAGE-3111–126 | MAGE-3115–130 | ||
| DR13 positive | pg/ml | ||
| LB 1555 | DR3+ DR52+ | >4,000 | >4,000 |
| LB 1118 | DR3+ DR52+ | 3,761 | 3,909 |
| LB 1622 | DR3− DR52+ | 1,731 | 1,349 |
| OMW | DR3− DR52+ | >4,000 | >4,000 |
| MZ2 | DR3− DR52− | 3,429 | 3,096 |
| DR13 negative | |||
| BM16 | DR3− DR52+ | 99 | 120 |
| BOB | DR3− DR52+ | 128 | 101 |
| BOLETH | DR3− DR52+ | 77 | 67 |
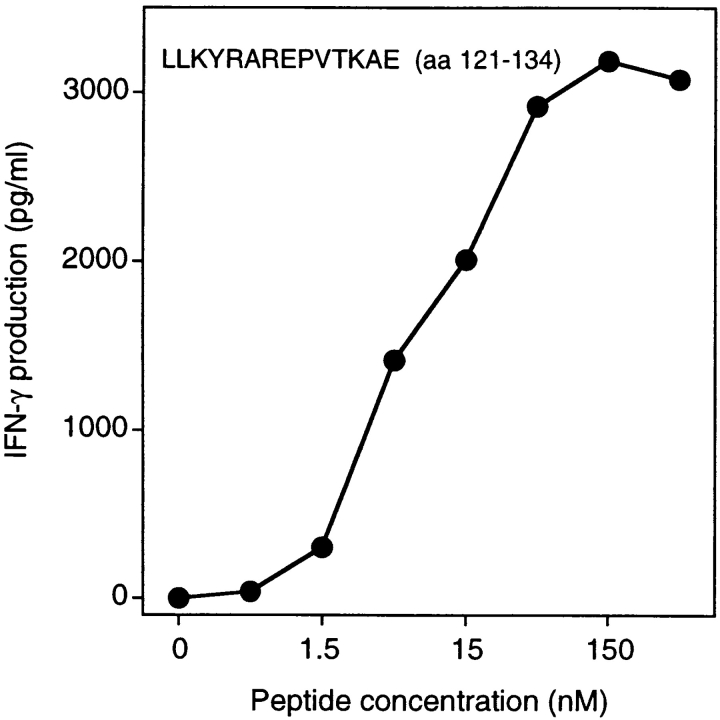
Unlike the peptides presented by class I molecules, those presented by class II usually vary in length and tolerate extensions at both the NH2 and COOH termini because they are not fixed by their ends in the groove (34). Therefore, it is difficult to define the length of those peptides precisely. We tested a large number of MAGE-3 peptides of different lengths (data not shown) and concluded that the shortest peptide well recognized by clone 37 is AELVHFLLLKYRAR (aa 114–127; Fig. 3 B). Half-maximum value of stimulation was obtained by incubating the stimulator cells with 10 nM of this peptide. This compares favorably with the results obtained with other epitopes recognized on HLA class II molecules by CD4+ T cells: a minimum of 30 nM of a bcr-abl peptide was needed to induce a significant proliferation of an anti–bcr-abl CD4+ cell line, whereas 330 nM of peptide p21-ras was required for anti–p21-ras CD4+ clones (22, 23).
Dendritic Cells Incubated with Cell Lysates Present the MAGE-3 Antigen.
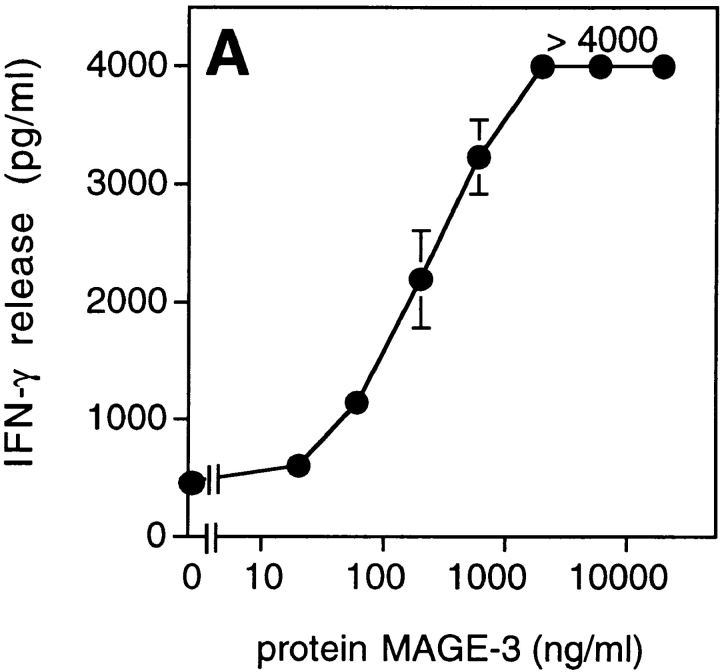
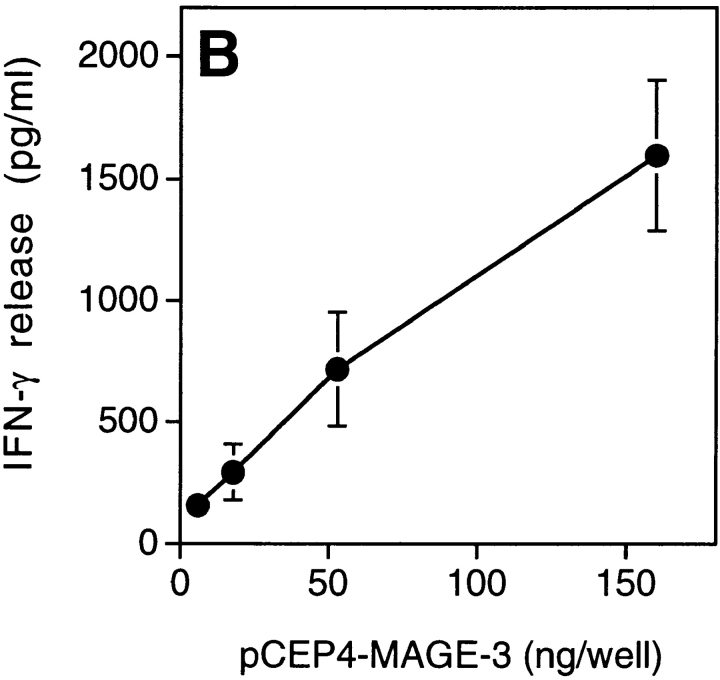
We tested dendritic cells, incubated with decreasing concentrations of MAGE-3 protein, for their ability to stimulate clone 37 (Fig. 4 A). Half-maximum production of IFN-γ was obtained when 104 dendritic cells were preincubated in a volume of 100 μl with 30 ng of the protein, which is approximately the amount present in 2 × 104 MZ2-MEL.43 tumor cells (Godelaine, D., personal communication). This suggested that a dendritic cell, having endocytosed debris of just one or a few tumor cells expressing MAGE-3, would be capable of stimulating anti–MAGE-3 CD4+ T cells. To test further the ability of dendritic cells to process debris of cells expressing MAGE-3, 293-EBNA cells transiently transfected with the MAGE-3 gene were lysed and incubated for 24 h with HLA-DR13 dendritic cells. These cells stimulated clone 37 (Fig. 4 B). The amount of IFN-γ released by clone 37 increased with the amount of plasmid used for the transfection.
Figure 4.
Presentation of the MAGE-3 antigen by dendritic cells incubated with purified protein or cell lysates. (A) HLA-DR13 dendritic cells were cultured for 24 h with different concentrations of MAGE-3 protein. The cells were washed, then incubated with 2,500 cells per well of clone 37. IFN-γ production was measured after 20 h by ELISA. The results shown represent the average of triplicate cultures. (B) 293-EBNA cells (5 × 105 cells per well) were transfected with different doses of pCEP4-MAGE-3 mixed with Lipofectamine®. 24 h after transfection, the transfected cells were lysed by freeze–thawing. HLA-DR13 dendritic cells (105 cells per well) were cultured with lysates at the equivalent of 5 293-EBNA cells per dendritic cell for 24 h. The experiment was pursued as in A.
Presentation of the Antigen by Cells Expressing the Ii. MAGE-3 Construct.
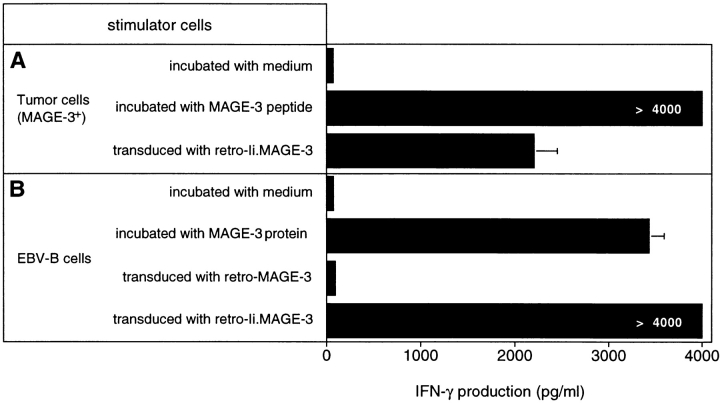
A melanoma cell line which expresses HLA-DR13 and MAGE-3 was unable to stimulate the release of IFN-γ by clone 37, unless it was pulsed with the MAGE-3115–130 peptide (Fig. 5 A). This suggests that the MAGE-3 protein synthesized endogenously does not reach the class II presentation pathway. It has been reported that signals within the Ii could be used to target endogenously synthesized protein to the class II antigen-processing compartments. The expression of several constructs, which encoded fusion proteins containing part of the mouse Ii followed by fragments of the OVA gene, generated OVA peptides recognized by murine CD4+ T cells on MHC class II molecules. The highest level of antigen presentation was obtained with a construct containing the sequence coding for the first 80 amino acids of the Ii (35). Accordingly, MZ2-MEL.43 cells were transduced with a retroviral construct encoding the first 80 amino acids of the human Ii fused with MAGE-3 (retro–Ii.MAGE-3). MZ2-MEL.43 cells transduced with retro–Ii.MAGE-3 stimulated a high production of IFN-γ by clone 37, indicating that the Ii.MAGE-3 protein was processed through the class II presentation pathway (Fig. 5 A). HLA-DR13 EBV-B cells transduced with the same construct also stimulated clone 37 (Fig. 5 B). In contrast, HLA-DR13 EBV-B cells transduced with retro–MAGE-3 alone were unable to stimulate clone 37.
Figure 5.
Recognition by clone 37 of cells expressing an Ii MAGE-3 construct. (A) MZ2-MEL.43 melanoma cells express both HLA-DR13 and MAGE-3. Cells were pulsed for 2 h with 5 μg/ml of peptide MAGE-3115–130 and washed, or transduced with a retroviral construct encoding Ii.MAGE-3. (B) MZ2-EBV cells express HLA-DR13. As a positive control for stimulating cells, we incubated them for 20 h with 20 μg/ml of protein MAGE-3. MZ2-EBV cells were transduced with a retroviral construct encoding MAGE-3 alone or Ii.MAGE-3. Tumor cells (104) and EBV-B cells (5 × 103) were distributed in microwells, and 2,500 cells of clone 37 were added. IFN-γ production was measured after 20 h of coculture by ELISA. The results shown represent the average of triplicate cultures.
Two Other CD4+ Clones Recognized a Neighboring MAGE-3 Peptide on HLA-DR13.
CD4+ T cells isolated from the blood of melanoma patient 7002, who had been immunized with the MAGE-3 protein, were stimulated with autologous dendritic cells loaded with MAGE-3 protein. After three restimulations, one microculture produced IFN-γ upon stimulation with autologous EBV-B cells loaded with the protein. CD4+ clone 2 was isolated, which produced IFN-γ upon stimulation with cells that were either loaded with MAGE-3 protein or transduced with Ii.MAGE-3, the latter proving that the clone was directed against MAGE-3 and not against a contaminant of the protein batch (Fig. 6 A).
Figure 6.
Clone 2 recognizes a different MAGE-3 epitope than clone 37. (A) Production of IFN-γ by clone 2 stimulated with autologous EBV-B cells incubated with protein MAGE-3, or transduced with a retroviral vector encoding the Ii.MAGE-3 fusion protein. Autologous EBV-B cells were incubated for 20 h with 20 μg/ml of the recombinant MAGE-3 protein. Clone 2 (2,500 cells) was incubated with 5,000 autologous EBV-B cells loaded with MAGE-3, or transduced with Ii.MAGE-3. IFN-γ production was measured after 20 h by ELISA. The results shown represent the average of triplicate cultures. (B) HLA-DR13 EBV-B cells were incubated for 2 h with different concentrations of overlapping MAGE-3 peptides. The peptides were purified by HPLC, to compare accurately the efficacy of the different peptides used at the same concentrations. Clones (2,500 cells) were incubated with 5,000 peptide-pulsed cells for 20 h. IFN-γ production was measured by ELISA. The results shown represent the average of triplicate cultures.
When the set of overlapping MAGE-3 peptides were tested for recognition by clone 2, peptide ELVHFLLLKYRAREPV (aa 115–130) scored positive (Fig. 6 B). This peptide was shown previously to be recognized by clone 37, isolated from hemochromatosis patient LB 1555. However, peptide FLLLKYRAREPVTKAE (aa 119–134) scored positive with clone 2 but was not recognized by clone 37 (Fig. 7 B). Moreover, peptide RKVAELVHFLLLKYRA (aa 111–126), previously shown to be recognized by clone 37, was not recognized by clone 2. This indicated clearly that clones 2 and 37 were directed against two adjacent but distinct epitopes. Several EBV-B cell lines were tested for their ability to present peptide MAGE-3119–134 to clone 2. All and only those expressing DR13 were capable of presenting the peptide (Table II). Patient 7002 expresses the DRB1*1302 allele. We tested a large number of MAGE-3 peptides at different concentrations to identify the shortest peptide efficiently recognized by clone 2 (data not shown). This proved to be LLKYRAREPVTKAE (aa 121–134; Fig. 7).
Figure 7.
Stimulation of CD4 clone 2 by the MAGE-3121–134 peptide. HLA-DR13 EBV-B cells were pulsed for 2 h with different concentrations of the peptide. Clones (2,500 cells) were incubated with 5,000 peptide-pulsed cells for 20 h. IFN-γ production was measured by ELISA.
Table II.
HLA-DR13 Cells Present Peptide MAGE-3119–134 to CD4+ Clones 2 and 22
| EBV-B cell line | Serological specificity | IFN-γ production after stimulation with pep- tide MAGE-3119–134 of | |
|---|---|---|---|
| Clone 2 | Clone 22 | ||
| DR13 positive | pg/ml | ||
| 7002 | DR1+ DR52+ | >4,000 | >4,000 |
| LB 1158 | DR1+ DR52+ | >4,000 | 3,967 |
| LB 1118 | DR1− DR52+ | >4,000 | 1,345 |
| LB 1622 | DR1− DR52+ | 3,993 | 1,250 |
| OMW | DR1− DR52+ | 3,153 | 3,651 |
| MZ2 | DR1+ DR52− | >4,000 | 2,030 |
| DR13 negative | |||
| LKT3 | DR1+ DR52+ | 0 | 0 |
| RSH | DR1+ DR52+ | 0 | 0 |
| BM16 | DR1− DR52+ | 12 | 7 |
Interestingly, peptide MAGE-3121–134 is also present in proteins MAGE-1, -2, and -6. Therefore, we loaded DR13 EBV-B cells with a MAGE-1 recombinant protein and used them to stimulate the CD4+ clones. Clone 2, but not clone 37, produced IFN-γ (data not shown).
From hemochromatosis patient LB 1158, yet another anti–MAGE-3 CD4+ clone was obtained, referred to hereafter as clone 22. Patient LB 1158 expresses the DRB1* 1301 allele. Like clone 2, clone 22 was stimulated by DR13 cells presenting peptide MAGE-3119–134 (Table II) and did not recognize peptide MAGE-3111–126 (data not shown). It was also stimulated by DR13 cells loaded with the MAGE-1 protein (data not shown).
Discussion
The procedure described here seems efficient for the activation of anti-MAGE CD4+ precursors present in the blood of individuals with or without cancer, and for the resulting isolation of anti-MAGE CD4+ permanent T cell clones. We are quite confident that it will prove to be applicable to other proteins for the identification of new antigens presented by HLA class II molecules. But our approach is not without disadvantages: a large number of the CD4+ clones obtained were apparently directed against contaminants of the protein batch. Therefore, it will be preferable to use one source of antigen for stimulation of the T cells and another to test their specificity. This may be achieved using two batches of protein produced in different organisms, such as bacteria and insect cells infected with baculovirus constructs, or, as shown here, by the use of presenting cells expressing the protein of interest fused to a truncated Ii.
Antigens can be processed through two different pathways that lead to presentation on HLA class II molecules. The endogenous pathway channels some proteins synthesized in the cell towards endosomal compartments, where they are cleaved into peptides that then associate with class II molecules (36). In the exogenous pathway, the APC takes up protein by endocytosis. Early endosomes undergo a progressive transition to late endosomes and then to lysosomes. The late endosomes have a relatively low pH, contain lysosomal hydrolases, and are enriched in HLA-DM and MHC class II molecules, which are associated with Ii in nonameric complexes. Therefore, they are also called MHC class II compartments (MIIC). Ii is rapidly degraded but class II–associated Ii peptide (CLIP), a small part of it, remains bound within the groove of the MHC molecule and blocks access of other peptides until HLA-DM catalyzes its dissociation from class II molecules. This permits the subsequent loading of potentially antigenic peptides. The peptide–HLA class II complexes are then directed to the cell surface (37).
Melanocyte-specific proteins such as tyrosinase contain a lysosomal targeting sequence that enables them to follow the endogenous class II processing pathway (38). Therefore, these differentiation antigens can be recognized directly by CD4+ T cells on those melanomas that express class II molecules (39). Constitutive expression of class II occurs frequently in melanoma. In line with their cytosolic and nuclear localizations, the MAGE proteins appear to lack the targeting sequences that would enable them to follow the endogenous class II pathway (5, 40, 41). This probably explains why the anti–MAGE-3 CD4+ clones obtained were unable to recognize an HLA-DR13 tumor cell line expressing MAGE-3.
The anti–MAGE-3 CD4+ clones could be stimulated by HLA-DR13 dendritic cells loaded with extracts of cells producing a large amount of MAGE-3 protein. However, they failed to recognize dendritic cells incubated with lysates of tumor cells expressing MAGE-3. This might be due to a lower quantity of protein MAGE-3 in these tumor cell extracts. However, this process may be more efficient in vivo, where loading of dendritic cells with apoptotic bodies from tumor cells might result in very efficient presentation of MAGE-3 epitopes (42). The notion that MAGE-3 antigens can be presented to CD4+ T cells in vivo is supported by the observation that some cancer patients produce anti-MAGE antibodies (11, 43, 44). This is probably due to destruction of some tumor cells followed by uptake of the debris by macrophages or dendritic cells.
In a recently completed clinical trial, 25 tumor-bearing HLA-A1 melanoma patients with advanced disease received 3 subcutaneous injections of a MAGE-3 peptide presented by HLA-A1 (45). Tumor regressions were observed in seven patients, and three of these were complete. No increase in anti-MAGE CTLs could be detected in the blood of these patients, including those whose tumor regressed. The regressions occurred very slowly, suggesting that they may have been caused by a weak immune response. A major limitation of such a class I peptide–based approach might be that the CTLs elicited by the peptide reach the tumor but fail to be restimulated properly at the tumor site so that CTL amplification does not occur. This could be due to the lack of help by tumor-specific CD4+ T cells. Vaccination strategies with MAGE-3 products may benefit from the induction of anti–MAGE-3 CD4+ T cells, despite the fact that tumor cells that express both class II molecules and MAGE-3 are unable to activate CD4+ T cells directly. Upon lysis of some tumor cells by a first CTL attack, tumor debris could be processed by tumor-infiltrating APCs. Activated tumor-specific CD4+ T cells located around the tumor could then be restimulated by these cells. These CD4+ T lymphocytes might then favor the activation and proliferation of the effector CD8+ T cells, provoke the maturation of dendritic cells via CD40–CD40L interactions, and mediate recruitment of additional immune cells at the tumor sites (46–48). The resulting amplification of the immune response might then lead rapidly to the complete destruction of the tumor mass. Immunization with MAGE-3 peptides presented by class I and class II molecules or with a purified protein may represent one possibility for inducing both MAGE-3–specific CD4+ and CD8+ T cells. Another possibility is the reinfusion into patients of autologous APCs loaded with MAGE-3 peptides binding to both HLA class I and class II molecules. This will require the characterization of several epitopes to cope with HLA restriction. Alternatively, autologous dendritic cells could be infected with recombinant viruses encoding an Ii.MAGE-3 fusion protein. Expression of Ii.MAGE-3 in EBV-transformed B cells was shown to result not only in the presentation of class II epitopes, but also in a very efficient presentation of a class I epitope recognized by an anti– MAGE-3.A1 CTL (Corthals, J., and K. Thielemans, manuscript in preparation). Patients could also be immunized directly with recombinant viruses encoding Ii.MAGE-3.
A number of results obtained in mice support the importance of immunization against antigens presented by class II molecules. Direct injection of a recombinant vaccinia encoding a human papilloma virus E7 protein linked to the sorting signal of the lysosomal-associated membrane protein (LAMP-1) was very effective in inducing protective immunity against a challenge with an E7+ class II+ tumor. In contrast, animals injected with a vaccinia coding for the normal E7 protein were not protected (49). In another mouse model, strong protection was achieved against highly aggressive tumor cells lacking MHC class II expression through a single vaccination with a tumor-specific T helper peptide encoded by the Moloney murine leukemia virus (21). In this model, the CD8+ CTLs were helped efficiently by peptide-primed tumor-specific CD4+ T cells.
A clinical trial was recently initiated with a MAGE-3 recombinant protein. In this trial as in others involving recombinant viruses harboring large MAGE sequences, it will be essential to have reliable monitoring of the anti– MAGE-3 CD4+ response. Our observations that CD4+ cells directed against a minor contaminant in the protein batch can easily be activated in vitro have implications for this monitoring. To avoid possible misinterpretation of in vitro assays and of delayed-type hypersensitivity (DTH) assays, a protein produced in another organism or autologous cells expressing the antigen endogenously should be used for these assays. Another possibility, which narrows the analysis to certain epitopes, is the use of a set of relevant peptides that can be used either to select and amplify peptide-specific T cells in vitro or to label directly T cell receptors with soluble HLA tetramers presenting the relevant peptide (50, 51). Multimeric soluble MHC class II molecules, complexed with a peptide attached covalently, were recently shown to bind with appropriate specificity and affinity to mouse-specific T cells (52). This approach requires prior identification of the antigenic peptides, as described in this report. Considering that several MAGE genes share the sequence coding for one of the two MAGE-3.DR13 epitopes reported here, these peptides are relevant to ∼16% of Caucasian melanoma patients, as 20% of Caucasians express HLA-DR13 and ∼80% of metastatic melanomas express MAGE-1, -2, -3, or -6.
Acknowledgments
We thank Ms. N. Krack and Mr. S. Mapp for help in the preparation of this manuscript.
Abbreviations used in this paper
aa
amino acid(s)
Ii
invariant chain(s)
Footnotes
P. Chaux and R. Luiten were supported by a postdoctoral fellowship from the Training and Mobility of Researchers Program of the European Commission. V. Vantomme was partially supported by the Fonds National de la Recherche Scientifique (TELEVIE grants), Brussels, Belgium. This work was partially supported by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming, and by grants from the Association contre le Cancer, Brussels, Belgium, from the BIOMED 2 programme of the European Community, from the Fonds J. Maisin, Belgium, and from the Caisse Générale d'Epargne et de Retraite (CGER)-Assurances and VIVA, Brussels, Belgium.
References
- 1.Anichini A, Fossati G, Parmiani G. Clonal analysis of the cytolytic T-cell response to human tumors. Immunol Today. 1987;8:385–389. doi: 10.1016/0167-5699(87)90215-5. [DOI] [PubMed] [Google Scholar]
- 2.Hérin M, Lemoine C, Weynants P, Vessière F, Van Pel A, Knuth A, Devos R, Boon T. Production of stable cytolytic T-cell clones directed against autologous human melanoma. Int J Cancer. 1987;39:390–396. doi: 10.1002/ijc.2910390320. [DOI] [PubMed] [Google Scholar]
- 3.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora J-P, De Smet C, Brasseur F, van der Bruggen P, Lethé B, Lurquin C, et al. Structure, chromosomal localization and expression of twelve genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 4.Van den Eynde B, van der Bruggen P. T cell defined tumor antigens. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Shichijo S, Noguchi M, Hirohata M, Itoh K. Identification of MAGE-1 and MAGE-4 proteins in spermatogonia and primary spermatocytes of testis. Cancer Res. 1995;55:3478–3482. [PubMed] [Google Scholar]
- 6.Haas GG, Jr, D'Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18:47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 7.Boël P, Wildmann C, Sensi M-L, Brasseur R, Renauld J-C, Coulie P, Boon T, van der Bruggen P. BAGE, a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 8.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas S, De Smet C, Arden KC, Viars CS, Lethé B, Lurquin C, Boon T. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 10.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. LAGE-1, a new gene with tumor specificity. Int J Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Sahin U, Türeci O, Pfreundschuh M. Serological identification of human tumor antigens. Curr Opin Immunol. 1997;9:709–716. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 12.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethé B, Brasseur F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celis E, Tsai V, Crimi C, DeMars R, Wentworth PA, Chesnut RW, Grey HM, Sette A, Serra HM. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc Natl Acad Sci USA. 1994;91:2105–2109. doi: 10.1073/pnas.91.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bruggen P, Bastin J, Gajewski T, Coulie PG, Boël P, De Smet C, Traversari C, Townsend A, Boon T. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur J Immunol. 1994;24:3038–3043. doi: 10.1002/eji.1830241218. [DOI] [PubMed] [Google Scholar]
- 15.Kawashima I, Hudson S, Tsai V, Southwood S, Takesako K, Appella E, Sette A, Celis E. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59:1–14. doi: 10.1016/s0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka F, Fujie T, Tahara K, Mori M, Takesako K, Sette A, Celis E, Akiyoshi T. Induction of antitumor cytotoxic T lymphocytes with a MAGE-3-encoded synthetic peptide presented by human leukocytes antigen-A24. Cancer Res. 1997;57:4465–4468. [PubMed] [Google Scholar]
- 17.Herman J, van der Bruggen P, Luescher I, Mandruzzato S, Romero P, Thonnard J, Fleischhauer K, Boon T, Coulie PG. A peptide encoded by the human gene MAGE-3 and presented by HLA-B44 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Immunogenetics. 1996;43:377–383. doi: 10.1007/BF02199806. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg PD, Kern DE, Cheever MA. Therapy of disseminated murine leukemia with cyclophosphamide and immune Lyt-1+,2−T cells. Tumor eradication does not require participation of cytotoxic T cells. J Exp Med. 1985;161:1122–1134. doi: 10.1084/jem.161.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grohmann U, Bianchi R, Fioretti MC, Fallarino F, Binaglia L, Uyttenhove C, Van Pel A, Boon T, Puccetti P. CD8+ cell activation to a major mastocytoma rejection antigen, P815AB: requirement for tum− or helper peptides in priming for skin test reactivity to a P815AB- related peptide. Eur J Immunol. 1995;25:2797–2802. doi: 10.1002/eji.1830251013. [DOI] [PubMed] [Google Scholar]
- 20.Pulaski BA, Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res. 1998;58:1486–1493. [PubMed] [Google Scholar]
- 21.Ossendorp F, Mengede E, Camps M, Filius R, Melief C. Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med. 1998;187:693–702. doi: 10.1084/jem.187.5.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch GJ, Joosten AM, Kessler JH, Melief CJ, Leeksma OC. Recognition of BCR-ABL positive leukemic blasts by human CD4+ T cells elicited by primary in vitro immunization with a BCR-ABL breakpoint peptide. Blood. 1996;88:3522–3527. [PubMed] [Google Scholar]
- 23.Fossum B, Breivik J, Meling GI, Gedde-Dahl T, III, Hansen T, Knutsen I, Rognum TO, Thorsby E, Gaudernack G. A K-ras 13Gly→ Asp mutation is recognized by HLA-DQ7 restricted T cells in a patient with colorectal cancer. Modifying effect of DQ7 on established cancers harbouring this mutation? . Int J Cancer. 1994;58:506–511. doi: 10.1002/ijc.2910580409. [DOI] [PubMed] [Google Scholar]
- 24.Topalian SL, Gonzales MI, Parkhurst M, Li YF, Southwood S, Sette A, Rosenberg SA, Robbins PF. Melanoma-specific CD4+T cells recognize nonmutated HLA-DR–restricted tyrosinase epitopes. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi H, Kokubo T, Sato K, Kimura S, Asano K, Takahashi H, Iizuka H, Miyokawa N, Katagiri M. CD4+T cells from peripheral blood of a melanoma patient recognize peptides derived from nonmutated tyrosinase. Cancer Res. 1998;58:296–301. [PubMed] [Google Scholar]
- 26.Li K, Adibzadeh M, Halder T, Kalbacher H, Heinzel S, Muller C, Zeuthen J, Pawelec G. Tumour-specific MHC-class-II-restricted responses after in vitro sensitization to synthetic peptides corresponding to gp100 and Annexin II eluted from melanoma cells. Cancer Immunol Immunother. 1998;47:32–38. doi: 10.1007/s002620050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mott JE, Grant RA, Ho Y-S, Platt T. Maximizing gene expression from plasmid vectors containing the λ PLpromoter: strategies for overproducing transcription termination factor ρ. Proc Natl Acad Sci USA. 1985;82:88–92. doi: 10.1073/pnas.82.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riviere I, Brose K, Mulligan RC. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci USA. 1995;92:6733–6737. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham F, Van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;54:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 30.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 31.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 32.Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 33.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Charron D, Dupont B, Erlich HA, Fauchet R, Mach B, et al. Nomenclature for factors of the HLA system, 1996. Tissue Antigens. 1997;49:297–321. doi: 10.1111/j.1399-0039.1997.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 34.Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson S, Frauwirth K, Shastri N. Expression of endogenous peptide-major histocompatibility complex class II complexes derived from invariant chain-antigen fusion proteins. Proc Natl Acad Sci USA. 1995;92:7217–7221. doi: 10.1073/pnas.92.16.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuchtern JG, Biddison WE, Klausner RD. Class II MHC molecules can use the endogenous pathway of antigen presentation. Nature. 1990;343:74–76. doi: 10.1038/343074a0. [DOI] [PubMed] [Google Scholar]
- 37.Geuze HJ. The role of endosomes and lysosomes in MHC class II functioning. Immunol Today. 1998;19:282–287. doi: 10.1016/s0167-5699(98)01269-9. [DOI] [PubMed] [Google Scholar]
- 38.Vijayasaradhi S, Xu Y, Bouchard B, Houghton AN. Intracellular sorting and targeting of melanosomal membrane proteins: identification of signals for sorting of the human brown locus protein, gp75. J Cell Biol. 1995;130:807–820. doi: 10.1083/jcb.130.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topalian SL, Rivoltini L, Mancini M, Markus NR, Robbins PF, Kawakami Y, Rosenberg SA. Human CD4+ T cells specifically recognize a shared melanoma-associated antigen encoded by the tyrosinase gene. Proc Natl Acad Sci USA. 1994;91:9461–9465. doi: 10.1073/pnas.91.20.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y-T, Stockert E, Chen Y, Garin-Chesa P, Rettig WJ, van der Bruggen P, Boon T, Old LJ. Identification of the MAGE-1 gene product by monoclonal and polyclonal antibodies. Proc Natl Acad Sci USA. 1994;91:1004–1008. doi: 10.1073/pnas.91.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurk M, Kremmer E, Schwarz U, Forster R, Winnacker EL. MAGE-11 protein is highly conserved in higher organisms and located predominantly in the nucleus. Int J Cancer. 1998;75:762–766. doi: 10.1002/(sici)1097-0215(19980302)75:5<762::aid-ijc16>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Rovere P, Vallinoto C, Bondanza A, Crosti MC, Rescigno M, Ricciardi-Castagnoli P, Rugarli C, Manfredi AA. Bystander apoptosis triggers dendritic cell maturation and antigen-presenting function. J Immunol. 1998;161:4467–4471. [PubMed] [Google Scholar]
- 43.Hoon DS, Yuzuki D, Hayashida M, Morton DL. Melanoma patients immunized with melanoma cell vaccine induce antibody responses to recombinant MAGE-1 antigen. J Immunol. 1995;154:730–737. [PubMed] [Google Scholar]
- 44.Chen YT, Gure AO, Tsang S, Stockert E, Jager E, Knuth A, Old LJ. Identification of multiple cancer/ testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci USA. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchand M, van Baren N, Weynants P, Brichard V, Dréno B, Tessier M-H, Rankin E, Parmiani G, Arienti F, Humblet Y, et al. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int J Cancer. 1999;80:219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 46.Ridge JP, Di'Rosa F, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature. 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 47.Schoenberger SP, Toes REM, van der Voort EIH, Offringa R, Melief C. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 48.Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 49.Lin KY, Guarnieri FG, Staveley-O'Carroll KF, Levitsky HI, August JT, Pardoll DM, Wu TC. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 50.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 51.Romero P, Rod P, Dunbar, Valmori D, Pittet M, Ogg GS, Rimoldi D, Chen JL, Liénard D, Cerottini JC, Cerundolo V. Ex vivo staining of metastatic lymph nodes by class I major histocompatibility complex tetramers reveals high numbers of antigen-experienced tumor-specific cytolytic T lymphocytes. J Exp Med. 1998;188:1641–1650. doi: 10.1084/jem.188.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]