A Human Histocompatibility Leukocyte Antigen (HLA)-G–specific Receptor Expressed on All Natural Killer Cells (original) (raw)
Abstract
Human natural killer (NK) cells express several killer cell immunoglobulin (Ig)-like receptors (KIRs) that inhibit their cytotoxicity upon recognition of human histocompatibility leukocyte antigen (HLA) class I molecules on target cells. Additional members of the KIR family, including some that deliver activation signals, have unknown ligand specificity and function. One such KIR, denoted KIR2DL4, is structurally divergent from other KIRs in the configuration of its two extracellular Ig domains and of its transmembrane and cytoplasmic domains. Here we show that recombinant soluble KIR2DL4 binds to cells expressing HLA-G but not to cells expressing other HLA class I molecules. Unlike other HLA class I–specific KIRs, which are clonally distributed on NK cells, KIR2DL4 is expressed at the surface of all NK cells. Furthermore, functional transfer of KIR2DL4 into the cell line NK-92 resulted in inhibition of lysis of target cells that express HLA-G, but not target cells that express other class I molecules including HLA-E. Therefore, given that HLA-G expression is restricted to fetal trophoblast cells, KIR2DL4 may provide important signals to maternal NK decidual cells that interact with trophoblast cells at the maternal–fetal interface during pregnancy.
Keywords: natural killer cell, human histocompatibility leukocyte antigen G, killer cell immunoglobulin-like receptor, pregnancy, trophoblast
Human NK cells express two types of receptors that bind to HLA class I molecules on target cells (1). The killer cell Ig-like receptors (KIRs)1 form a family including members that inhibit NK cells upon recognition of specific classical HLA class I molecules, in particular HLA-C and HLA-B. In addition, the lectin-like CD94/NKG2A heterodimer is an inhibitory receptor specific for the nonclassical class I molecule HLA-E (2–4). Another nonclassical HLA class I molecule, HLA-G, has been reported to inhibit NK cells (5–8). However, the protection of HLA-G–expressing target cells from NK-mediated lysis can be explained by the recognition of HLA-E on these cells by the CD94/NKG2A receptor. Cell surface expression of HLA-E depends on the binding of a specific peptide derived from the leader sequence of other class I molecules, including HLA-G (9, 10).
The KIR and NKG2 families include members that activate, rather than inhibit, NK cells (1, 11). These activating receptors, such as KIR2DS and NKG2C, have short cytoplasmic tails that lack immunoreceptor tyrosine-based inhibition motifs (ITIMs). In addition, a lysine residue in their transmembrane domain contributes to their association with DAP12 (12), a homodimer of a 12-kD molecule with a cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM). In contrast, the inhibitory receptors, such as KIR2DL, KIR3DL, and NKG2A, have long cytoplasmic tails with two ITIM sequences. In their case, inhibition is achieved by the recruitment of the tyrosine phosphatase SHP-1 to phosphorylated ITIMs. Expression of the different KIR and NKG2 family members, including activating and inhibitory forms, is clonally distributed among NK cells. Individual NK cells express several such receptors at once, in no particular combination. Besides the few inhibitory KIRs with established specificities for HLA class I, all other KIR family members have poorly defined ligand specificities.
HLA-G is expressed only on fetal trophoblast cells that invade the maternal decidua (13). These invading trophoblast cells are surrounded by maternal stromal cells and NK cells that are abundant in the decidua basalis during early pregnancy. It has been proposed that HLA-G on trophoblast cells interacts with a receptor on NK cells (13). Such an interaction may serve to inhibit NK cytotoxicity, but may also provide signals that result in a positive NK response.
In a search for an HLA-G–specific receptor, soluble forms of several KIRs were produced and tested for binding to transfected cells expressing HLA-G. One of the KIRs of unknown function, called KIR2DL4, is structurally unique among the KIR family members. Complementary DNA clones encoding KIR2DL4 predict a molecule with two Ig domains in a unique D0–D2 configuration (14, 15). Other KIRs have either two or three Ig domains in the order D1–D2 or D0–D1–D2, respectively. The predicted transmembrane region that includes an arginine residue and the long cytoplasmic tail with a single ITIM are also unique. Therefore, KIR2DL4 has features typical of both activating and inhibitory receptors, leaving some uncertainty as to what type of signal it may deliver to NK cells. In this study, we describe expression of KIR2DL4 at the surface of all NK cells and identify it as an HLA-G–specific receptor.
Materials and Methods
Cells and Antibodies.
The NK cell lines NK3.3 (a gift from J. Kornbluth, St. Louis University School of Medicine, St. Louis, MO) and NK-92 (obtained from H.-G. Klingemann, Rush University, Chicago, IL) were cultured as previously described (16, 17). HLA class I transfectants of the 721.221 cell line were obtained from J. Gumperz and P. Parham (Stanford University, Stanford, CA), except for the HLA-G transfectant, which was obtained from L. Lanier (DNAX, Palo Alto, CA). The T leukemic cell line Jurkat and the monocytic cell line HL60 were obtained from the American Type Culture Collection. RMA-S cells transfected with HLA-E (RMA-S-E) cells were obtained from E.H. Weiss and M. Ulbrecht (Institut für Anthropologie and Humangenetik der Universität München, Munich, Germany) and used as previously described (3). NK clones were produced from the peripheral blood of normal donors as previously described (18). Anti-CD3 and anti-CD56 antibodies conjugated to PE were purchased from Becton Dickinson. The HLA-specific mAb B9.12.1, the KIR2DL1-reactive mAb EB6, and isotype-matched control Abs were obtained from Immunotech. Rabbit antisera to the NH2-terminal region of KIR2DL4 (referred to as anti-2DL4) were raised by immunization with the synthetic peptide VGGQDLPFC, and were affinity purified on the same peptide (Research Genetics). The mAb VV1-IG10, specific for the A33 early/intermediate vaccinia protein, was a gift of A. Schmaljohn (U.S. Army, Fort Detrick, Frederick, MD). The MHC class I–specific mAb DX17 was a gift of L. Lanier. The HLA-G–specific mAb (G233) was a gift of A. King (Cambridge University, Cambridge, UK). This mAb does not react with classical HLA molecules and reacts with HLA-G in extravillous trophoblasts (19).
KIR2DL4–Ig Fusion Proteins and Binding Assay.
The KIR2DL4– Ig fusion protein was produced by the same strategy used for other KIR–Ig proteins (20). The extracellular region was PCR amplified from a cDNA clone of KIR2DL4 obtained from A. Selvakumar and B. Dupont (Memorial Sloan Kettering Cancer Center, New York, NY) with the forward primer CAGAGTGTGCTAGCGCACGTGGGTGGTCAGGACAAGCC containing an NheI site, and the reverse primer GAGTACCTAGGATCCGCATGCAGGTGTCTGGCGATACC containing a BamHI site. These PCR fragments were cloned into the expression vector Cd5lneg1 (obtained from B. Seed, Massachusetts General Hospital, Charlestown, MA). SDS-PAGE analysis of the purified recombinant protein identified a species of ∼65 kD under reducing conditions. The binding assay was performed as previously described (20), except that the cells were incubated with goat IgG (50 μg/ml) for 30 min after incubation with the 2DL4–Ig fusion protein and before addition of the FITC-conjugated goat anti–human Fc. Binding was assessed by flow cytometry.
Vaccinia Virus Infection, Cytotoxicity Assays, and Peptide Loading.
cDNAs encoding KIR2DL4 and NKG2A (obtained from J. Houchins, R&D Systems, Minneapolis, MN) were subcloned into the plasmid pSC65 and used to generate recombinant vaccinia viruses as previously described (21). Purified viruses encoding KIR2DL4 or NKG2A were used to infect the human cell line NK-92, as previously described (20). Vaccinia virus infections were monitored by flow cytometry with the mAb VV1-IG10. Infected and uninfected control cells were simultaneously plated for standard 51Cr-release assays and for Ab staining followed by flow cytometry as previously described (20). Peptide loading was done as previously described (3). In brief, 500 μM of the HLA-G signal sequence peptide (VMAPRTFL) was incubated overnight with RMA-S-E cells plated at 5 × 105 cells/ml. Cells were washed and used for antibody staining followed by flow cytometry.
Results and Discussion
Direct Binding of Soluble KIR2DL4 to 721.221 Cells Expressing HLA-G.
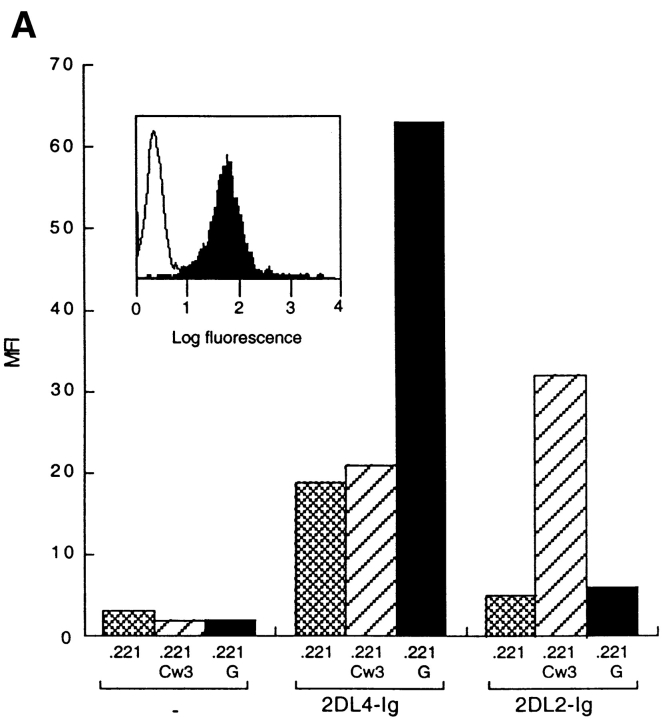
A soluble recombinant protein containing the extracellular portion of KIR2DL4 fused to the Fc region of human IgG1 (KIR2DL4–Ig) was produced in order to search for its ligand. Binding of KIR2DL4–Ig to a panel of HLA-transfected 721.221 cells was analyzed by flow cytometry. KIR2DL4–Ig displayed a uniform binding to all the 721.221 transfectants tested, as well as untransfected 721.221 cells (Fig. 1 A). This HLA class I–independent binding of KIR2DL4 to 721.221 cells may be due to the first Ig domain (D0), as similar results have been reported with soluble KIR3DL1 (22) and KIR3DL2 (23), both of which contain a D0 domain, and with a soluble D0–Ig fusion protein (22). In contrast, KIR2DL4–Ig bound strongly to 721.221 cells expressing HLA-G (221–G). As expected, KIR2DL2–Ig bound to its ligand HLA-Cw3 but not to HLA-G expressed on 721.221 cells (Fig. 1 A). Unlike previous studies describing weak and heterogeneous binding of the similar p49 KIR (15) and the Ig-like transcript (ILT)-2 and ILT-4 members of the ILT inhibitory receptor family expressed mainly on monocytic cells (24, 25), binding of KIR2DL4–Ig to HLA-G was detected by flow cytometry as a bright and uniform peak (Fig. 1 A, inset).
Figure 1.
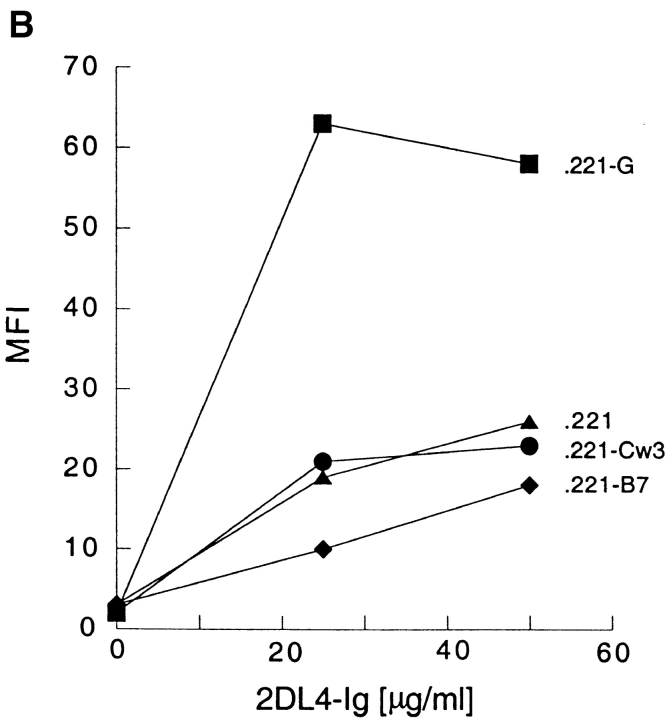
Binding of soluble KIR2DL4–Ig to 221–G cells. (A) 721.221 (221) and 721.221 transfectants expressing HLA-Cw*0304 (221-Cw3) or HLA-G (221-G) were incubated with no fusion protein, 25 μg/ml of 2DL2–Ig, or 25 μg/ml of 2DL4–Ig. The bound fusion proteins were detected by flow cytometry after reaction with FITC-conjugated goat anti– human Fc specific antibodies. Data are expressed as median fluorescence intensity (MFI). Inset, histogram profile (filled) of ungated 221-G cells stained with 2DL4–Ig. Open histogram is control staining with secondary antibodies alone. (B) Binding of KIR2DL4–Ig to 721.221 transfectants was detected as described in A. Cell surface expression of HLA class I on the different cells, as detected by staining with the mAb DX17, was as follows (MFI in parenthesis): 221 (21); 221–B7 (1,731); 221–Cw3 (588); and 221–G (1,540).
The panel of HLA transfectants included HLA-A1, -A2, -B7, -Cw3 and -G, all permissive for HLA-E expression, and HLA-B46, -B51, and -B58, which are not permissive for HLA-E expression (Fig. 1, and data not shown). Thus, it is unlikely that the KIR2DL4–Ig binds to the HLA-E molecules that reach the cell surface upon binding the peptide derived from the HLA-G leader sequence. The comparison of KIR2DL4–Ig binding to 221–B7 and 221–G cells (Fig. 1 B) provides further evidence that KIR2DL4 is not binding to HLA-E. The peptide from the HLA-B7 leader sequence binds HLA-E fivefold better than the HLA-G–derived leader peptide in an in vitro peptide binding assay, resulting in higher HLA-E surface expression (2). Yet despite a very high surface level of HLA-B7 expression on 221–B7 cells, there was no binding of KIR2DL4–Ig to 221–B7 above the HLA-independent binding (Fig. 1 B).
Cell Surface Expression of KIR2DL4 on All NK Cells.
The KIR2DL4 gene is transcribed in every NK cell tested (15, 26), but there is no information on protein expression of KIR2DL4 in NK cells. An antiserum against a peptide corresponding to a unique NH2-terminal sequence of KIR2DL4 was produced to examine cell surface expression of KIR2DL4 on a panel of NK, T, B, and monocyte/macrophage cell lines. The anti-KIR2DL4 (anti-2DL4) antiserum reacted with the NK cell lines NK-92 and NK3.3. (Fig. 2 A). There was negligible staining of cell lines such as the T leukemic line Jurkat, the B cell line 721.221, and the monocytic cell line HL-60. All NK clones in a random panel tested (n = 14) expressed uniformly high cell surface KIR2DL4 (Fig. 2 A). To test whether KIR2DL4 was expressed on all NK cells, CD3−CD56+ cells were isolated from the peripheral blood lymphocytes of four donors. All CD56+ cells (>99%) in these cultures reacted with the anti-2DL4 antiserum (Fig. 2 B). In contrast, only 2% of the CD3+ lymphocytes reacted with the anti-2DL4 antiserum (Fig. 2 B). The proportion of KIR2DL4+ cells within the CD3+ population varied between 1 and 9% among six donors tested. Three-color analysis of the CD3+2DL4+ subpopulation by flow cytometry showed that the majority of these cells also expressed the NK marker CD56 (data not shown).
Figure 2.
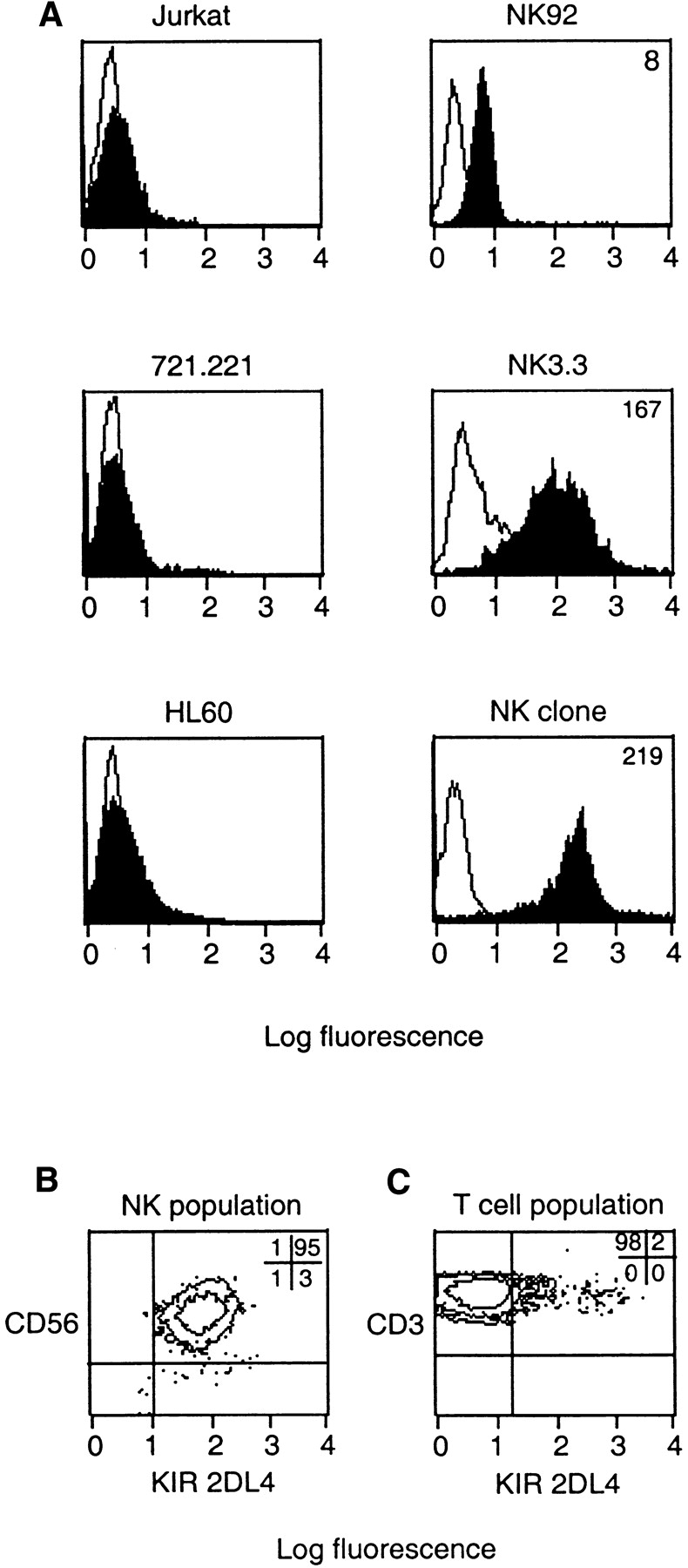
Cell surface expression of KIR2DL4 on NK cells. (A) Flow cytometry profiles of the T leukemia cell line Jurkat, the B lymphoblastoid cell line 721.221, the monocytic cell line HL60 and the NK cell lines NK-92 and NK3.3 as well as a CD3−CD56+CD94− NK clone. Cells were incubated with anti-2DL4 antiserum followed by FITC anti–rabbit IgG. (B) Flow cytometry analysis of CD3−CD56+ NK cells isolated from the peripheral blood of a normal donor and double stained with PE-conjugated CD56 and rabbit anti-2DL4 antiserum followed by FITC-conjugated goat anti–rabbit IgG. The percentages of total cells in each quadrant are listed. (C) Flow cytometry analysis of a primary T cell culture from a normal donor gated on CD3+ cells and double stained for PE-conjugated CD3 and rabbit anti-2DL4 followed by FITC-conjugated goat anti–rabbit IgG. The percentages of total cells in each quadrant are listed.
Functional Transfer of KIR2DL4 in NK-92 Cells.
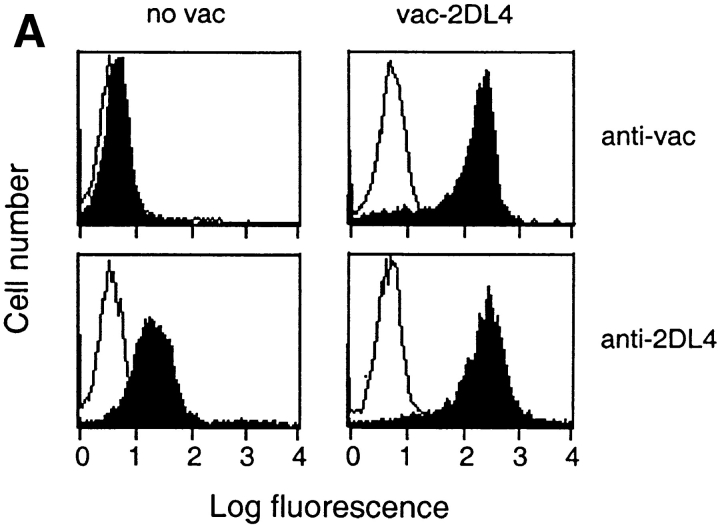
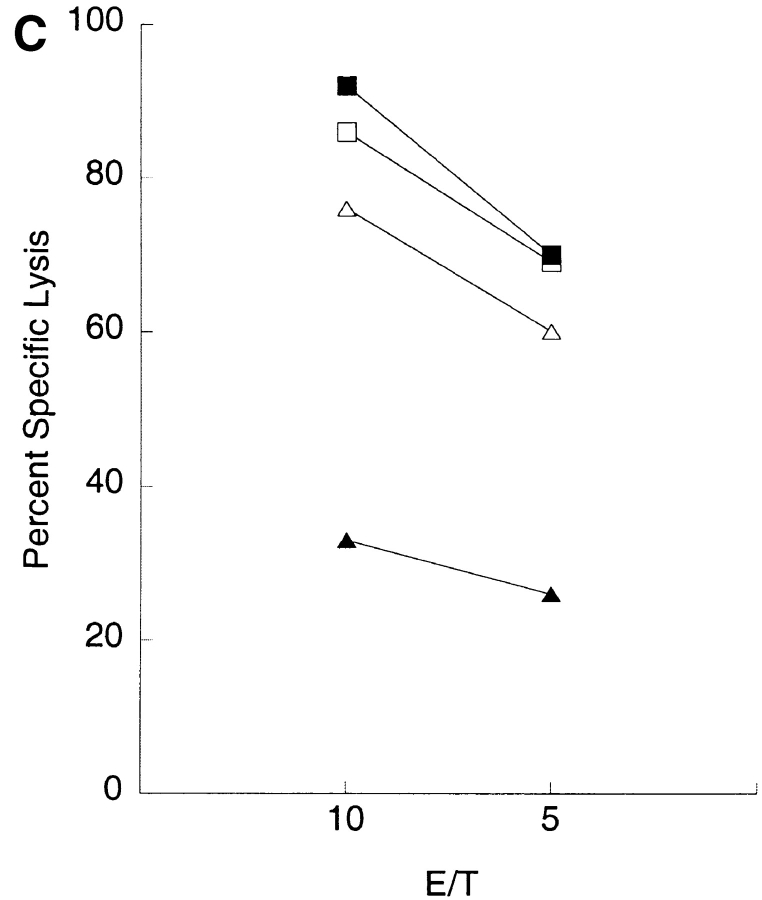
A recombinant vaccinia virus encoding KIR2DL4 (vac-2DL4) was produced for functional transfer experiments in order to confirm the ligand specificity seen in binding studies and to test whether KIR2DL4 can inhibit the lysis of HLA-G– bearing targets. The highly lytic NK tumor cell line NK-92, chosen because it has been used successfully for vaccinia virus-mediated expression (18, 20), was tested for the inhibition of lysis of 721.221 target cells expressing different HLA class I molecules. NK-92 cells express a low endogenous level of KIR2DL4 as determined by flow cytometry (Figs. 2 A and 3 A). In multiple experiments (n = 30), lysis of the HLA-G–expressing target cell 221–G was reduced to 73 ± 19% of the lysis observed with the HLA class I–deficient parental cell 721.221. This low but reproducible inhibition would be consistent with recognition of HLA-G by endogenous KIR2DL4. The level of expression of KIR2DL4 on infected NK-92 (Fig. 3 A) was very similar to that of endogenous KIR2DL4 on NK clones (Fig. 2 A). Vaccinia virus–infected NK-92 cells expressing KIR2DL4 lysed 721.221 cells and 721.221 cells transfected with HLA-Cw4 (Fig. 3 B). In contrast, there was striking inhibition of lysis of 221–G cells. A number of other 721.221 transfectants expressing HLA class I genes, such as HLA-A1, -A2, -B7, -B58, -Cw3, and -Cw7, were not protected from lysis by NK-92 cells expressing KIR2DL4 (Table I, and data not shown). If 221–G cells are protected from lysis through recognition of HLA-G by KIR2DL4, lysis should be restored in the presence of anti–HLA class I antibodies such as DX17 (27). KIR2DL4-expressing NK-92 cells incubated with target cells in the presence of DX17 mAb lysed the 221–G cells to the same extent as untransfected 721.221 cells (Fig. 3 C).
Figure 3.
Functional transfer of KIR2DL4 in NK-92 cells results in HLA-G–mediated inhibition. (A) Flow cytometry analysis of cell surface KIR2DL4 in NK-92 cells either uninfected, or infected with vac-2DL4 (15 PFU/cell). The cells were stained with either anti-2DL4 rabbit antiserum followed by FITC goat anti–rabbit IgG or with the mAb VV1-1G10 specific for a vaccinia cell surface protein. The histograms show log fluorescence of ungated cells. (B) Aliquots of uninfected or vac-2DL4–infected cells in A were tested for their ability to lyse 721.221 cells and 721.221 cells transfected with HLA-Cw*0401 and HLA-G, in a 4-h 51Cr-release assay. Lysis is shown for an E/T ratio of 5. Similar data was obtained at other E/T ratios and in five independent experiments. (C) Aliquots of NK-92 cells infected with vac-2DL4 were tested for their ability to lyse 721.221 (squares) or 221-G cells (triangles) in the presence of 5 μg/ml of the anti-class I mAb DX17 (open symbols) or control IgG1 (filled symbols). Similar results were obtained in two independent experiments.
Table I.
Inhibition of Lysis of 721.221 Cells by NK92 Cells Expressing NKG2A and KIR2DL4
| Transfected HLA class I | Surface level of class I* | Permissive for surface HLA-E‡ | Percentage of specific lysis§ | ||
|---|---|---|---|---|---|
| Uninfected | vac–2DL4 | vac–NKG2A | |||
| None | 10 | − | 101 | 103 | 102 |
| A*0101 | 125 | + | 98 | 88 | 45 |
| B*5801 | 322 | − | 100 | 103 | 100 |
| Cw*0304 | 155 | + | 86 | 83 | 41 |
| Cw*0401 | 114 | + | 87 | 85 | 28 |
| G | 259 | + | 62 | 21 | 0 |
Several HLA class I molecules, including HLA-G, are permissive for the expression of HLA-E (2–4). However, lysis of the HLA-E–permissive 221–Cw4 cells by NK cells expressing KIR2DL4 (Fig. 2) suggests that HLA-E is not recognized by KIR2DL4. To further distinguish between HLA-G– and HLA-E–mediated protection, NK-92 cells expressing either KIR2DL4 or the HLA-E–specific inhibitory receptor CD94/NKG2A were tested against a panel of 721.221 cells transfected with individual HLA class I genes. The endogenous CD94/NKG2A present on NK-92 cells is not sufficient to provide inhibition of lysis upon recognition of HLA-E–expressing target cells (Table I). However, increasing the cell surface expression of NKG2A using a recombinant vaccinia virus (as detected by flow cytometry; data not shown) resulted in inhibition of lysis that correlated with HLA-E expression (Table I). Thus, there was inhibition of lysis of target cells expressing HLA-A1, -Cw3, -Cw4, and -G, but not HLA-B58, an allele not permissive for HLA-E expression (Table I). It is worth noting that the complete inhibition of 221–G target cell lysis by NK-92 infected with vac-NKG2A may reflect the combined inhibition mediated by recognition of HLA-E by CD94/NKG2A and of HLA-G by endogenous KIR2DL4. In contrast, as reported above, NK-92 cells expressing KIR2DL4 were inhibited only by HLA-G–expressing cells and not by cells expressing other HLA class I molecules along with HLA-E. These data show that functional transfer of KIR2DL4 into NK-92 cells conferred specificity for HLA-G, leading to inhibition of target cell lysis.
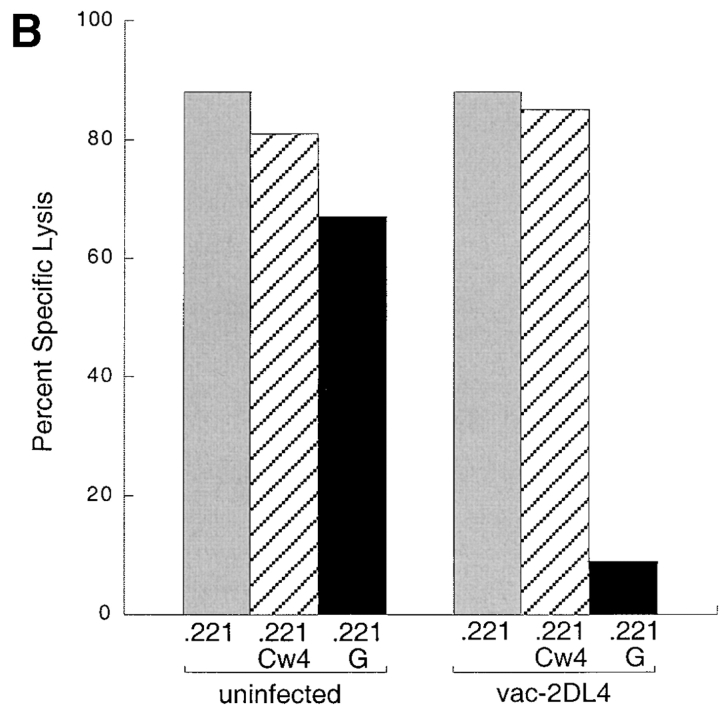
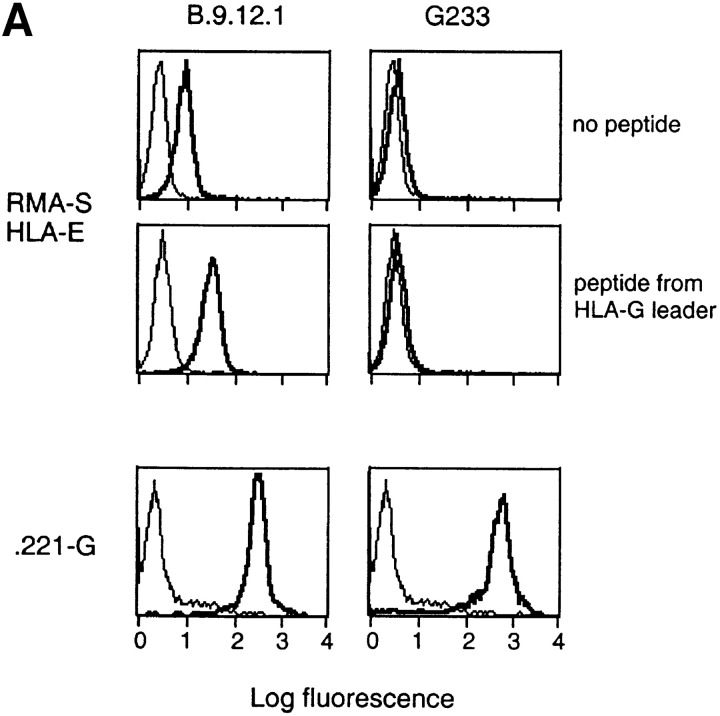
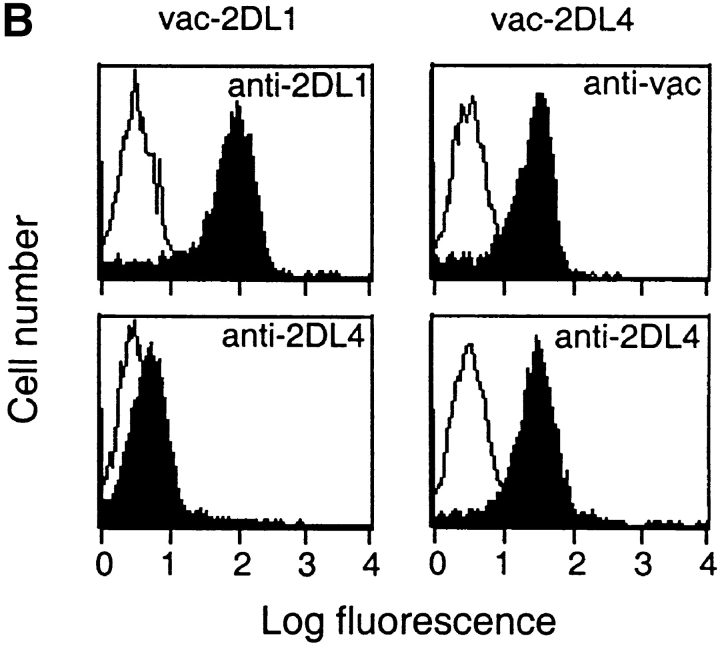
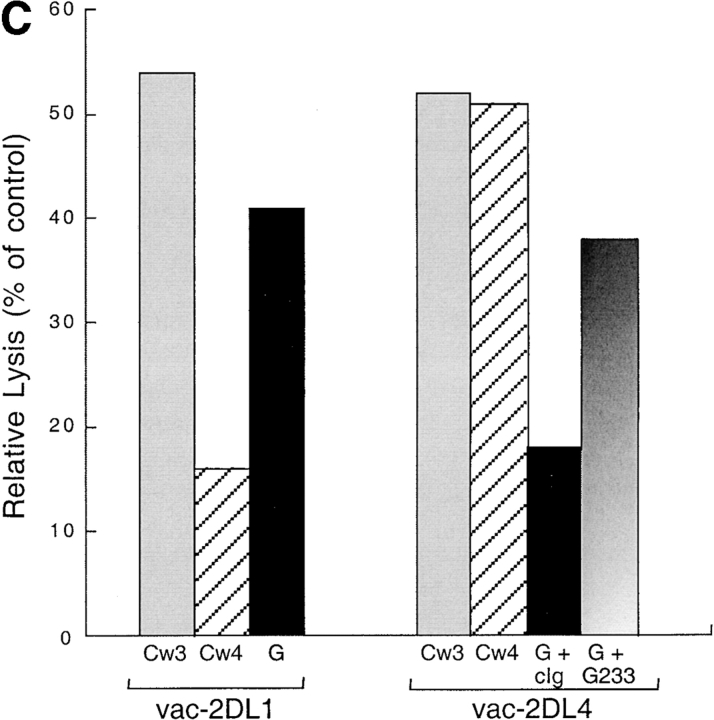
Finally, to obtain independent evidence that HLA-G and not HLA-E is recognized by KIR2DL4, we tested the effect of an HLA-G–specific mAb on the KIR2DL4-mediated inhibition of NK-92. To test whether this mAb may recognize HLA-E in the context of a bound peptide derived from the HLA-G signal peptide, the transporter for antigen presentation (TAP)-deficient mouse RMA-S cell transfected with HLA-E (denoted RMAS-E) was loaded with the HLA-G signal sequence peptide VMAPRTLFL at 26°C. This resulted in increased surface stabilization of HLA-E as detected with the anti-HLA mAb B9.12.1. However, no binding of mAb G233 was detected (Fig. 4 A). Thus, mAb G233 does not recognize HLA-E. NK-92 cells were infected with recombinant vaccinia viruses encoding KIR2DL1 or KIR2DL4, resulting in high surface expression of these receptors (Fig. 4 B). Lysis of 221–G target cells was tested in the presence of G233 mAb or an isotype-matched IgG2a. Only G233 restored the lysis of 221–G cells by KIR2DL4-expressing NK-92 cells to the level observed with KIR2DL1-expressing NK-92 (Fig. 4 C). mAb G233 had no effect on the inhibition of lysis of 221–Cw4 cells by KIR2DL1- expressing NK-92 cells (data not shown).
Figure 4.
KIR2DL4-mediated recognition of HLA-G is reversed using a HLA-G–specific mAb that does not recognize HLA-E. (A) mAb G233 does not recognize HLA-E. RMAS-E cells were loaded with 500 μM of the HLA-G signal sequence–derived peptide VMAPRTLFL at 26°C and stained with either the anti-HLA reactive mAb B9.12.1 or the HLA-G specific mAb G233. The staining of 221-G cells using both antibodies is shown in the bottom panel. (B) Flow cytometry analysis of cell surface KIR2DL1 and KIR2DL4 in NK-92 cells infected with either vac-2DL1 or vac-2DL4. Cells were stained with mAb EB6 (specific for KIR2DL1), mAb VV1-1G10 (anti-vac), or anti-2DL4 rabbit antiserum followed by FITC-labeled species-specific secondary reagents. (C) Aliquots of infected cells expressing KIR2DL1 or KIR2DL4 were tested for their ability to lyse 221-Cw3, 221-Cw4, or 221-G target cells. The interaction between vac-2DL4 expressing NK-92 cells and those expressing 221-G was tested in the presence of either 5 μg/ml of isotype-matched control IgG2a (cIg) or HLA-G–specific mAb G233. Similar results were obtained in three independent experiments.
These data demonstrate that the single ITIM in the context of the KIR2DL4 molecule can deliver an inhibitory signal in NK-92 cells. Experiments to test the possibility that KIR2DL4 may interact with other proteins via the positively charged arginine residue in the transmembrane domain have been hampered by the lack of an anti-KIR2DL4 antibody suitable for immunoprecipitations. It is possible that the inhibitory activity of KIR2DL4, clearly evident in the cell line NK-92, could be modulated in other cells by association with ITAM-bearing molecules such as DAP12 (which associates with KIR2DS, CD94/ NKG2C, and Ly49D/H; references 12, 28, 29) or FcRγ (which associates with ILT-1 and NKR-P1; references 30, 31). In this regard, it is interesting that the cell line NK3.3 and some NK clones are not inhibited by HLA-G despite expression of KIR2DL4 (6, 8, 32, and our unpublished observations).
In conclusion, these results clearly identify KIR2DL4 as a specific receptor for HLA-G, by both direct binding and functional transfer. Previously, the only NK receptor that reproducibly inhibited the lysis of HLA-G–expressing cells by NK cells was the CD94/NKG2A heterodimer (6–8). However, this inhibition can be explained by the binding of CD94/NKG2A to the class Ib molecule HLA-E. Moreover, CD94/NKG2A is not expressed by all NK cells (26, 33), and CD94− NK cells that are inhibited by HLA-G have been reported (6–8), suggesting the existence of yet another NK receptor specific for HLA-G. The ILT2 receptor expressed on monocytes and on a subset of NK cells can also inhibit lysis of target cells expressing HLA-G (34). However, in contrast to KIR2DL4, ILT2 and CD94/ NKG2A are not expressed by all NK cells.
The basis for the immune privilege of the fetus, which is a hemi-allogeneic graft, represents an interesting immunological puzzle. Trophoblast cells do not express HLA-A or HLA-B molecules on their cell surface, a feature thought to confer protection from T cell responses. In contrast, expression of HLA-G on trophoblast cells may result in a functional interaction with KIR2DL4 on maternal decidual NK cells. CD16−CD56bright NK cells constitute the major population of lymphocytes in the decidua (13). The outcome of this interaction in situ remains to be established and might involve the regulation of a number of NK functions such as cytotoxicity, cytokine production, or proliferation. For example, the high expression of HLA-G on cytotrophoblasts may play a role in preventing local activation of maternal NK cells. This may provide a basis for earlier observations showing that cultured fetal trophoblast cells are resistant to lysis by NK cells isolated from either human decidua or peripheral blood (35). Alternatively, recognition of HLA-G by KIR2DL4-expressing NK cells might regulate trophoblast differentiation or invasion into the maternal decidua. Delineating the biological significance of the HLA-G–KIR2DL4 interaction at the maternal–fetal interface will be a step towards resolving the apparent immunological paradox of a successful pregnancy.
Acknowledgments
We thank A. King for useful discussions; M. Weston and C. Winter for expert technical assistance; F. Borrego, J. Gumperz, A. King, H.-G. Klingemann, L. Lanier, P. Parham, and A. Schmaljohn for cell lines and antibodies; and J. Houchins, B. Seed, and A. Selvakumar for cDNAs and plasmids.
Abbreviations used in this paper
ITAM
immunoreceptor tyrosine-based activation motif
ITIM
immunoreceptor tyrosine-based inhibition motif
KIR
killer cell immunoglobulin-like receptor
References
- 1.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 2.Braud VM, Allan DSJ, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 3.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence–derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty DE. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/ NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Villar JJ, Melero I, Navarro F, Carretero M, Bellón T, Llano M, Colonna M, Geraghty DE, López-Botet M. The CD94/NKG2-A inhibitory receptor complex is involved in natural killer cell-mediated recognition of cells expressing HLA-G1. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 7.Soderstrom K, Corliss B, Lanier LL, Phillips JH. CD94/NKG2 is the predominant inhibitory receptor involved in recognition of HLA-G by decidual and peripheral NK cells. J Immunol. 1997;159:1072–1075. [PubMed] [Google Scholar]
- 8.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le Bouteiller P, Moretta L, Moretta A. HLA-G recognition by human natural killer cells. Involvement of CD94 both as inhibitory and as activating receptor complex. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 9.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 10.Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 11.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 12.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 13.Loke YW, King A. Immunology of human placental implantation: clinical implications of our current understanding. Mol Med Today. 1997;3:153–159. doi: 10.1016/s1357-4310(97)01011-3. [DOI] [PubMed] [Google Scholar]
- 14.Selvakumar A, Steffens U, Dupont B. NK cell receptor gene of the KIR family with two Ig domains but highest homology to KIR receptors with three Ig domains. Tissue Antigens. 1996;48:285–295. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 15.Cantoni C, Verdiani S, Falco M, Pessino A, Cilli R, Conte R, Pende D, Ponte M, Mikaelsson MS, Moretta L, Biassoni R. p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol. 1998;28:1980–1990. doi: 10.1002/(SICI)1521-4141(199806)28:06<1980::AID-IMMU1980>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Kornbluth J, Flomenberg N, Dupont B. Cell surface phenotype of a cloned line of human natural killer cells. J Immunol. 1982;129:2831–2837. [PubMed] [Google Scholar]
- 17.Gong JH, Maki G, Klingemann HG. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8:652–658. [PubMed] [Google Scholar]
- 18.Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loke YW, King A, Burrows T, Gardner L, Bowen M, Hiby S, Howlett S, Holmes N, Jacobs D. Evaluation of trophoblast HLA-G antigen with a specific monoclonal antibody. Tissue Antigens. 1997;50:135–146. doi: 10.1111/j.1399-0039.1997.tb02852.x. [DOI] [PubMed] [Google Scholar]
- 20.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 21.Earl, P.L., and B. Moss. 1988. Generation of recombinant vaccinia viruses. In Current Protocols in Molecular Biology. F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, editors. John Wiley & Sons, New York. 16.17.1–16.17.16.
- 22.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 23.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 24.Colonna M, Navarro F, Bellón T, Llano M, García P, Samaridis J, Angman L, Cella M, López-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O'Callaghan CA, Dunbar R, Ogg GS, Cerundolo V, Relink A. Cutting edge: human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 26.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 27.Phillips JH, Chang CW, Mattson J, Gumperz JE, Parham P, Lanier LL. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 28.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 29.Smith KM, Wu J, Bakker ABH, Phillips JH, Lanier LL. Cutting edge: Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 30.Arase N, Arase H, Park SY, Ohno H, Ra C, Saito T. Association with FcRγ is essential for activation signal through NKR-P1 (CD161) in natural killer (NK) cells and NK1.1+T cells. J Exp Med. 1997;186:1957–1963. doi: 10.1084/jem.186.12.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima H, Samaridis J, Angman L, Colonna M. Cutting edge: human myeloid cells express an activating ILT receptor (ILT1) that associates with Fc receptor gamma-chain. J Immunol. 1999;162:5–8. [PubMed] [Google Scholar]
- 32.Mandelboim O, Pazmany L, Davis DM, Valés-Gómez M, Reyburn HT, Rybalov B, Strominger JL. Multiple receptors for HLA-G on human natural killer cells. Proc Natl Acad Sci USA. 1997;94:14666–14670. doi: 10.1073/pnas.94.26.14666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantoni C, Biassoni R, Pende D, Sivori S, Accame L, Pareti L, Semenzato G, Moretta L, Moretta A, Bottino C. The activating form of CD94 receptor complex: CD94 covalently associates with the Kp39 protein that represents the product of the NKG2-C gene. Eur J Immunol. 1998;28:327–338. doi: 10.1002/(SICI)1521-4141(199801)28:01<327::AID-IMMU327>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 34.Navarro F, Llano M, Bellon T, Colonna M, Geraghty DE, López-Botet M. The ILT2 (LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur J Immunol. 1999;29:277–283. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 35.King A, Birkby C, Loke YW. Early human decidual cells exhibit NK activity against the K562 cell line but not against first trimester trophoblast. Cell Immunol. 1989;118:337–344. doi: 10.1016/0008-8749(89)90382-1. [DOI] [PubMed] [Google Scholar]