A Critical Role for the RelA Subunit of Nuclear Factor κB in Regulation of Multiple Immune-response Genes and in Fas-induced Cell Death (original) (raw)
Abstract
Binding sites for the nuclear factor (NF)-κB transcription factor have been identified within control regions of many genes involved in inflammatory and immune responses. Such κB sites are often found adjacent to those of interferon (IFN)-γ–inducible transcription factors, suggesting a requirement for multiple signaling pathways for gene regulation. Using fibroblasts from RelA (p65)-deficient mice generated by gene targeting, we have investigated the role of this subunit of NF-κB in gene activation by microbial lipopolysaccharide, tumor necrosis factor α, and in possible synergism with the IFN-γ–signaling pathway. Our results indicate not only that RelA is required for activation of key genes involved in adaptive (acquired) immune responses, including major histocompatibility complex class I, CD40, and the Fas death receptor, but also that both NF-κB–inducing signals and IFN-γ are necessary for maximal activation. In contrast, neutrophil-specific chemokine genes KC and MIP-2, which can function as nonspecific mediators in innate immune responses, were strongly induced by RelA in the absence of IFN-γ. Our results show that RelA plays a critical role in activation of immune system genes in response to nonspecific stimuli and demonstrate a novel proapoptotic function for this protein in Fas-induced cell death.
Keywords: nuclear factor κB, apoptosis, Fas, gene expression, immune regulation
Innate immune responses are dependent on the presence of receptors that are capable of recognizing highly conserved features present on microorganisms (1, 2). Thus, LPS present in cell walls of gram-negative bacteria is recognized by cell surface receptors present on many different types of cells, and viral products such as double-stranded (ds)RNA molecules can activate intracellular kinases (1, 2). As a result of interaction of host cells with these foreign substances, defense mechanisms may be initiated to neutralize the invading infectious agents. Such innate defense mechanisms have been highly conserved. Thus, both plants and animals possess such mechanisms and in some cases may even use similar mediators (3, 4). In contrast, an adaptive immune response exists only in vertebrates and is dependent on the presence of antigen receptors displayed on specialized cells of the immune system. However, an effective adaptive response in vertebrates generally requires participation of innate response mediators. Signals produced by the innate immune system may provide information about the origin or harmfulness of foreign substances and thus determine the kind of adaptive response that is generated (1, 2).
Microbial LPS and viral dsRNA are potent inducers of nuclear factor (NF)-κB transcription factors, which have been implicated in regulation of host defense mechanisms in diverse species ranging from insects to mammals (3). Although NF-κB can be activated in response to both nonspecific and antigen-specific signals generated during an immune response in vertebrates, only nonspecific signals may be inducers of NF-κB proteins in insects (5, 6), which lack an adaptive immune system. The possible involvement of NF-κB factors in both innate and adaptive responses suggests they may function as an important link between these two systems. NF-κB proteins exist as dimers which typically reside in an inactive form in the cytoplasm complexed with the inhibitory IκB proteins (3, 7). Treatment of cells with inducers of NF-κB results in phosphorylation and degradation of IκB proteins, which allows free NF-κB proteins to translocate to the nucleus and regulate the expression of target genes (7). Activation of NF-κB target genes often occurs within minutes of receiving an inducing signal, making this system optimally responsive to many harmful stimuli, including invading microbes, DNA damaging agents, and oxidative stress (3, 7). The predominant dimeric form of NF-κB in most cells is a complex of a 50-kd protein (p50) and a 65-kd protein called RelA or p65 (7). However, studies on mice deficient in p50 and RelA have revealed distinct functions for these proteins (8, 9) and have identified a requirement for RelA for embryonic survival (9, 10). Gene disruption of other members of the NF-κB family, p52, c-Rel, and RelB, have indicated key roles for these proteins in various tissues (11–14).
Among the best characterized endogenously produced inducers of NF-κB in mammals are the proinflammatory cytokines TNF-α and IL-1 (3). These cytokines can be produced by nonspecific mechanisms such as macrophage activation after phagocytosis or exposure to microbial products such as LPS. In addition, interaction of macrophages with the T cell–derived cytokine IFN-γ can also result in production of TNF-α and IL-1. Activation of NF-κB by these cytokines may activate expression of genes involved in regulation of an inflammatory response (3). Indeed, our previous studies with RelA−/− mice have shown that RelA is required for the activation of GM-CSF and IκB genes in fibroblasts treated with TNF-α (9). Recent studies have also shown that this subunit of NF-κB can inhibit TNF-α–induced cell death, an adaptation that may allow TNF-α–responsive cells to function without induction of cell death (15). Such an antiapoptotic function of RelA is likely the result of activation of genes that inhibit TNF-α–induced cell death.
Many genes containing potential NF-κB binding elements have been identified (7). Interestingly, such genes often contain binding sites for other transcription factors as well, including AP-1, NF-IL6, and members of the IFN-γ–activated factors, signal transducer and activator of transcription (STAT)1 and IFN-regulatory factor (IRF)-1 (16). Thus, transcription factor interactions within control regions may be used for optimal regulation of gene expression, as demonstrated biochemically for the IFN-β gene (17). Of particular interest are synergistic interactions between TNF-α–inducible and IFN-γ–inducible factors. These two cytokines cooperate to activate many target genes, and such cooperation is often manifested in functional interactions between monocytes/macrophages (source of TNF-α) and T lymphocytes (source of IFN-γ) in inflammatory and immune responses.
Although NF-κB factors have been implicated in regulation of many important genes, the relatively ubiquitous nature of these proteins has precluded determination of specific contributions of different subunits in gene activation. One approach to determine the function of individual subunits is gene targeting in embryonic stem cells. Using this approach, we have now analyzed the role of the RelA subunit in activation of genes containing κB-like binding sites in fibroblasts, cells that not only are important for production of chemokines and cytokines but which also function as target cells for cytotoxic T cells and in some cases as APCs (18). We find that RelA activation by LPS or TNF-α and activation of the IFN-γ pathway are both required for activation of key genes involved in adaptive immune responses. In contrast, RelA activation alone is sufficient for potent activation of genes involved in innate responses. Our results indicate a dual role for RelA in regulation of genes involved in both kinds of immune responses and in potentiating adaptive responses by nonspecific stimuli.
Materials and Methods
Cell Culture and Treatments.
Mouse embryonic fibroblasts (MEFs) derived from RelA-deficient mice (9) were cultivated in DMEM supplemented with 10% calf serum and antibiotics. MEFs were stimulated in the presence of 10 ng/ml of TNF-α (Genzyme), 10 ng/ml of IFN-γ (Genzyme), or 10 μg/ml of LPS (Sigma) for the time periods indicated in the figure legends.
Antibodies.
mAb against Fas receptor (Jo2) and PE-conjugated hamster Jo2 were purchased from PharMingen. Jo2 was used at 1 μg/ml for Fas-induced cell killing.
Northern Blot Analysis.
RNA was isolated from cells grown under normal conditions (untreated cells) or stimulated for 6 h with LPS and/or cytokines. Total RNA was extracted using the TRIzol reagent (Molecular Research Center) as recommended by the manufacturer. 10 μg of total RNA was size fractionated on denaturing formaldehyde gels for 4–5 h and transferred overnight to a nylon membrane. Different mRNAs were detected by hybridization to specific 32P probes (reverse transcription PCR products from mouse fibroblast cDNA) in the presence of salmon sperm DNA (Sigma) for 1 h at 68°C. Final washes (twice for 15 min) were performed in 0.2% SSC, 0.1% SDS at 25°C. RNA loading was controlled by normalization to a β-actin probe. The different mRNA levels were quantified by phosphorimaging.
Flow Cytometric Analysis.
Cells were treated with LPS and/or cytokines, harvested, and stained with PE-conjugated hamster anti–murine Fas mAb Jo2 for 30 min on ice. Cells were washed twice, fixed in 4% paraformaldehyde, and analyzed using a Becton Dickinson flow cytometer.
Analysis of Fas-mediated Cell Death.
MEFs were cultured for 12 h in the presence or absence of LPS, TNF-α, and/or IFN-γ. After treatments, cells were washed with fresh medium and anti– mouse Jo2 was added overnight at 1 μg/ml. Cells were collected and counted in the presence of trypan blue. Data are expressed as a percentage of viable cells in the untreated population.
Results and Discussion
The RelA Component of NF-κB Is Critically Required for Activation of Key Genes Involved in Adaptive Immune Responses.
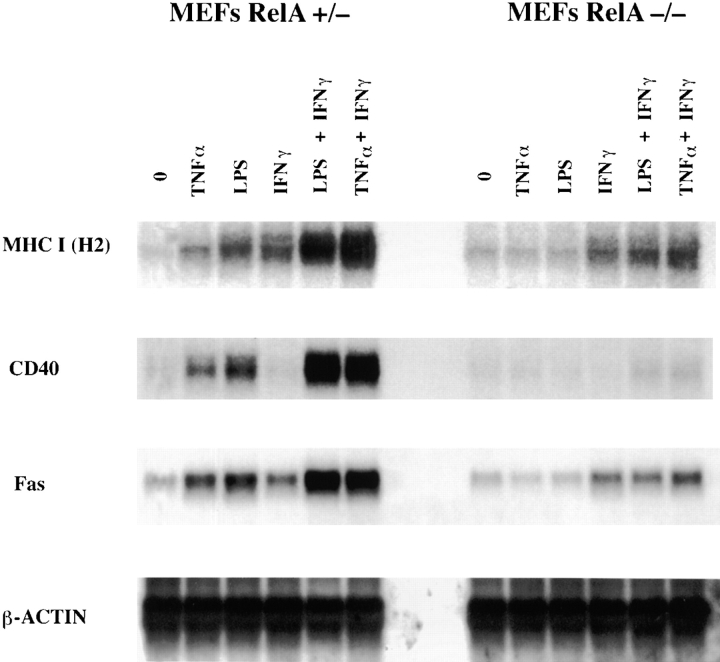
The gene encoding for the MHC class I H-2 molecule can be activated by TNF-α and IFN-γ (19). Although NF-κB sites have been found within transcriptional control regions of this gene (20, 21), the role of NF-κB proteins in its activation is not known. Furthermore, transcription factors that do not belong to the NF-κB family have also been shown to bind specifically to the MHC κB site (22). Therefore, we wished to determine whether RelA participates in activation of this gene. To this end, primary MEFs derived from RelA+/− or RelA−/− mice were first treated with TNF-α or LPS. We have previously shown that LPS is a potent inducer of NF-κB in MEFs, although the effect of such treatment on gene induction was not determined (23). Treatments were carried out for 6 h, since this time period is sufficient for maximal activation of the genes we have studied and does not result in significant death of RelA−/− MEFs by TNF-α. Northern blot analysis of RNA obtained from TNF-α– or LPS-treated RelA+/− MEFs showed a moderate increase in H-2 expression (two- to threefold). However, this increase was significantly potentiated in the presence of IFN-γ (10.5-fold with TNF-α). In contrast, no induction of expression was seen in the RelA−/− cells after TNF-α or LPS treatment, whereas IFN-γ–mediated activation of H-2 expression (threefold) was unaltered in these cells (Fig. 1). These results demonstrate that RelA is required for activation of the H-2 gene after TNF-α or LPS and for potentiation of expression in the presence of IFN-γ.
Figure 1.
The RelA subunit of NF-κB is required for activation of genes involved in adaptive immune responses. RNA was isolated from RelA+/− and RelA−/− MEFs after 6 h of different treatments as indicated. 10 μg of total RNA was hybridized to specific labeled probes for H-2, CD40, Fas, and β-actin.
MHC class I is critical for the initial interaction between T lymphocytes and target cells. We next tested whether other genes functionally important in interactions between T lymphocytes and target cells or APCs are also under RelA control. The CD40 gene can be inducibly expressed on APCs and is important for activation of both T cells which express the CD40 ligand and the APCs (24). Once again, TNF-α or LPS treatment increased expression of CD40 mRNA in RelA+/− MEFs, and the presence of IFN-γ synergistically increased the amount of CD40 mRNA. In contrast, RelA−/− MEFs showed virtually no increase in the amount of CD40 mRNA in the presence of TNF-α or LPS alone or in combination with IFN-γ (Fig. 1).
Next we tested the effect of these inducers on expression of the Fas death receptor. Coupling of Fas expressed on target cells with Fas ligand expressed on T cells generally results in apoptotic demise of the target cell (25). Fas expression is generally low in most cells, although certain cells such as hepatocytes constitutively express high levels of Fas (25). As shown in Fig. 1, Fas expression was dramatically induced by TNF-α plus IFN-γ or LPS plus IFN-γ (∼10-fold), but less strongly by these inducers alone (2–3-fold). Importantly, RelA−/− MEFs showed greatly diminished induction of Fas mRNA (approximately twofold) under the same conditions (Fig. 1). These results provide the first direct proof for a role of NF-κB in regulation of the MHC class I, CD40, and Fas death receptor genes, and suggest that NF-κB may play a key role in activation of genes involved in specific immune responses by nonspecific stimuli such as LPS. Activation of NF-κB by such stimuli may thus allow synchronous expression of multiple genes that carry out similar or complementary functions.
IFN-γ–dependent and –independent Gene Activation by RelA.
IFN-γ is a potent activator of macrophages, especially in combination with LPS. Two genes critically involved in macrophage function that can also be activated in fibroblasts are those encoding the inducible form of nitric oxide synthase (iNOS) and the proinflammatory cytokine IL-6. Therefore, we tested whether activation of these genes was dependent on RelA. As above, both iNOS and IL-6 genes were potently induced by the combined effects of LPS and IFN-γ in RelA+/− MEFs (Fig. 2 A). Interestingly, activation of these genes was considerably less after TNF-α or TNF-α and IFN-γ treatment (Fig. 2 A). Induction was again significantly reduced in RelA−/− MEFs (Fig. 2 A), demonstrating that RelA is specifically required for activation of these genes. Next we tested the potential involvement of RelA in regulation of chemokine gene expression. Chemokines produced at sites of inflammation, often by fibroblasts, play an important role in both initiation and potentiation of an inflammatory response. Thus IFN-inducible protein (IP)-10, a chemokine specific for macrophages and T lymphocytes (26), was also synergistically induced by LPS or TNF-α and IFN-γ in RelA+/− MEFs, and induction was reduced in RelA−/− MEFs (Fig. 2 A). Taken together, our results demonstrate that the RelA component of NF-κB is important for gene induction by LPS and TNF-α alone and for synergistic activation in the presence of IFN-γ.
Figure 2.
IFN-γ–dependent and –independent gene activation by RelA. Northern blot analysis was performed as described in the legend to Fig. 1. (A) IFN-γ–dependent gene activation by NF-κB. The blot was hybridized to labeled probes for iNOS, IL-6, and IP-10. (B) IFN-γ–independent gene activation by NF-κB. The blot was hybridized to labeled probes for KC and MIP-2.
An inflammatory response can occur without the involvement of antigen-specific lymphocytes, often providing the first line of defense against invading pathogens. Key leukocytes involved in such “immediate” responses are neutrophils, which are attracted to infected or inflamed sites by chemokines. Two such neutrophil-specific chemokines, KC (27) and macrophage-inflammatory protein (MIP)-2 (28), were found to be potently induced by LPS in RelA+/− MEFs (and to a lesser degree by TNF-α), whereas induction was significantly reduced in RelA−/− MEFs (Fig. 2 B). Importantly, and in contrast to the examples described above, IFN-γ neither induced expression of these chemokines nor potentiated expression by LPS or TNF-α treatments, in both RelA+/− and RelA−/− MEFs. These results indicate a potentially dual role for RelA in target gene regulation. In the absence of specific immune effectors such as IFN-γ produced by T cells, RelA alone can function as a potent inducer of innate response genes, e.g., neutrophil chemokines. However, IFN-γ is required for optimal induction by RelA of genes involved in an adaptive immune response.
Interestingly, significant differences in induction of specific genes in response to LPS or TNF-α and in synergy with IFN-γ were noticed. For example, although MHC class I, CD40, and Fas were activated to a comparable extent by TNF-α or LPS, the induction of IP-10, KC, and MIP-2 is dramatically more by LPS than by TNF-α. The basis for such differences is presently unclear, but they indicate that different NF-κB inducers may activate distinct heterodimers of RelA-containing complexes, and κB sites in different genes may preferentially bind different heterodimers. Induction of iNOS expression in macrophages requires c-Rel (29), suggesting that both RelA and c-Rel are important for activation of this gene.
Increased Resistance of RelA-deficient MEFs to Fas-induced Cell Death.
We next wished to determine the consequence of RelA-dependent activation of Fas expression on induction of cell death by this receptor. First, we determined whether the observed induction of Fas mRNA correlated with increased surface expression of this molecule after LPS, IFN-γ, or LPS plus IFN-γ treatment of RelA+/− or RelA−/− MEFs. Only LPS was used as the NF-κB activator, since it results in potent induction of Fas mRNA in RelA+/− cells in the presence of IFN-γ and is not cytotoxic to RelA−/− cells. Combined treatment of RelA+/− MEFs with LPS and IFN-γ resulted in an ∼10-fold increase in Fas surface expression, as measured by FACS® analysis, whereas LPS and IFN-γ treatments alone resulted in less significant increases (Fig. 3 A). In contrast, Fas expression was increased only twofold in RelA−/− cells after LPS plus IFN-γ treatment (Fig. 3 B). Thus, potent mRNA induction of Fas results in increased cell surface expression of Fas in RelA+/− but not in RelA−/− MEFs. We have also found that IFN-γ treatment is moderately cytostatic to both RelA+/− and RelA−/− cells (see Fig. 4, A and B, below); in RelA−/− cells, some cytotoxicity through likely production of TNF-α or TNF-β was also noticed after LPS and IFN-γ treatment.
Figure 3.

Cell surface expression of Fas in RelA+/− (A) and RelA−/− MEFs (B). Cells were grown for 12 h in the presence or absence of LPS, IFN-γ, or LPS and IFN-γ, washed, and stained with PE-conjugated hamster anti–murine Fas antibody for analysis of Fas cell surface expression. Unstained cells were used as negative controls.
Figure 4.
RelA−/− MEFs are resistant to Fas-mediated cell death. RelA+/− (A) and RelA−/− (B) MEFs were pretreated for 12 h with LPS, IFN-γ, or LPS and IFN-γ and washed, followed by addition of the anti-Fas antibody Jo2 for 12 h. Cells were collected, and cell viability was determined by trypan blue exclusion.
We then tested whether cross-linking with the Fas-specific Jo2 antibody (30) resulted in death of RelA+/− or RelA−/− fibroblasts. Significant cell death was observed in RelA+/− cells by trypan blue staining when Jo2 was added after LPS or IFN-γ treatment, and combined treatment resulted in even greater killing (85% compared with similar treatments without Jo2; Fig. 4 A). No cell death was observed in the absence of LPS or IFN-γ treatment. In contrast, Fas-induced cell death was significantly reduced in RelA−/− cells after LPS or combined treatment with LPS and IFN-γ (5% compared with similar treatments without Jo2; Fig. 4 B). Our results indicate that the level of Fas expression may be critical in determining whether a cell will undergo apoptosis after ligand binding, and that the RelA component of NF-κB is required for activation of Fas expression and thus for induction of cell death by this receptor. Furthermore, activation of Fas expression appears to be directly mediated by RelA, since consensus NF-κB binding sites have been found in the human Fas promoter (31) and induction of Fas expression was found to be independent of new protein synthesis (data not shown).
Previous studies have demonstrated an antiapoptotic function for RelA in TNF-α and DNA damage–induced cell death pathways (15, 32–34). However, the results presented here indicate that RelA is involved in regulation of both proapoptotic and antiapoptotic genes. Recent studies suggest that Fas expression may be responsible for tissue destruction in mouse autoimmune disease models for experimental autoimmune encephalomyelitis (EAE) and insulin-dependent diabetes (IDD) (35–38). Indeed, elevated Fas expression has been found in β cells of the pancreas (36). Our results indicate that Fas expression may be elevated by cytokines such as IFN-γ, TNF-α, TNF-β, and IL-1 produced by infiltrating T cells and macrophages. A role for NF-κB in regulation of Fas expression suggests that inhibition of this transcription factor may have therapeutic potential for the treatment of autoimmune diseases.
Acknowledgments
We wish to thank P. Bruzzo for technical assistance and Dr. J. Manley for discussion and for comments on this manuscript. We are also indebted to Dr. D. Baltimore, in whose laboratory RelA-deficient mice were generated.
This work was supported by National Institutes of Health grant RO1 CA074982 to A.A. Beg.
References
- 1.Medzhitov R, Janeway CA., Jr Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 4.Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 5.Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, Tatei K, Levine M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. . Cell. 1993;75:753–764. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 6.Ip YT, Levine M. Molecular genetics of Drosophilaimmunity. Curr Opin Genet Dev. 1994;4:672–677. doi: 10.1016/0959-437x(94)90133-n. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle PA, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 8.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 9.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 10.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-κB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caamano JH, Rizzo CA, Durham SK, Barton DS, Raventos-Suarez C, Snapper CM, Bravo R. Nuclear factor (NF)-κB2 (p100/p52) is required for normal splenic microarchitecture and B cell–mediated immune responses. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, Grinberg A, Tran T, Scharton-Kersten T, Anver M, et al. Mice deficient in nuclear factor (NF)-κB/ p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck R-P, Lira SA, Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/ Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 14.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 15.Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 16.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 17.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 18.Kundig TM, Bachmann MF, DiPaolo C, Simard JJ, Battegay M, Lother H, Gessner A, Kuhlcke K, Ohashi PS, Hengartner H, et al. Fibroblasts as efficient antigen-presenting cells in lymphoid organs. Science. 1995;268:1343–1347. doi: 10.1126/science.7761853. [DOI] [PubMed] [Google Scholar]
- 19.Ting JP, Baldwin AS. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin AS, Jr, Sharp PA. Two transcription factors, NF-kappa B and H2TF1, interact with a single regulatory sequence in the class I major histocompatibility complex promoter. Proc Natl Acad Sci USA. 1988;85:723–727. doi: 10.1073/pnas.85.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban MB, Kourilsky P, Baeuerle PA, Israël A. The DNA binding subunit of NF-κB is identical to factor KBF1 and homologous to the reloncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin AS, Jr, LeClair KP, Singh H, Sharp PA. A large protein containing zinc finger domains binds to related sequence elements in the enhancers of the class I major histocompatibility complex and kappa immunoglobulin genes. Mol Cell Biol. 1990;10:1406–1414. doi: 10.1128/mcb.10.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 24.van Kooten C, Banchereau J. Functional role of CD40 and its ligand. Int Arch Allergy Immunol. 1997;113:393–399. doi: 10.1159/000237614. [DOI] [PubMed] [Google Scholar]
- 25.Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 26.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 27.Frevert CW, Huang S, Danaee H, Paulauskis JD, Kobzik L. Functional characterization of the rat chemokine KC and its importance in neutrophil recruitment in a rat model of pulmonary inflammation. J Immunol. 1995;154:335–344. [PubMed] [Google Scholar]
- 28.Driscoll KE, Hassenbein DG, Howard BW, Isfort RJ, Cody D, Tindal MH, Suchanek M, Carter JM. Cloning, expression, and functional characterization of rat MIP-2: a neutrophil chemoattractant and epithelial cell mitogen. J Leukocyte Biol. 1995;58:359–364. doi: 10.1002/jlb.58.3.359. [DOI] [PubMed] [Google Scholar]
- 29.Grigoriadis G, Zhan Y, Grumont RJ, Metcalf D, Handman E, Cheers C, Gerondakis S. The Rel subunit of NF-kappaB-like transcription factors is a positive and negative regulator of macrophage gene expression: distinct roles for Rel in different macrophage populations. EMBO (Eur Mol Biol Organ) J. 1996;15:7099–7107. [PMC free article] [PubMed] [Google Scholar]
- 30.Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 31.Behrmann I, Walczak H, Krammer PH. Structure of the human APO-1 gene. Eur J Immunol. 1994;24:3057–3062. doi: 10.1002/eji.1830241221. [DOI] [PubMed] [Google Scholar]
- 32.Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 33.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 34.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 35.Kang SM, Schneider DB, Lin Z, Hanahan D, Dichek DA, Stock PG, Baekkeskov S. Fas ligand expression in islets of Langerhans does not confer immune privilege and instead targets them for rapid destruction. Nat Med. 1997;3:738–743. doi: 10.1038/nm0797-738. [DOI] [PubMed] [Google Scholar]
- 36.Chervonsky AV, Wang Y, Wong FS, Visintin I, Flavell RA, Janeway CA, Jr, Matis LA. The role of Fas in autoimmune diabetes. Cell. 1997;89:17–24. doi: 10.1016/s0092-8674(00)80178-6. [DOI] [PubMed] [Google Scholar]
- 37.Waldner H, Sobel RA, Howard E, Kuchroo VK. Fas- and FasL-deficient mice are resistant to induction of autoimmune encephalomyelitis. J Immunol. 1997;159:3100–3103. [PubMed] [Google Scholar]
- 38.Benoist C, Mathis D. Cell death mediators in autoimmune diabetes—no shortage of suspects. Cell. 1997;89:1–3. doi: 10.1016/s0092-8674(00)80174-9. [DOI] [PubMed] [Google Scholar]