Premature Expression of T Cell Receptor (Tcr)αβ Suppresses Tcrγδ Gene Rearrangement but Permits Development of γδ Lineage T Cells (original) (raw)
Abstract
The T cell receptor (TCR)γδ and the pre-TCR promote survival and maturation of early thymocyte precursors. Whether these receptors also influence γδ versus αβ lineage determination is less clear. We show here that TCRγδ gene rearrangements are suppressed in TCRαβ transgenic mice when the TCRαβ is expressed early in T cell development. This situation offers the opportunity to examine the outcome of γδ versus αβ T lineage commitment when only the TCRαβ is expressed. We find that precursor thymocytes expressing TCRαβ not only mature in the αβ pathway as expected, but also as CD4−CD8− T cells with properties of γδ lineage cells. In TCRαβ transgenic mice, in which the transgenic receptor is expressed relatively late, TCRγδ rearrangements occur normally such that TCRαβ+CD4−CD8− cells co-express TCRγδ. The results support the notion that TCRαβ can substitute for TCRγδ to permit a γδ lineage choice and maturation in the γδ lineage. The findings could fit a model in which lineage commitment is determined before or independent of TCR gene rearrangement. However, these results could be compatible with a model in which distinct signals bias lineage choice and these signaling differences are not absolute or intrinsic to the specific TCR structure.
Keywords: lineage commitment, TCR transgenic mice, thymus, differentiation, positive selection
Introduction
The thymus is able to generate distinct types of mature T cells that are differentiated for specific TCR recognition and effector functions. Early in development, precursor thymocytes rearrange and express the genes encoding TCRs and mature as either αβ or γδ lineage T cells (for reviews, see references 1, 2). The first T cells are γδ lineages that arise only in the fetal thymus. Each of these bears a unique, canonical TCR and colonizes distinct epithelial tissues of the periphery. The γδ T cells that populate the lymphoid organs have more diverse receptors and develop in both the fetal and adult thymus. Lymphoid γδ T cells and precursors to the αβ T cell lineage (bearing the pre-TCR) appear roughly around the same time in the adult thymus and are thought to derive from a common CD4−CD8− precursor. The productive rearrangement and expression of the TCRγδ or of the pre-TCR (a heterodimer of TCRβ with invariant pTα) is critical for survival and further differentiation of these early thymocytes 3. Of major interest is whether these receptors play a role in αβ versus γδ lineage determination or only in the progression of already committed precursors 4 5.
The pathways of αβ and γδ T cell development are quite distinct. Although discrete stages of γδ development have not been identified, most γδ lineage T cells never express the CD4 or CD8αβ coreceptors and have no requirement for MHC for maturation 6 7. In contrast, precursor CD4−CD8− thymocytes expressing the pre-TCR proliferate, upregulate TCRα rearrangement, and progress to a CD4+CD8+ intermediate stage 3. If rearrangement of TCRα is productive, TCRα replaces pTα to form the mature TCRαβ. Recognition of MHC by TCRαβ is required for the development of mature αβ lineage T cells, expressing either CD4 or CD8. The development of an additional subset of αβ T cells, the so-called NK T cells, is β2-microglobulin (β2m) dependent 8 9. This minor population of T cells expresses either CD4 or no coreceptor, a restricted TCR repertoire, and is not detected until after birth. Although the lineage relationship of NK T cells to conventional αβ T cells is somewhat controversial, NK T cells have characteristic phenotypic and functional properties that clearly distinguish them from other T cell subsets 9.
With the advent of TCRαβ transgenic mice, a novel population of TCRαβ+CD4−CD8− (TCRαβDN) T cells was observed 10 11 12 13. These cells appear early in the fetal thymus, colonize both epithelial and lymphoid tissues, and are especially prominent in TCRαβ transgenic mice undergoing strong negative selection. Naturally, questions arose as to their origin and lineage relationship to other T cells. There was speculation that these cells could be related to the TCRαβ1CD4−CD8− cells of wild-type mice (NK T cells) or to the abnormal TCRαβ1CD4−CD8− cells observed in lpr mutant mice 14. Others suggested that they derive from conventional αβ T cells after the downregulation of CD4 or CD8 14 15 or that they mature in the αβ lineage without ever expressing the CD4/CD8 coreceptors 16.
Evidence that the TCRαβDN T cells mature in a lineage separate from conventional αβ T cells came from studies of transgenic HY TCR mice. In contrast to the CD8 T cells of these mice, the TCRαβDN cells do not express endogenous TCRα genes, their TCRδ gene segments are not deleted 17, and they do not develop in mice deficient for the common cytokine receptor γ chain 18. TCRαβDN cells mature in the absence of the selecting MHC and, most noteworthy, in HY TCR mice with a pTα null mutation (pTα−/−), a few TCRαβDN cells coexpress endogenous TCRγδ and the transgenic TCRαβ 17. Given these characteristics, it was proposed that TCRαβDN cells of TCRαβ transgenic mice belong to the γδ lineage. In this model, the transgenic TCRαβ replaces TCRγδ while still allowing γδ lineage development. This model was contested, however, in an additional report using DO11.10 TCR transgenic mice 16. Since TCRαβDN cells required specific MHC for development, the authors hypothesized that these cells were αβ lineage T cells that mature without passing through the CD4+CD8+ intermediate stage of development.
In previous studies, there was only limited characterization of TCRαβDN cells of TCRαβ transgenic mice, making it difficult to determine their relationship to conventional T cell subsets. As no single marker can distinguish γδ lineage T cells (with the exception of the TCR itself), we examined TCRαβDN cells using a number of criteria (phenotype, function, development, and localization). An analysis of several strains of TCRαβ transgenic mice reveals that TCRαβDN cells clearly exhibit characteristics of γδ lineage T cells. The MHC requirements for maturation and the regulation of TCR gene rearrangement are distinctly different in TCRαβDN cells than in conventional αβ lineage T cells. The results indicate that the premature expression of TCRαβ allows thymocyte precursors to mature in the γδ lineage. These findings have implications for models of γδ/αβ lineage determination.
Materials and Methods
Mice.
C57BL/6 (B6), C57BL/10 (B10), B10.A, B10.Q, and B10.D2 mice were obtained from a National Institutes of Allergy and Infectious Diseases contract to Taconic Farms, Inc., and B10.BR and BALB/c, from The Jackson Laboratory. TCRαβ transgenic mice were backcrossed, intercrossed, and selected as described previously 19 to obtain H-2b, H-2k, H-2d, H-2q, H-2b recombination activating gene (RAG)-2−/−, H-2q RAG-2−/−, or H-2b MHC class II+/−CD4+/− AND TCR mice 20 21 22 23; H-2d and H-2b class II−/− DO11.10 TCR mice 24; and H-2d HA TCR mice 25. H-2b and H-2d HY TCR mice 26 were obtained by backcrossing 12 times to B10 and then to B10.D2; H-2b and H-2k 5CC7 TCR mice, by crossing B6 5CC7 TCR mice 27 to B10 or B10.A; and H-2b and H-2b class I−/− P14 TCR mice 28, by backcrossing 10 times to B6 and then to β2m−/− 29. Except where noted, all TCRαβ transgenic mice were on the positive-selecting MHC background: AND TCR (H-2b or H-2k), 5CC7 TCR (H-2k), DO11.10 TCR (H-2d), HY TCR (H-2b), and P14 TCR (H-2b). TCRγδ transgenic mice included the G8 TCR mice (H-2b β2m−/−) crossed and selected as described 7, or H-2b TG78 TCR mice 30, backcrossed eight times to B6.
Fetal mice were obtained from timed matings. The day of finding a vaginal plug was designated as day 0 of embryonic development. Mice were bred and maintained in a National Institutes of Allergy and Infectious Diseases Research Animal Facility or on a National Institutes of Allergy and Infectious Diseases contract to Taconic Farms, Inc., according to American Association of Accreditation of Laboratory Animal Care specifications. All protocols for animal studies were approved by the National Institutes of Allergy and Infectious Diseases Animal Care and Use Committee.
Cell Preparation, Antibodies, and Flow Cytometry.
Cultured cell lines used for these studies included: DN7.3 (TCRVγ2/Vδ5), a mouse CD4−CD8− T cell/BW5147 hybridoma, and DCEK, a mouse L cell fibroblast line transfected with EαEbk. Thymocytes, LNs, and LN T cells were prepared in single cell suspensions as described previously 31. For enrichment of heat stable antigen (HSA)lo (CD24lo) thymocytes, a culture supernatant of anti-HSA (J11d) antibody was used with a 1:10 dilution of Lo-Tox–M rabbit complement (Cedarlane) and DNase (106 U/ml; Calbiochem). For magnetic bead isolation of CD4−CD8− thymocytes or LN T cells, 107 cells were reacted with 250 μl of purified H129.19 and 53-6.7 (and RA3-6B2, for LN T cells) antibodies (30 min, 4°C). CD4+CD8+ cells were removed by treatment with sheep anti–rat IgG-coated magnetic beads (30 min, 4°C) at a 5:1 bead to cell ratio, using an MPC-1 magnetic particle concentrator (Dynal). This process was repeated at a 10:1 bead to cell ratio. Epidermal lymphocytes were isolated and prepared in a single cell suspension as described 32. Trypsinized surface antigens were resynthesized in overnight culture with 20 U/ml recombinant IL-2 (Genzyme). To enrich for viable cells, harvested cells were incubated with biotin-labeled goat anti–hamster IgG (Caltag) (30 min, 4°C), washed twice, and bound to streptavidin-coated magnetic beads (Miltenyi Biotech) (30 min, 4°C). Cells were passed over a MACS column (Miltenyi Biotech) and the nonadherent fraction was collected.
Antibodies and staining reagents included: anti–TCRβ–FITC, –PE or –allophycocyanin (APC) (H57-597), anti–γδ TCR-FITC, -PE, or unlabeled (GL3), anti-CD4–FITC, –PE, –APC, or –CyChrome (RM4-5), anti-CD8α–CyChrome or unlabeled (53-6.7), anti-CD8β.2–FITC (53-5.8), anti–IL-2Rβ–FITC (TM-β1), anti-NK1.1–PE (PK136), anti-CD5–FITC (53-7.3), anti-Vα11 TCR–FITC or unlabeled (RR8-1), anti-Vα2–FITC or –PE (B20.1), anti-Ab–FITC or –PE (AF6-120.1), anti-Ek–FITC (14-445), anti–H-2Kd–FITC (SF1-1.1), anti–H-2Kk–FITC (36-75), anti–H-2Kb–FITC (AF6-88.5), anti–H-2Kq–FITC (KH-114), and anti-CD45R/B220–FITC, –PE, or unlabeled (RA3-6B2), all obtained from BD PharMingen; anti-CD8α–FITC, –PE, or –biotin (CT-CD8a), anti-CD4–biotin (YTS 191.1), Thy 1.2–FITC or –PE (5a-8), streptavidin-APC or -TriColor, goat anti–mouse IgG1–PE, goat anti–mouse IgG2a–FITC, all obtained from Caltag; goat anti–rat IgG–FITC (Kirkegaard & Perry); rat anti–mouse IgG1–FITC and streptavidin-FITC (Zymed Laboratories); and anti-CD24 (J11d), anti-HY TCR (T3.70), anti-HA TCR (6.5), and anti-DO11.10 TCR (KJ-126) culture supernatants.
Cells were stained and analyzed by flow cytometry and/or electronically sorted using standard protocols 33. For some analyses, cells were pretreated with an unlabeled anti-FcRγ culture supernatant (24G2) to block Fc receptor binding of the labeled antibodies. Multicolor flow cytometry was performed on a FACS® 440, FACSCalibur™, FACStarPlus™, or FACS Vantage™ (Becton Dickinson). Dead cells were excluded by light scatter and propidium iodide gating. 150,000 events were collected for three- and four-color analyses. For live-gated samples, 10,000–20,000 CD4−CD8− events were collected. Isolation of thymocyte and LN T cell subsets by electronic cell sorting was performed on a FACStarPlus™ (Becton Dickinson) or an EPICS 753 (Beckman Coulter).
For typing of transgenic or mutant mice, peripheral blood lymphocytes were stained with labeled antibody to the appropriate surface antigen, counterstained with Thy1.2 or B220 (used for live gating for T or B cells, respectively). After staining, samples were depleted of red blood cells with ACK lysing buffer (pH 7.4) and analyzed by flow cytometry.
In Vitro TCR Stimulation for Proliferation, Induction of CD8αα Expression, and IL-4 Secretion.
For TCR stimulation, cells were added to U-bottomed 96-well plates coated with anti-TCR antibodies as described 31. Proliferation was determined on day 3 of culture, measuring [3H]thymidine incorporation (1 μCi/ml pulse for 18 h). Coexpression of CD8α and CD8β was assessed on day 4 of culture by flow cytometry. IL-4 production was assayed by specific ELISA 34 using 100 μl of supernatant collected at day 3 of culture and stored at −20°C.
Radiation Bone Marrow Chimeras.
Bone marrow chimeras were made as described by reconstituting irradiated recipients (1,000 rads, Cs source) with T-depleted bone marrow 19. For the cyclosporine A (CsA) experiments, reconstituted mice received daily intraperitoneal injections of 0.4 or 0.6 mg Sandimmune™ CsA (Sandoz) in 100 μl olive oil (Bertolli Classico) or of 100 μl olive oil only, starting on day 3 after reconstitution.
Quantitative PCR.
T cell subsets were isolated by electronic cell sorting. 105 sorted cells were digested using 1× PCR Buffer (PerkinElmer), 2.5 mM MgCl2, 20 mg/ml proteinase K, 0.05% Tween 20, and 20% InstaGene Matrix (Bio-Rad Laboratories) at 56°C (2 h), followed by boiling (10 min). PCR was performed using a reaction mixture containing 1× PCR Buffer (PerkinElmer), 2.5 mM MgCl2, 200 μM each dNTP, 12.5 pmol each primer, and 0.25 U native Taq polymerase (PerkinElmer) bound to anti-Taq (CLONTECH Laboratories, Inc.). The total reaction volume was 50 μl with 5 μl of DNA. Samples were incubated at 95°C (5 min); amplified for 40 cycles at 94°C (30 s), 56°C (1 min), and 72°C (1.5 min), and incubated at 72°C (10 min) using a 96-well plate in a PTC-100 thermocycler (MJ Research, Inc.). Aliquots of 5 μl were removed every three cycles beginning at cycle 18.
The following primers and probes were used: Vγ2, TGTCCTTGCAACCCCTACCC; Jγ1, TGTTCCTTCTGCAAATACCTTG; Vγ2 probe, GAGGAAGAAGACGAAGCTATC; 5′ Cγ1, TTACAGACAAAAGGCTTGAGTC; 3′ Cγ1, GTTCTCATGTTTGACAATACATCTG; and Cγ1 probe, CTGAAGACTAACGACACATAC.
Quantitation was performed using a modified ELISA as described 35. In brief, one primer for each gene was labeled with a 5′ biotin moiety allowing capture of the PCR product on an avidin-coated plate. The second strand was denatured with 0.1 M NaOH and an FITC-labeled probe was bound to the captured strand. Bound probe was detected with an anti-FITC labeled with alkaline phosphatase in the presence of substrate, CSPD (Tropix). Chemiluminescence was measured using a luminometer (Dynatech).
To estimate the relative frequency of Vγ2–Jγ1 rearrangements in the experimental populations, a standard curve generated by titrating DN7.3 cells (containing three Vγ2–Jγ1 rearrangements per cell 36) with DCEK fibroblast cells and amplifying the serially diluted samples in the same PCR. For each sample of 105 cells, a PCR ELISA was performed and the quantity of PCR product (in light units) was determined as a function of cycle number (18–39 cycles). Primers and probes specific for Cγ1 were used to normalize the amount of DNA present. Data from the luminometer were fit to a logistic equation, and the parameters were used to calculate the cycle value at half-maximum (C 50) of amplification 35. C 50 values were plotted against the corresponding log10 cell number of DN7.3 cells in each input sample and a best-fit line was generated. C 50 values for experimental samples obtained in the same assay could be matched to this best-fit line to estimate the relative frequency of Vγ2–Jγ1 rearrangements.
Results
CD4−CD8− T Cells of TCRαβ Transgenic Mice Have Properties of γδ Lineage T Cells.
Wild-type mice bear two CD4−CD8− subpopulations of mature T cells, one bearing TCRαβ (referred to as NK T cells) and the other, TCRγδ. In contrast, an analysis for TCR on CD4−CD8− T cells of HY TCR (TCRαβ) transgenic mice reveals no TCRγδ+ and a larger than usual population of TCRαβ+ cells 37. Also in contrast to CD4 or CD8 αβ lineage T cells, the CD4−CD8− T cells of HY TCR and 2B4 TCR transgenic mice express only the transgenic TCRα and no endogenous TCRα 17 38. Because of these unusual features, we further characterized the TCRαβDN subset of AND TCR and other TCR transgenic mice to assess lineage properties relative to normal T cell subsets.
TCRαβDN cells were analyzed for phenotype and function and compared with the NK T, γδ T, and the major CD4 and CD8 αβ T cell subsets of wild-type mice, as well as CD4−CD8−TCRγδ+ cells of TCRγδ transgenic mice (TG78) 30. As shown previously 8, freshly isolated NK T cells of B6 mice (TCRαβDN) express IL-2R (CD122) and NK 1.1, and produce high amounts of IL-4 in response to in vitro TCR stimulation (Fig. 1A and Fig. B, and Fig. 2). In contrast, the TCRαβDN population of AND TCR mice expresses lower levels of these markers and produces no IL-4 (39; Fig. 1A and Fig. B, and Fig. 2). The TCRαβDN cells also express relatively lower levels of CD5, delineating this subset from mature CD4 and CD8 T cells, but not from TCRγδ+ T cells (Fig. 1 C). Together these phenotypic and functional properties distinguish TCRαβDN cells from NK T and the major αβ lineage, but not from γδ lineage T cells.
Figure 1.
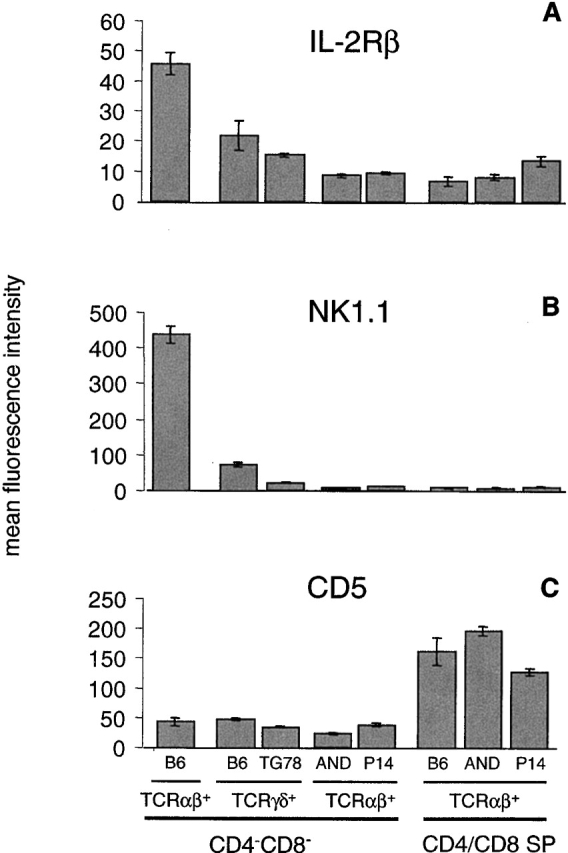
Phenotypic markers distinguish the TCRαβDN T cell subset of TCRαβ transgenic mice from αβ lineage T cells (CD4 and CD8), NK T, but not from γδ lineage T cells. Thymocytes (A and B) or B cell–depleted lymph node T cells (C) from B6, transgenic TCRγδ (TG78), or transgenic TCRαβ (AND and P14) mice were each stained for TCRαβ, TCRγδ, CD4 and/or CD8, and a fifth marker (IL-2Rβ, NK1.1, or CD5), and analyzed by flow cytometry. The mean fluorescence intensity for the specified markers was determined for CD4/CD8 single positive (SP) T cells by software gating for CD4+CD8−TCRαβhi and CD4−CD8+TCRαβhi, or for CD4−CD8− T cells, by live gating for CD4−CD8− followed by software gating for TCRαβ+ or TCRγδ+. Each bar represents the means (with SE bars) collected from analysis of three individual mice with the exception of B6 (two mice).
Figure 2.
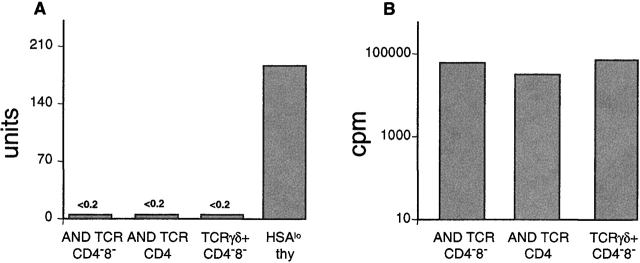
TCRαβDN T cells respond to anti-TCR stimulation by proliferating but do not produce an IL-4 response. Lymph node CD4−CD8− or CD4+ T cells from AND TCR (Vα11/Vβ3), CD4−8−TCRγδ+ T cells from G8 TCR mice (purified using magnetic beads and electronic cell sorting), or HSAlo B6 thymocytes (an enriched source of NK T cells), plated at 5 × 104 cells/well, were stimulated with 10 μg/ml immobilized anti-TCR antibody (anti-Vα11 for AND TCR, anti-TCRγδ for G8 TCR, and anti-TCRαβ for B6 TCR) and assayed for (A) IL-4 production where 1 unit = 0.5 pg of IL-4, and for (B) proliferation. Data are representative of two experiments, averaging values from triplicate wells, and are derived from dose–response curves using 0.1–100 μg/ml of antibody.
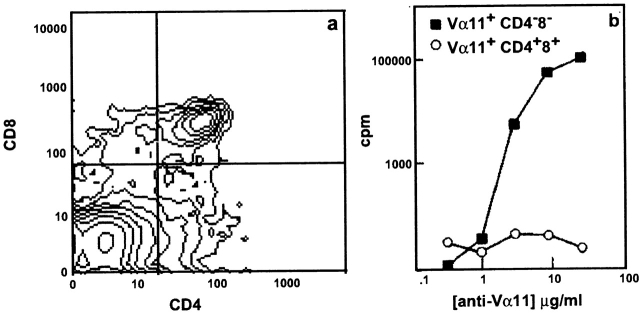
γδ T cells appear early in adult T cell development, before the αβ lineage T cells 40 41 42. To assess when the TCRαβDN cells arise in thymic development, we generated hematopoietic stem cell chimeras using bone marrow from AND TCR mice to reconstitute irradiated recipients. Between days 10 and 15 after reconstitution, we observed a population of Vα11+CD4−CD8− followed by Vα11+ CD4+CD8+ thymocytes. By days 18–20, mature CD4+ CD8− thymocytes develop (data not shown). On day 15, transgenic TCR+ (Vα11) CD4−CD8−, and CD4+CD8+ thymocytes were sorted and stimulated in vitro using anti-Vα11 antibody (Fig. 3, a and b). The Vα11+CD4−CD8− thymocytes are competent to incorporate [3H]thymidine in response to anti-Vα11 cross-linking while Vα11-bearing CD4+CD8+ thymocytes are not. Therefore, like γδ T cells, TCRαβDN T cells appear well before the CD4+ CD8− thymocytes and much earlier than NK T cells that arise after the CD4+CD8− and CD4−CD8+ thymocytes in wild-type mice 8.
Figure 3.
TCRαβDN cells appear early in thymic development. Thymocytes were harvested day 15 after reconstituting B10.BR or B10.A RAG-2−/− irradiated recipients with T-depleted H-2k AND TCR bone marrow. (a) Cells were stained for CD4, CD8, and Vα11 TCR and analyzed by three-color flow cytometry. (b) Thymocytes were electronically sorted for Vα11+CD4+CD8+ and Vα11+CD4−CD8−. Sorted cells (12 × 104/well) were tested in a proliferation assay for response to plate-bound anti-Vα11 (RR8-1) antibody. Proliferation data are representative of two sorting experiments, and the cytometric analysis on day 15 is representative from several series of analyses performed on thymocytes from chimeric mice on days 10–20 after reconstitution.
Previous studies 43 44 45 have indicated that the development of the major αβ T cell subsets (CD4/CD8), as well as the minor TCRαβ+CD4−CD8− (NK T) subset of wild-type mice, are inhibited by CsA. γδ lineage T cells are relatively less sensitive. To further assess lineage properties, TCRαβDN cells of AND TCR mice were tested for sensitivity to CsA, administered over the course of adult T cell development. Irradiated recipients reconstituted with AND TCR bone marrow were treated daily with CsA for 5 wk, after reconstitution. As shown in Table , the development of Vα11+CD4−CD8− TCR+ is up to 75-fold less sensitive to CsA than are Vα11+CD4+CD8− thymocytes. These data indicate that TCRαβ+DN cells are relatively resistant to CsA administered during development, as are γδ lineage T cells.
Table 1.
Effect of CsA on Developing Thymocyte Subsets
| No. of thymocytes | ||||
|---|---|---|---|---|
| Treatment | Total | CD4−CD8− | CD4+CD8+ | CD4+CD8− |
| Oil | 3.1 × 106 | 2.25 × 106 | 3.8 × 105 | 3.2 × 105 |
| 0.4 mg CsA/oil | 1.4 × 106 | 1.1 × 106 | 1.7 × 105 | 3.2 × 104 |
| Fold reduction | 2× | 2× | 2× | 10× |
| Oil | 16.2 × 106 | 5.2 × 106 | 2.9 × 106 | 7.6 × 106 |
| 0.6 mg CsA/oil | 1.5 × 106 | 1.1 × 106 | 2.6 × 105 | 2.0 × 104 |
| Fold reduction | 11× | 5× | 11× | 380× |
Since CD4−CD8− TCRγδ+ thymocytes of wild-type mice can be induced to express CD8αα after in vitro activation (46; Fig. 4 c), we tested the ability of mature TCRαβDN cells to make this response. As shown in Fig. 4, a and b, Vα11+CD4−CD8−, but not Vα11+CD4+ T cells, are induced to express CD8αα in response to anti-TCR stimulation. Similar responses have been obtained from TCRαβDN splenocytes of TCRα transgenic mice 39. Thus, by all of the criteria we examined, TCRαβDN cells are clearly distinguished from conventional αβ lineage and NK T cells, and most resemble γδ lineage T cells.
Figure 4.
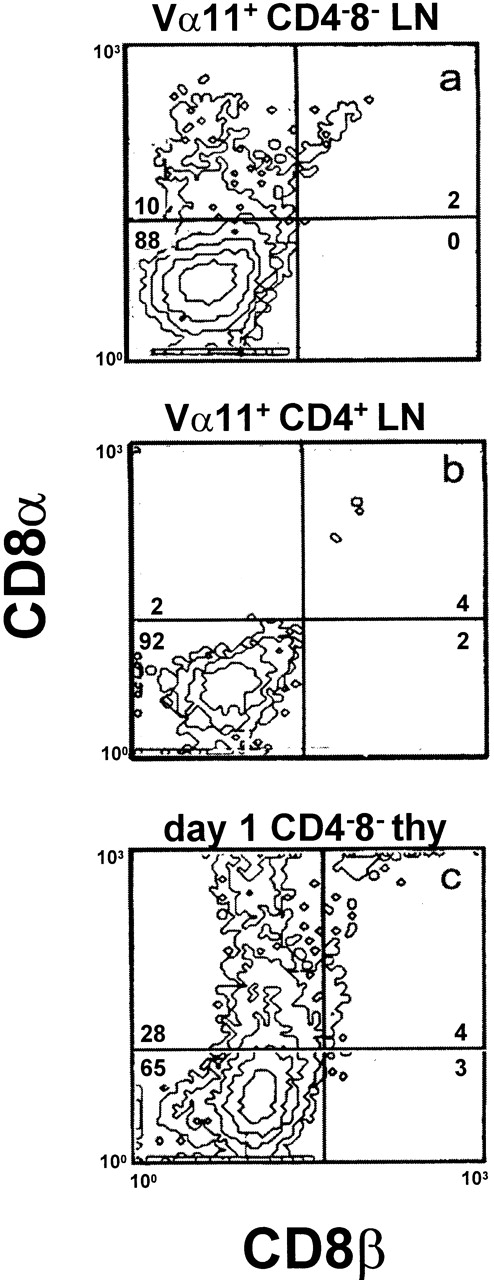
TCR stimulation can induce CD8αα expression on TCRαβ DN T cells. (a) Vα11+ CD4−CD8−, (b) Vα11+CD4+ lymph node T cells from AND TCR mice (isolated by electronic cell sorting and cultured at 4 × 104/well on 30 μg/ml plate-bound anti-Vα11, RR8-1, in the presence of recombinant IL-1 and IL-2, 100 U/ml, each), and (c) CD4−CD8− thymocytes of day 1 neonatal mice (isolated by magnetic bead depletion and cultured at 10 × 104/well on 24 μg/ml immobilized anti-γδ, GL-3, in the presence of rIL-1 and rIL-2, 20 U/ml each) were assayed for expression of CD8α and CD8β by flow cytometry. The data are representative of three or more experiments. B6 LN T cells were used as a positive control for CD8β staining (data not shown).
TCRαβ DN Cells of TCRαβ Transgenic Mice Do Not Require MHC for Development.
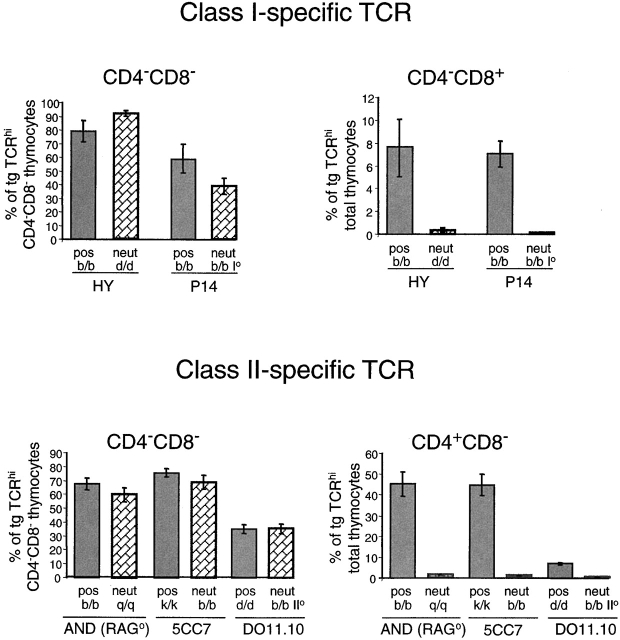
One of the hallmarks of αβ T cell development is the requirement for MHC-specific positive selection 47. In contrast, γδ T cells fully mature in the absence of MHC 6 7. Since there are conflicting reports on the selection requirements of TCRαβDN cells 10 12 16, we tested several strains of TCRαβ transgenic mice, bearing MHC class I– or class II–specific TCRs. As shown in Fig. 5, the TCRαβDN cells of five different strains of TCRαβ mice develop equally well in the positively selecting or in the neutral (nonselecting) MHC background. Development is comparable both in percentage (Fig. 5) and in absolute number (data not shown). Thus, TCRαβDN cells show no MHC dependence for development, in clear contrast to mainstream αβ lineage T cells (CD4−CD8+ or CD4+CD8−) of the same mice that show an absolute requirement for specific MHC. These findings argue against the view that TCRαβDN cells derive from conventional CD4 or CD8 T cells by the downregulation of a coreceptor.
Figure 5.
TCRαβDN cells do not require MHC-dependent positive selection for development. Thymocytes were isolated from MHC class I–specific (HY and P14) or class II–specific (AND, 5CC7, and DO11.10) TCR transgenic mice bred onto a positively selecting (pos) or nonselecting, neutral (neut) MHC background. Cells were stained for CD4, CD8, and TCR (using antibodies: T370 for HY, anti-Vα2 for P14, anti-Vα11 for AND and 5CC7, and KJ-126 for DO11.10 TCR). The percent of transgenic (tg) TCRhi cells was determined from analysis of total thymocytes by software gating for CD4−CD8−, CD4−8+, or CD4+CD8−. Each bar represents the mean percentage (with SE bars) of TCRhi of CD4−CD8− or of total thymocytes from analyses of three to six individual mice per group.
TCRαβDN Cells of Some Strains Coexpress Endogenous TCRγδ and Transgenic TCRαβ.
The analyses above indicate that TCRαβDN cells have γδ lineage properties. Therefore, CD4−CD8− T cells of several strains of TCRαβ transgenic mice were analyzed for expression of TCRγδ. An obvious population of CD4−CD8− thymocytes and peripheral T cells bearing only the transgenic TCRαβ was apparent in all of the mice analyzed. In some strains, however, there existed a second subset of CD4−CD8− T cells coexpressing the transgenic TCRαβ and endogenous TCRγδ (Fig. 6 and Table ). This latter subset bearing both TCRs was most prominent in the P14 TCR mice. It is noteworthy that like the TCRαβDN subset of AND TCR mice, both TCRαβDN–bearing subsets of P14 TCR mice exhibited properties of γδ lineage T cells (Fig. 1 and data not shown). It was previously reported that TCRγδ+ cells develop in P14 TCR mice 48; however, it was not appreciated that these T cells coexpress the transgenic TCRαβ.
Figure 6.
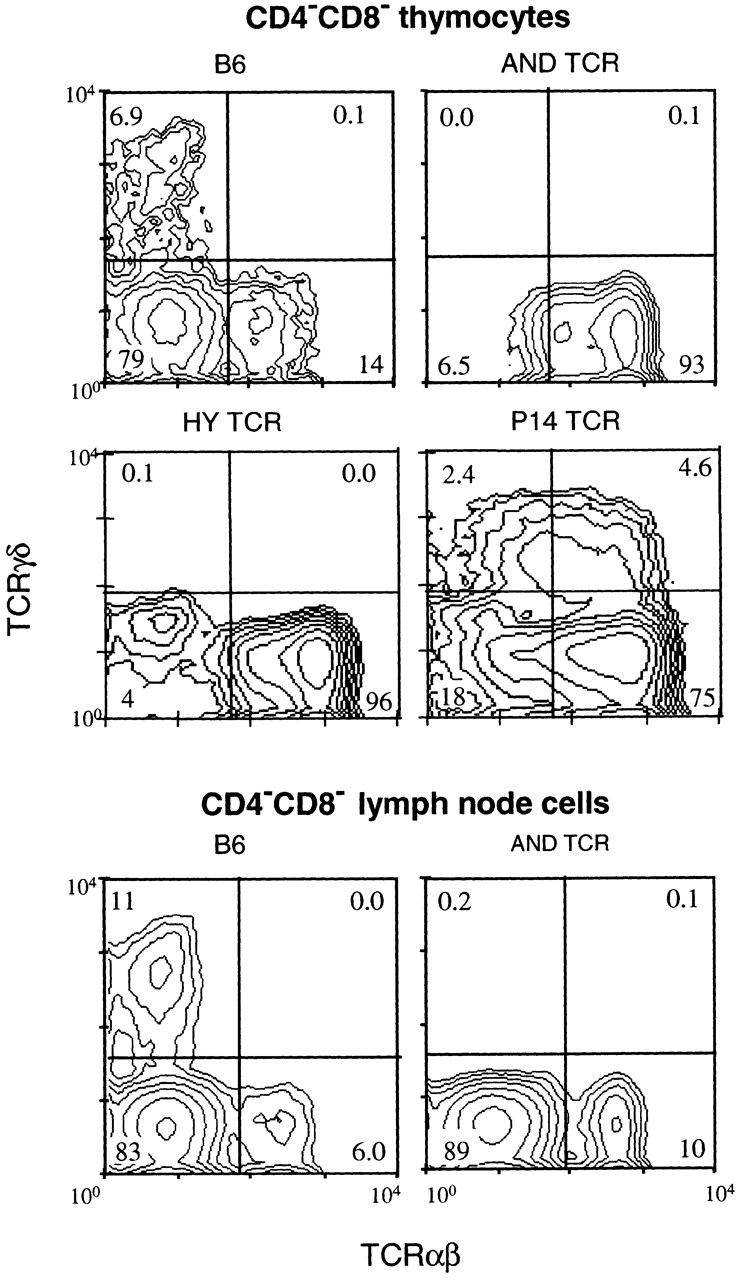
CD4−CD8− T cells of AND and HY TCR mice express transgenic TCRαβ but no TCRγδ, while those of P14 TCR mice coexpress transgenic TCRαβ and endogenous TCRγδ. Thymocytes and lymph node cells, stained for TCRαβ (H57-597), TCRγδ (GL3), CD4, and CD8, were analyzed by flow cytometry using live gating to collect data only from CD4−CD8− cells. The numbers inside the quadrants represent the percentage of CD4−CD8− thymocytes in each population. Statistics are given in Table .
Table 2.
Coexpression of Transgenic TCRαβ and Endogenous TCRγδ in CD4−CD8− Thymocytes
| Percentage of CD4−CD8− thymocytes | |||
|---|---|---|---|
| Mice | TCRαβ1TCRγδ− | TCRαβ1TCRγδ+ | n |
| B6 | 12.5 (2.0) | 0.06 (0.02) | 8 |
| AND | 94.2 (9.2) | 0.05 (0.01) | 7 |
| HY | 94.5 (3.0) | 0.06 (0.02) | 7 |
| HA | 74.8 (9.6) | 0.08 (0.04) | 5 |
| DO11.10 | 81.8 (4.9) | 0.15 (0.04) | 7 |
| 5CC7 | 80.7 (6.6) | 0.56 (0.26) | 6 |
| P14 | 71.3 (5.4) | 2.85 (0.54) | 7 |
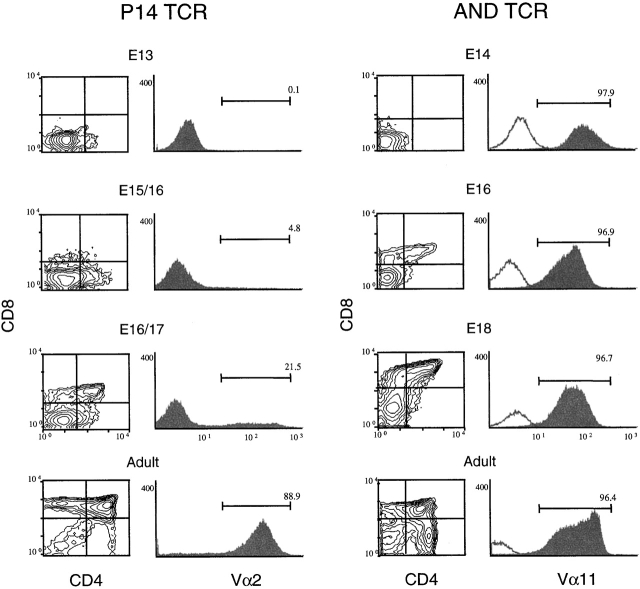
These different patterns of TCR expression prompted us to investigate the timing of transgenic TCRαβ expression during fetal thymic ontogeny, using the AND and P14 TCR mice as prototypes. As shown in Fig. 7, AND TCR is expressed early on a majority of E14 thymocytes. In contrast, the P14 TCR is first detected around E15–16, and then only on a minor subset of fetal thymocytes. These data, considered together with the data from adult thymocytes in Fig. 6, suggest that very early expression of the transgenic TCRαβ inhibits endogenous TCRγδ gene rearrangement and/or expression.
Figure 7.
The transgenic AND TCR is expressed much earlier than the P14 TCR in fetal development. Thymocytes from P14 TCR or AND TCR embryos, harvested on the days indicated (E13–18), were stained for Thy 1.2, CD4, CD8, and TCR (Vα2 for P14 and Vα11 for AND) and analyzed by four color flow cytometry. Distributions, gated for total Thy1.2+ thymocytes, display dual parameter, CD4 and CD8, or single parameter, TCR (shaded), overlaid with the negative control for background fluorescence (unshaded). Numbers indicate the percentage of cells within the indicated gates. Data are representative of two such experiments with similar time courses.
Endogenous TCRγδ Gene Rearrangements Are Suppressed in TCRαβDN Cells of AND, but Not in TCRαβDN Cells of P14 TCR Mice.
To determine the basis for differences in TCR expression in TCRαβDN cells of AND and P14 TCR mice, TCRγδ gene rearrangements were examined using a quantitative PCR assay. Since TCRVγ2 is commonly used by lymphoid γδ T cells 49, the frequency of TCRVγ2→Jγ1 rearrangement was determined in mature T cell subsets (Fig. 8). The analyses indicate that this gene rearrangement is much more suppressed in TCR+CD4− CD8− (TCRαβDN) cells of AND TCR than of P14 TCR mice. Similar differences between AND TCR and P14 TCR mice were observed with other Vγ and Vδ gene segments, although the rearrangement frequencies were much lower (data not shown). Of note, the occurrence of Vγ2→Jγ1 rearrangement in TCR+CD4−CD8− T cells of P14 TCR mice is equivalent to those of TCRγδ+ cells of G8 TCRγδ (Vγ2+) transgenic mice and of B6 wild-type mice (Fig. 8 a). Thus, in the P14 TCR mice that express the transgenic receptor relatively late, TCRγδ rearrangement is uninhibited and TCRαβDN cells bearing TCRγδ are observed (Fig. 6 Fig. 7 Fig. 8).
Figure 8.
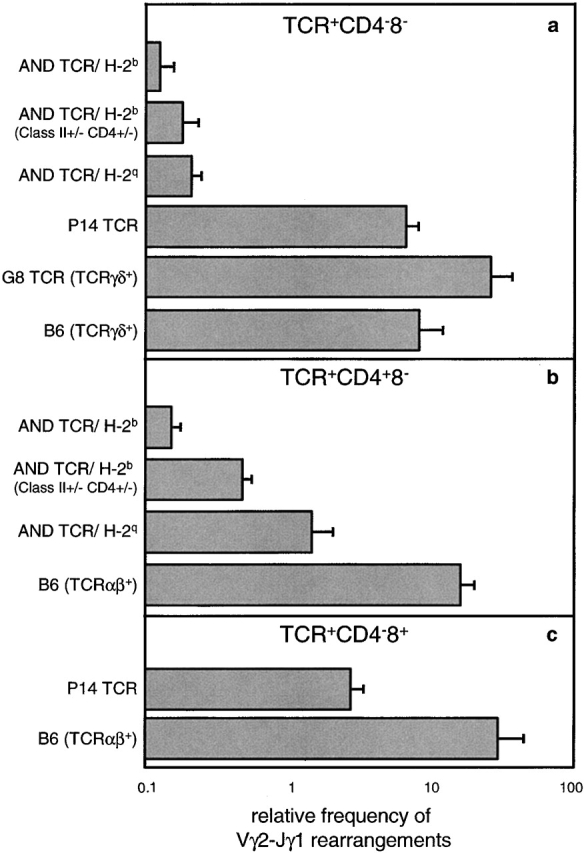
Vγ2–Jγ1 gene rearrangements are suppressed in TCR+ CD4−CD8− (TCRαβDN) cells of AND TCR but not of P14 TCR mice. Rearrangements are suppressed, independent of MHC haplotype, in the TCRαβDN but not the TCR+CD4+CD8− subset of AND TCR mice. Samples of 105 each of TCR+ (Vα11+ for AND, Vα2+ for P14, TCRγδ+ for G8 TCR, and TCRγδ+ or TCRαβ+ for B6) (a) CD4−CD8−, (b) CD4+CD8−, and (c) CD4−CD8+ lymph node T cells from AND TCR/H-2b, AND TCR/H-2b (MHC class II+/−, CD4+/−), AND TCR/H-2q, P14 TCR, G8 TCR, and B6 mice were isolated by electronic sorting. The relative frequency of Vγ2–Jγ1 rearrangements per sample was determined using a PCR ELISA as described in Materials and Methods. Bars represent the mean values (with SEs) of three individual sorts, using a total of five to eight mice per sort.
Interestingly, an analysis of the frequency of Vγ2→Jγ1 rearrangements in CD4 T cells of AND TCR mice is increased in the semiselecting (class II+/−, CD4+/−) or nonselecting (H-2q) MHC background in comparison over the frequency in the selecting MHC (H-2b) background (Fig. 8 b). These results fit with the notion that MHC engagement terminates RAG expression during αβ development 50. In contrast, the TCRαβDN cells developing in the CD4−CD8− (γδ) pathway follow different rules since rearrangement frequency is independent of MHC (Fig. 8 a). These findings suggest that TCR gene rearrangement is differentially regulated in the γδ and αβ lineages.
TCRγδ Is Expressed by Skin Lymphocytes of AND TCR Mice.
TCRγδ gene rearrangements in thymocyte precursors that localize to skin epithelium occur much earlier in fetal development than those destined for migration and residence in the lymphoid tissues 2. Therefore, there was the possibility that the dendritic epidermal T lymphocytes of AND TCR mice would express TCRγδ since some of their thymic precursors may have rearranged TCRγδ before transgenic TCRαβ expression. In contrast to the lymphoid CD4−CD8− T cells that fail to express TCRγδ (Fig. 6), skin lymphocytes express two subsets of T cells (Fig. 9), one expressing TCRαβ alone and the second expressing both TCRαβ and TCRγδ. Thus, when TCRαβ transgene expression occurs after endogenous TCRγδ rearrangements, rearrangement is not suppressed, and TCRγδ and TCRαβ can be expressed by the same cells. We determined in parallel analyses that these cells are CD4−CD8−, Vα11+, Vβ3+, and Vα3+.
Figure 9.
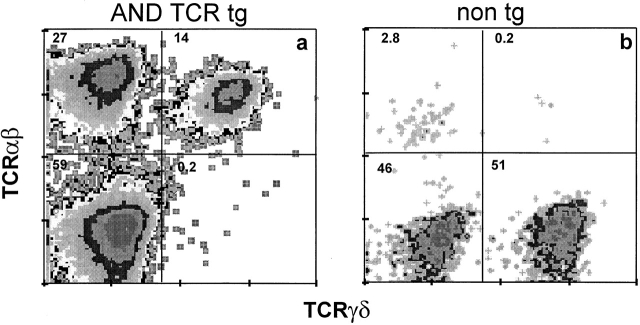
In contrast to lymphoid T cells, skin dendritic epithelial lymphocytes of AND TCR mice contain two subsets of T cells, one bearing only the transgenic TCRαβ and a second coexpressing the transgenic TCRαβ with endogenous TCRγδ. Isolated epidermal lymphocytes of (a) H-2d AND TCR transgenic (tg) or (b) nontransgenic (non tg) B10.D2 mice were stained for TCRαβ (H57-597) and TCRγδ (GL3) and analyzed by flow cytometry. The numbers inside the quadrant represent the percentage of cells in each population. The data are representative of several analyses of AND TCR mice of H-2d or other H-2 haplotypes.
In the periphery of normal mice, the canonical Vγ3+ TCR is expressed exclusively on skin lymphocytes 2. The finding that T cells, bearing the AND TCR and coexpressing the expected TCRγδ, can home at the right time to what is normally a γδ-specific site, provides additional evidence that TCRαβDN cells are γδ lineage T cells. Presumably, the skin lymphocytes expressing only the transgenic TCRαβ have an out of frame TCRγδ or, alternatively, some cells express the transgenic TCR early enough to suppress endogenous TCRγδ rearrangements. In any case, the finding that even the TCRαβ+TCRγδ− subpopulation is able to traffic to this traditionally γδ-specific site demonstrates that skin homing is not dependent on the canonical TCR. These data, like those above, reveal that when the TCR is expressed early (regardless of whether it is TCRγδ or TCRαβ), the receptor allows γδ lineage commitment and maturation in the γδ lineage.
Discussion
These studies examine T cell development in transgenic mice with premature expression of TCRαβ. An interesting feature of the mice is a population of mature CD4−CD8− thymocytes and peripheral T cells, expressing only the transgenic TCRαβ (TCRαβDN). To determine whether TCRαβDN cells belong to the αβ or γδ lineage, we analyzed these cells in several TCR transgenic strains and compared them to the T cell subsets of normal mice. By all criteria examined, the TCRαβDN cells clearly exhibit characteristics of γδ lineage T cells. The lack of a coreceptor, the level of CD5, and the early maturation delineate TCRαβDN cells from the major TCRαβ+ CD4 and CD8 T cell subsets. TCRαβDN cells do not express NK1.1 or IL-2Rβ (CD122) or produce IL-4, distinguishing them from the NK T cells of wild-type mice. In contrast, TCRαβDN cells are similar to γδ T cells since their development is early, is relatively insensitive to CsA, and is MHC independent. Also, like γδ lineage cells, TCRαβDN cells can be induced to express CD8αα homodimers in response to anti-TCR stimulation. Most notable, in TCRαβ strains where the transgenic receptor is expressed later in development, CD4−CD8− T cells arise coexpressing the transgenic TCRαβ and endogenous TCRγδ (Table , and Fig. 6 and Fig. 7). TCRαβDN cells with both receptors exhibit the same phenotype and properties as those lacking TCRγδ expression. These findings provide the most direct evidence that TCRαβDN cells are γδ lineage T cells.
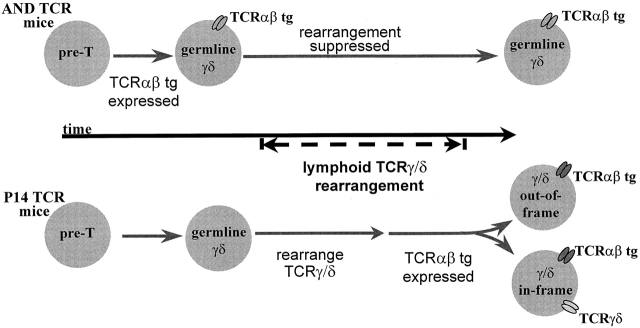
The different patterns of TCR expression in CD4−CD8− T cells of TCRαβ mice appear to be related to the timing of TCRαβ transgene expression with respect to endogenous TCRγδ gene rearrangement. As modeled in Fig. 10, the early expression of transgenic TCRαβ in precursor thymocytes of AND TCR mice causes suppression of endogenous TCRγδ gene rearrangement; nevertheless, the transgenic receptor allows continued maturation in the CD4−CD8− (γδ) pathway. In P14 TCR mice, the transgenic receptor is expressed later such that TCRγδ gene rearrangements occur normally. If rearrangements are productive, mature CD4−CD8− T cells emerge coexpressing the TCRαβ (P14 TCR) and endogenous TCRγδ (Fig. 6 Fig. 7 Fig. 8, and Table ). The different TCR expression patterns in skin versus lymph node CD4−CD8− T cells of AND TCR mice also can be explained by this model. A subset of epidermal lymphocytes coexpresses the transgenic TCRαβ and endogenous TCRγδ (Fig. 9), but lymph node T cells bear only the transgenic TCRαβ (Fig. 6). Thus, the rearrangements of genes encoding the lymphoid type TCRγδ are suppressed by AND TCR expression, whereas rearrangements that occur early in the fetal thymus, encoding the TCRγδ of skin lymphocytes, are not suppressed. Of significance, either TCR expression pattern allows development in the CD4−CD8− (γδ) pathway.
Figure 10.
A model to explain the different patterns of TCR expression in CD4−CD8− thymocytes of TCRαβ transgenic mice. Lymphoid-type TCRγδ gene rearrangements occur at a distinct time (stage) in thymocyte development. In AND TCR mice, the transgenic TCRαβ is expressed early with respect to TCRγδ gene rearrangement and rearrangement is suppressed. Since transgenic TCRαβ expression occurs somewhat later in P14 TCR mice, there is no interference with rearrangement and cells with in-frame TCRγδ rearrangements can express both the transgenic TCRαβ and endogenous TCRγδ. Irrespective of the TCR expression pattern, CD4−CD8− cells mature with properties of γδ lineage T cells.
We have considered these and previous results for understanding the role of the TCR in αβ versus γδ lineage determination. Evidence exists for an instructional model in which successful rearrangement of TCRγδ or TCRβ genes biases the decision of a precursor to become a γδ or αβ lineage T cell. Of note, αβ lineage T cells are depleted of productive TCRγ and -δ rearrangements, suggesting that the production of a functional TCRγδ favors a γδ lineage decision 51 52 53. In addition, mice deficient for the pTα component of the pre-TCR show an increase in the number of γδ lineage T cells, implying that normally pre-TCR signals inhibit γδ lineage development 54. Other studies, however, have prompted speculation that γδ/αβ lineage determination may occur before or independent of TCR gene rearrangement 55 56 57 58. Of relevance, a few CD4+CD8+ thymocytes arise in TCRβ−/− null mutant mice 59, and these cells are enriched for in-frame TCRγδ rearrangements 60 61, indicating that TCRγδ, in some circumstances, can promote αβ development. Moreover, CD4+CD8+ cells develop, although inefficiently, in TCRγδ transgenic mice when endogenous TCRβ recombination is diminished or suppressed 56 62. Even in normal mice, a minor population of TCRγδ-bearing CD4+CD8+ cells has been observed 63. Complicating the issue further are reports that the majority of TCRβ rearrangements are productive in TCRγδ+ T cells 51 64. Others disagree, finding that these rearrangements are predominantly out of frame 65. Clearly, the data on this question are mixed and the issue is unresolved.
Since a transgenic TCRαβ permits both γδ and αβ development, our results and those of others 17 38 39 could fit a model in which γδ/αβ fate is predetermined, before or independent of TCR rearrangement/expression 4 66. In this scenario, the TCR plays no role in lineage commitment but is needed only for survival and/or lineage progression. While this model would not always couple the appropriate TCR with lineage commitment, it is noteworthy that additional mechanisms operate to correct TCR expression in the wrong lineage. In the αβ lineage, TCRγ is downregulated at the CD4+CD8+ stage 67 and TCRα rearrangement results in the deletion of the TCRδ locus. In the γδ pathway, pTα is turned off 68 and TCRα rearrangement is not upregulated 69.
At first glance, the finding that premature expression of TCRαβ can permit both a γδ and αβ cell fate appears to be inconsistent with an instructional mechanism for lineage commitment. However, one version of an instructional model proposes that TCRγδ and pre-TCR signals influence lineage commitment, but does not necessarily imply that signaling differences are absolute or inherent in the TCR structure. Thus, quantitative differences in TCRγδ and pre-TCR signaling could bias lineage choice. Perhaps signals generated by the prematurely expressed transgenic TCRαβ quantitatively mimic TCRγδ signals. An additional possibility is that the timing of TCR expression influences the lineage decision. Recent evidence indicates that TCRγδ rearrangements occur slightly ahead of TCRβ in adult thymopoiesis 41 42. Conceivably, these ordered rearrangements could be coordinated with developmentally regulated changes in TCR signal transduction such that the earliest TCR signals promote a γδ fate, whereas later TCR signals favor an αβ fate. Our data could fit with such a sequential model since distinct TCR signals regulating lineage choice would be generated as a function of time, irrespective of TCR substitutions. In some sense, this sequential model can be seen as both predetermined and instructional: predetermined, since changes in intracellular TCR signals over time are developmentally preprogrammed, and instructional, since distinct signals mediate lineage commitment. However, such signals are not inherent to the TCR structure. In any case, the previous results demonstrating that αβ T cells are depleted of in-frame TCRγδ rearrangements 51 52 53 and the low frequency of productive TCRβ rearrangements in γδ T cells 65 support a sequential model.
TCRαβ transgenic mice are widely used to study antigen-specific immune responses in vivo. The studies reported here should send a note of caution regarding the use of such mice for this purpose. If, as we conclude, the transgenic TCRαβ receptor can substitute for the TCRγδ in γδ lineage T cells, cells that would normally be immunologically silent can now participate in an antigen-specific response. Because γδ T cells have unique developmental, functional, and homing properties, they could contribute to the response in nonphysiological ways. Thus, difficulties with these mice could be related to the large number of mature T cells expressing a single TCR, but also because γδ lineage cells (bearing transgenic TCRαβ) contribute to the antigenic response in unpredictable ways. Even sorting for CD4+ cells may not help, since a few γδ T cells express CD4 57. A new generation of TCR transgenic mice, with delayed TCRαβ expression, may provide a solution to this problem.
Acknowledgments
The authors thank S. Hedrick, H. von Boehmer, D. Loh, F. Alt, L. Glimcher, B. Koller, H. Pircher, M. Davis, J. Bluestone, and G. Sims for gifts of transgenic or gene-targeted mutant mice, and A. Kruisbeek and Ron Germain for cell lines. We thank our colleagues R. Swofford and C. Eigsti for flow cytometry and sorting, and A. Bendelac, S. Gurunathan, E. Schweighoffer, E. Robey, and Juan Zuniga-Pflucker for advice, assistance, and/or critical comments on the manuscript.
K. Terrence and C. Pavlovich performed this work as Howard Hughes Medical Institute–National Institutes of Health Research Scholars.
Footnotes
Abbreviations used in this paper: APC, allophycocyanin; β2m, β2-microglobulin; B6, C57BL/6; B10, C57BL/10; CsA, cyclosporine A; HSA, heat stable antigen; RAG, recombination activating gene; TCRαβDN, TCRαβ+CD4−CD8−.
K. Terrence and C.P. Pavlovich contributed equally to this work.
References
- Bluestone J.A., Cron R.Q., Barrett T.A., Houlden B., Sperling A.I., Dent A., Hedrick S., Rellahan B., Matis L.A. Repertoire development and ligand specificity of murine TCRγδ cells. Immunol. Rev. 1991;120:5–33. doi: 10.1111/j.1600-065x.1991.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Allison J. γδ T-cell development. Curr. Opin. Immunol. 1993;5:241–246. doi: 10.1016/0952-7915(93)90011-g. [DOI] [PubMed] [Google Scholar]
- von Boehmer H., Aifantis I., Azogui O., Feinberg J., Saint-Ruf C., Zober C., Garcia C., Buer J. Crucial function of the pre-T-cell receptor (TCR) in TCRβ selection, TCRβ allelic exclusion and αβ versus γδ lineage commitment. Immunol. Rev. 1998;165:111–119. doi: 10.1111/j.1600-065x.1998.tb01234.x. [DOI] [PubMed] [Google Scholar]
- Kang J., Raulet D.H. Events that regulate differentiation of αβ TCR+ and γδ TCR+ T cells from a common precursor. Semin. Immunol. 1997;9:171–179. doi: 10.1006/smim.1997.0069. [DOI] [PubMed] [Google Scholar]
- Robey E., Fowlkes B.J. The αβ versus γδ T-cell lineage choice. Curr. Opin. Immunol. 1998;10:181–187. doi: 10.1016/s0952-7915(98)80247-1. [DOI] [PubMed] [Google Scholar]
- Correa I., Bix M., Liao N.S., Zijlstra M., Jaenisch R., Raulet D. Most γδ T cells develop normally in β2- microglobulin-deficient mice. Proc. Natl. Acad. Sci. USA. 1992;89:653–657. doi: 10.1073/pnas.89.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighoffer E., Fowlkes B.J. Positive selection is not required for thymic maturation of transgenic γδ T cells. J. Exp. Med. 1996;183:2033–2041. doi: 10.1084/jem.183.5.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Rivera M.N., Park S.H., Roark J.H. Mouse CD1-specific NK1 T cellsdevelopment, specificity, and function. Annu. Rev. Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- MacDonald H.R. NK1.1+ T cell receptor-α/β+ cellsnew clues to their origin, specificity, and function. J. Exp. Med. 1995;182:633–638. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H., Kirberg J., Rocha B. An unusual lineage of αβ T cells that contains autoreactive cells. J. Exp. Med. 1991;174:1001–1008. doi: 10.1084/jem.174.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh H.S., Kishi H., Scott B., Borgulya P., von Boehmer H., Kisielow P. Early deletion and late positive selection of T cells expressing a male-specific receptor in T-cell receptor transgenic mice. Dev. Immunol. 1990;1:1–10. doi: 10.1155/1990/18208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J.H., Meleedy-Rey P., McCulley D.E., Sha W.C., Nelson C.A., Loh D.Y. Evidence for CD8-independent T cell maturation in transgenic mice. J. Immunol. 1990;144:3318–3325. [PubMed] [Google Scholar]
- Rocha B., von Boehmer H., Guy-Grand D. Selection of intraepithelial lymphocytes with CD8 α/α coreceptors by self-antigen in the murine gut. Proc. Natl. Acad. Sci. USA. 1992;89:5336–5340. doi: 10.1073/pnas.89.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd R.C., Mixter P.F. The origin of CD4−CD8−TCRαβ+ thymocytesa model based on T-cell receptor avidity. Immunol. Today. 1995;16:428–431. doi: 10.1016/0167-5699(95)80019-0. [DOI] [PubMed] [Google Scholar]
- Teh H.S., Kishi H., Scott B., von Boehmer H. Deletion of autospecific T cells in T cell receptor (TCR) transgenic mice spares cells with normal TCR levels and low levels of CD8 molecules. J. Exp. Med. 1989;169:795–806. doi: 10.1084/jem.169.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.P., Kappler J.W., Marrack P. Thymocytes can become mature T cells without passing through the CD4+CD8+, double-positive stage. J. Exp. Med. 1996;184:1619–1630. doi: 10.1084/jem.184.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L., Fehling H.J., von Boehmer H. The αβ T cell receptor can replace the γδ receptor in the development of γδ lineage cells. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- DiSanto J.P., Guy-Grand D., Fisher A., Tarakhovsky A. Critical role for the common cytokine receptor γ chain in intrathymic and peripheral T cell selection. J. Exp. Med. 1996;183:1111–1118. doi: 10.1084/jem.183.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matechak E.O., Killeen N., Hedrick S.M., Fowlkes B.J. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- Kaye J., Hsu M.L., Sauron M.E., Jameson S.C., Gascoigne N.R., Hedrick S.M. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Grusby M.J., Johnson R.S., Papaioannou V.E., Glimcher L.H. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- Killeen N., Littman D.R. Helper T-cell development in the absence of CD4-p56lck association. Nature. 1993;364:729–732. doi: 10.1038/364729a0. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K., Nakayama K., McCoy R.L., Wang F., Nishimura T., Habu S., Murphy K.M., Loh D.Y. Cellular and peptide requirements for in vitro clonal deletion of immature thymocytes. Proc. Natl. Acad. Sci. USA. 1992;89:9000–9004. doi: 10.1073/pnas.89.19.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirberg J., Baron A., Jakob S., Rolink A., Karjalainen K., von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex–restricted receptor. J. Exp. Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P., Bluthmann H., Staerz U.D., Steinmetz M., von Boehmer H. Tolerance in T cell receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Seder R.A., Paul W.E., Davis M.M., de St. Groth B.F. The presence of interleukin-12 acts directly on CD4+ T cells to enhance priming for interferon-γ production and diminishes interleukin-4 inhibition of such priming. J. Exp. Med. 1992;176:1091–1098. [Google Scholar]
- Pircher H., Burki K., Lang R., Hengartner H., Zinkernagel R.M. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- Koller B.H., Marrack P., Kappler J.W., Smithies O. Normal development of mice deficient in β2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- Sim G.K., Olsson C., Augustin A. Commitment and maintenance of the αβ and γδ T cell lineages. J. Immunol. 1995;154:5821–5831. [PubMed] [Google Scholar]
- Ramsdell F., Jenkins M., Dinh Q., Fowlkes B.J. The majority of CD4+8− thymocytes are functionally immature. J. Immunol. 1991;147:1779–1785. [PubMed] [Google Scholar]
- Nixon-Fulton J.L., Bergstresser P.R., Tigelaar R.E. Thy-1+ epidermal cells proliferate in response to concanavalin A and interleukin 2. J. Immunol. 1986;136:2776–2786. [PubMed] [Google Scholar]
- Coligan, J.E., A. Kruisbeek, D.H. Margulies, E.M. Shevach, and W. Strober. 1995. Current Protocols in Immunology. John Wiley and Sons, Inc., New York. 5.0.3–5.4.13
- Seder R.A., Gazazinelli R., Sher A., Paul W.E. Interleukin-12 acts directly on CD4+ T-cells to enhance priming for interferon-γ production and diminishes interleukin-4 inhibition of such priming. Proc. Natl. Acad. Sci. USA. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf S.W., Beverly B., Lantz O., Schwartz R.H. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clonesboth nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol. Cell. Biol. 1995;15:3197–3205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman A.J., Marusic-Galesic S., Spencer D., Kruisbeek A., Raulet D.H. Predominant variable region gene usage by γδ T cell receptor–bearing cells in the adult thymus. J. Exp. Med. 1988;168:1021–1040. doi: 10.1084/jem.168.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu. Rev. Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- Fritsch M., Andersson A., Petersson K., Ivars F. A TCR α chain transgene induces maturation of CD4−CD8− αβ+ T cells from γδ T cell precursors. Eur. J. Immunol. 1998;28:828–837. doi: 10.1002/(SICI)1521-4141(199803)28:03<828::AID-IMMU828>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Fritsch M., Ivars F. γδ T-cell precursor-derived CD4− CD8− αβ T cells retain γδ cell function. Scand. J. Immunol. 1998;48:8–14. doi: 10.1046/j.1365-3083.1998.00377.x. [DOI] [PubMed] [Google Scholar]
- Matsuzaki G., Yoshikai Y., Kishihara K., Nomoto K. Expression of T cell antigen receptor genes in the thymus of irradiated mice after bone marrow transplantation. J. Immunol. 1988;140:384–387. [PubMed] [Google Scholar]
- Capone M., Hockett R.D., Zlotnik A. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic developmentT cell receptor rearrangements are present in CD44+CD25+ Pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak F., Tourigny M., Schatz D.G., Petrie H.T. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- Jenkins M.K., Schwartz R.H., Pardoll D.M. Effects of cyclosporine A on T cell development and clonal deletion. Science. 1988;241:1655–1658. doi: 10.1126/science.241.4873.1655. [DOI] [PubMed] [Google Scholar]
- Gao E.K., Lo D., Cheney R., Kanagawa O., Sprent J. Abnormal differentiation of thymocytes in mice treated with cyclosporin A. Nature. 1988;336:176–179. doi: 10.1038/336176a0. [DOI] [PubMed] [Google Scholar]
- Takahama Y., Kosugi A., Singer A. Phenotype, ontogeny, and repertoire of CD4−CD8− T cell receptor αβ+ thymocytes. Variable influence of self-antigens on T cell receptor Vβ usage. J. Immunol. 1991;146:1134–1141. [PubMed] [Google Scholar]
- MacDonald H.R., Schreyer M., Howe R.C., Bron C. Selective expression of CD8α (Ly-2) subunit on activated thymic γδ cells. Eur. J. Immunol. 1990;20:927–930. doi: 10.1002/eji.1830200431. [DOI] [PubMed] [Google Scholar]
- Robey E., Fowlkes B.J. Selective events in T cell development. Annu. Rev. Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- Wilson A., Pircher H., Ohashi P., MacDonald H.R. Analysis of immature (CD4−CD8−) thymic subsets in T-cell receptor αβ transgenic mice. Dev. Immunol. 1992;2:85–94. doi: 10.1155/1992/45150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling A.I., Cron R.Q., Decker D.C., Stern D.A., Bluestone J.A. Peripheral T cell receptor γδ variable gene repertoire maps to the T cell receptor loci and is influenced by positive selection. J. Immunol. 1992;149:3200–3207. [PubMed] [Google Scholar]
- Brandle D., Muller C., Rulicke T., Hengartner H., Pircher H. Engagement of the T-cell receptor during positive selection in the thymus down-regulates RAG-1 expression. Proc. Natl. Acad. Sci. USA. 1992;89:9529–9533. doi: 10.1073/pnas.89.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E.C., Girardi M., Owen M.J., Hayday A.C. αβ and γδ T cells can share a late common precursor. Curr. Biol. 1995;5:659–669. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- Livak F., Petrie H.T., Crispe I.N., Schatz D.G. In-frame TCR δ gene rearrangements play a critical role in the αβ/γδ T cell lineage decision. Immunity. 1995;2:617–627. doi: 10.1016/1074-7613(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Kang J., Baker J., Raulet D.H. Evidence that productive rearrangements of TCR γ genes influence the fate of progenitor cells to differentiate into αβ or γδ T cells. Eur. J. Immunol. 1995;9:2706–2709. doi: 10.1002/eji.1830250946. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. Separate lineages of T cells expressing the αβ and γδ receptors. Nature. 1989;338:430–432. doi: 10.1038/338430a0. [DOI] [PubMed] [Google Scholar]
- Livak F., Wilson A., MacDonald H.R., Schatz D.G. αβ lineage-committed thymocytes can be rescued by the γδ T cell receptor (TCR) in the absence of TCRβ chain. Eur. J. Immunol. 1997;27:2948–2958. doi: 10.1002/eji.1830271130. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Iritani B.M., Krotkova A., Forbush K.A., Laplace C., Perlmutter R.M., von Boehmer H. Restoration of thymopoiesis in pTα−/− mice by anti-CD3ε antibody treatment or with transgenes encoding activated Lck or tailless pTα. Immunity. 1997;6:703–714. doi: 10.1016/s1074-7613(00)80446-x. [DOI] [PubMed] [Google Scholar]
- Mertsching E., Wilson A., MacDonald H.R., Ceredig R. T cell receptor α gene rearrangement and transcription in adult thymic γδ cells. Eur. J. Immunol. 1997;27:389–396. doi: 10.1002/eji.1830270208. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R., Hooper M.L., Tonegawa S. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- Passoni L., Hoffman E.S., Kim S., Crompton T., Pao W., Dong M.Q., Owen M.J., Hayday A.C. Intrathymic δ selection events in γδ cell development. Immunity. 1997;7:83–95. doi: 10.1016/s1074-7613(00)80512-9. [DOI] [PubMed] [Google Scholar]
- Washburn T., Schweighoffer E., Gridley T., Chang D., Fowlkes B.J., Cado D., Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Kersh G.J., Hooshmand F.F., Hedrick S.M. Efficient maturation of αβ lineage thymocytes to the CD4+CD8+ stage in the absence of TCR-β rearrangement. J. Immunol. 1995;154:5706–5714. [PubMed] [Google Scholar]
- Kang J., Coles M., Cado D., Raulet D.H. The developmental fate of T cells is critically influenced by TCRγδ expression. Immunity. 1998;8:427–438. doi: 10.1016/s1074-7613(00)80548-8. [DOI] [PubMed] [Google Scholar]
- Burtrum D.B., Kim S., Dudley E.C., Hayday A.C., Petrie H.T. TCR gene recombination and αβ-γδ lineage divergenceproductive TCR-β rearrangement is neither exclusive nor preclusive of γδ cell development. J. Immunol. 1996;157:4293–4296. [PubMed] [Google Scholar]
- Aifantis L., Azogui O., Feinberg J., Saint-Ruf C., Buer J., von Boehmer H. On the role of the pre-T cell receptor in αβ versus γδ T lineage commitment. Immunity. 1998;9:649–655. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- MacDonald H.R., Wilson A. The role of the T-cell receptor (TCR) in αβ/γδ lineage commitmentclues from intracellular TCR staining. Immunol. Rev. 1998;165:87–94. doi: 10.1111/j.1600-065x.1998.tb01232.x. [DOI] [PubMed] [Google Scholar]
- Kang J.S., Fehling H.J., Laplace C., Malissen M., Cado D., Raulet D.H. T cell receptor γ gene regulatory sequences prevent the function of a novel TCRγ/pTα pre-T cell receptor. Immunity. 1998;8:713–721. doi: 10.1016/s1074-7613(00)80576-2. [DOI] [PubMed] [Google Scholar]
- Bruno L., Rocha B., Rolink A., von Boehmer H., Rodewald H.R. Intra- and extra-thymic expression of the pre-T cell receptor α gene. Eur. J. Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- Nikolic-Zugic J., Moore M.W. T cell receptor expression on immature thymocytes with in vivo and in vitro precursor potential. Eur. J. Immunol. 1989;19:1957–1960. doi: 10.1002/eji.1830191030. [DOI] [PubMed] [Google Scholar]