Immune Cell Activation by Bacterial Cpg-DNA through Myeloid Differentiation Marker 88 and Tumor Necrosis Factor Receptor–Associated Factor (Traf)6 (original) (raw)
Abstract
Transition of immature antigen presenting cells (APCs) to the state of professional APCs is essential for initiation of cell-mediated immune responses to pathogens. Signal transduction via molecules of the Toll-like receptor (TLR)/interleukin 1 receptor (IL-1R) pathway is critical for activation of APCs either by pathogen-derived pattern ligands like lipopolysaccharides (LPS) or by CD40 ligation through T helper cells. The capacity of bacterial DNA (CpG-DNA) to induce APCs to differentiate into professional APCs represents an interesting discovery. However, the signaling pathways involved are poorly understood. Here we show that CpG-DNA activates the TLR/IL-1R signaling pathway via the molecules myeloid differentiation marker 88 (MyD88) and tumor necrosis factor receptor–associated factor 6 (TRAF6), leading to activation of kinases of the IκB kinase complex and the c-jun NH2-terminal kinases. Moreover, cells of TLR2- and TLR4-deficient mice are activated by CpG-DNA, whereas cells of MyD88-deficient mice do not respond. The data suggest that CpG-DNA initiates signaling via the TLR/IL-1R pathway in APCs in a manner similar to LPS and to T helper cell–mediated CD40 ligation. Activation of the TLR/IL-1R signaling pathway by foreign bacterial DNA may be one way to initiate innate defense mechanisms against infectious pathogens in vivo.
Keywords: Toll; signal transduction; CpG-DNA; interleukin 12; mice, knockout
Introduction
Key features of the innate immune system include the ability to limit an infectious challenge as the first line of host defense and to control the initiation of adaptive immune responses via antigen presentation in the context of costimulatory molecules and cytokines. Cells of the innate immune system such as macrophages and dendritic cells discriminate between “self” and infectious “non-self” via constitutive receptors that identify pattern ligands synthesized exclusively by pathogens, so-called pattern recognition factors (PRFs). Defined examples are LPS, bacterial lipoproteins (BLPs) and bacterial DNA (CpG-DNA). Recently it has been shown that immune cell activation by LPS and BLP requires Toll-like receptors (TLRs; 1 2 3). Through their cytoplasmic Toll/IL-1R homology domain (TIR domain), TLR2 and TLR4, as well as members of the IL-1R family, recruit the adapter molecule myeloid differentiation marker 88 (MyD88) to initiate signal transduction 4 5 6. In case of the IL-1R complex it has been shown that MyD88 and the IL-1R–associated kinase (IRAK)-1 are transiently recruited to the receptor complex, followed by interaction of IRAK-1 (and possibly other IRAKs such as IRAK-2 and IRAKm; references 7, 8) with TNF receptor–associated factor (TRAF)6 6. TRAF6 has also been found to interact directly with the cytoplasmic tail of CD40 9. This might explain the similar downstream events of TLR- and CD40-dependent signaling such as activation of kinases of the IKK IκB kinase (IKK) complex and the stress kinases c-Jun NH2-terminal kinase (JNK)1/2 and p38. That this pathway is essential in LPS signaling and Th cell–mediated CD40 ligation is supported by the LPS “non responder” phenotype of MyD88-deficient mice 10 and the defective LPS- and CD40-dependent signaling in TRAF6-deficient mice 11.
Recently, it has been shown that CpG-DNA, like LPS and BLP, induces gene expression via classic, receptor-driven signaling cascades such as the nuclear factor (NF)-κB and stress kinase pathways and the extracellular signal–regulated kinase pathway 12 13. In spite of their structural diversity, the immunobiology of LPS or BLP shows remarkable similarity to that of CpG-DNA 14. Together, these observations led us to test whether CpG-DNA signals via the TLR/IL-1R signaling pathway.
Materials and Methods
Cell Culture, Expression Plasmids, and Reagents.
The murine macrophage cell line RAW264.7 was grown in LowTox Clicks/RPMI (Biochrom) supplemented with 10% FCS (Seromed®; Biochrom). Peritoneal macrophages were cultured in RPMI 1640 medium (GIBCO BRL) supplemented with 10% fetal bovine serum (GIBCO BRL). To obtain peritoneal macrophages, mice were intraperitoneally injected with 2 ml of 4% thioglycollate. 3 d later, peritoneal exudate cells were isolated by peritoneal lavage with ice-cold Hanks' buffered salt solution. Cells were cultured for 2 h and washed extensively with Hanks' buffered salt solution to remove nonadherent cells. Adherent cells were used as peritoneal macrophages for stimulation with CpG-DNA. In the experiments where Poly I:C was used, cell culture medium was supplemented with 10 μg/ml polymyxin B (Sigma-Aldrich) during the time period of stimulation to exclude effects of trace amounts of LPS in the Poly I:C preparation.
The expression vectors for human TRAF6-C (AS289-522; reference 4) and TRAF6 wild-type were gifts from Tularik, Inc. A 5′ Flag epitope–tagged COOH terminus of murine MyD88 (AS158-302) was amplified by reverse transcription PCR from murine spleen RNA and cloned into a modified pcDNA3 vector (Invitrogen), containing an untranslated intervening sequence from the mouse IgG heavy chain for improved expression in eukaryotic cells (pCX).
Phosphothioate stabilized CpG-ODN (TCC-ATG-ACG-TTC-CTG-ATG-CT) and GpC-ODN (TCC-ATG-AGC-TTC-CTG-ATG-CT) 15 were purchased from TIB MOLBIOL, LPS (Salmonella minnesota Re 595) was purchased from Sigma-Aldrich, and Poly I:C was from Amersham Pharmacia Biotech.
Mice.
Generation of the mutant mice (TLR2-, TLR4-, and MyD88-deficient) has been described previously 2 3 16. Age-matched groups of wild-type and TLR4- and MyD88-deficient mice were used for the experiments.
ELISA.
Production of TNF from peritoneal macrophages was measured by ELISA according to the instructions of the manufacturer (Genzyme). Each value shown represents the mean of two independent stimulations.
Luciferase Reporter Plasmid Transfection and Luciferase Assay.
To investigate transcriptional activity of the IL-12 p40-promoter in transient transfection assays, we used a plasmid containing the luciferase gene under control of the −703 bp region of the murine IL-12 p40 gene, a gift from K. Murphy (Harvard University, Boston, MA 17). 5–10 × 106 RAW264.7 cells were transfected by electroporation in a 400 μl final volume (RPMI/25% FCS) at 280 V/960 μF in a Bio-Rad Laboratories gene pulser. 7 μg of reporter plasmid was used together with different amounts of specific expression vectors as indicated in the figure legends. The overall amount of plasmid DNA was held constant at 20 μg per electroporation by addition of the appropriate empty expression vector. After electroporation, cells were washed and split to into 6-well plates, 106 cells per well. 24 h after transfection, cells were stimulated in 2 ml of cell culture medium with CpG-ODN (1 μM) or Poly I:C (10 μg/ml) for 8 h. Preparation of cell extracts and luciferase assays were performed according to the manufacturer's instructions (Promega).
In Vitro Kinase Assay and Western Blotting.
For immune complex kinase assays with hemagglutinin (HA)-tagged kinases, RAW264.7 cells were transfected with expression vectors for HA-tagged IKKα and HA-tagged JNK-1. The cDNA for HA epitope–tagged human IKKα, a gift from R. Schmid (University of Ulm, Ulm, Germany), was expressed in pCX (see above), and HA–JNK-1 was a gift from M. Karin (University of California at San Diego, La Jolla, CA 18). 10 (HA-IKKα) and 6 μg (HA-JNK-1) of these vectors were used together with different amounts of specific expression vectors as indicated in the figure legends. The overall amount of plasmid DNA was held constant at 30 μg per electroporation by adding the appropriate empty expression vector. After electroporation, cells were washed and split to 6-well plates, 106 cells per well. 18 h after transfection, cells were stimulated in 2 ml of cell culture medium with 2 μM of phosphothioate-stabilized CpG-ODN or 100 μg of Poly I:C. Next, immune complex kinase assays were performed using antibodies to the HA tag (clone 12CA5; Boehringer). Kinase assays and Western blotting were performed as previously described 13. As substrate for the kinase assays, GST-IκBα(54) was used for HA-IKKα and GST-(79)-c-Jun for HA–JNK-1.
Results and Discussion
TRAF6, originally cloned as a CD40-interacting molecule, is composed of a highly conserved COOH-terminal TRAF domain and an NH2-terminal effector domain 9 19. Receptor-independent oligomerization of the NH2 terminus is sufficient to induce IKK, whereas JNK and p38 19, the COOH terminus (TRAF6-C) that seems to serve as the receptor docking and oligomerization domain of TRAF6, acts as a dominant negative molecule in CD40- and TLR/IL-1R–dependent signaling 5 20 21. To investigate, whether TRAF6 is involved in CpG-DNA–induced signaling, we transiently transfected cells of the macrophage cell line RAW264.7 with an IL-12 p40 promoter reporter vector and TRAF6-C. Previously, we had shown that IL-12 p40 is induced in different APCs and macrophage cell lines by CpG-DNA 13.
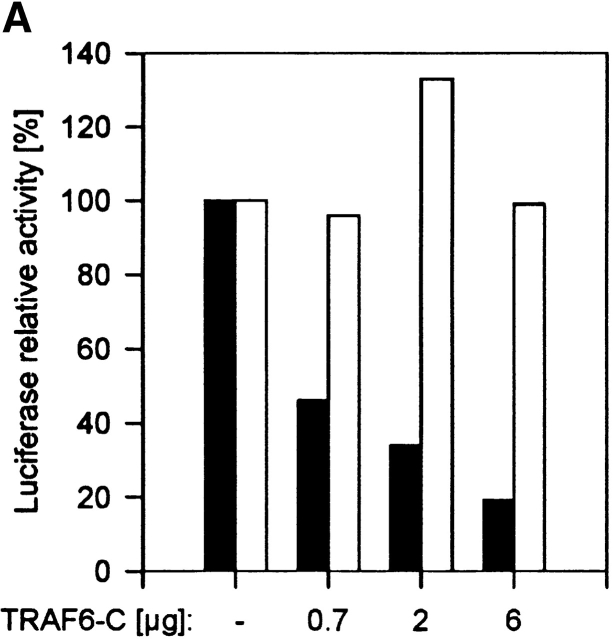
As shown in Fig. 1 A, TRAF6-C reduced CpG-DNA–induced activation of the IL-12 p40 promoter in a concentration-dependent manner. Importantly, dominant-negative TRAF6 did not affect induction of the IL-12 p40-promoter by double-stranded RNA (poly I:C). Although poly I:C is a weaker inducer of IL-12 p40 promoter activity than is CpG-DNA (9- and 32-fold activation, respectively), it shows that the activity of dominant-negative TRAF6 is specific for the inhibition of CpG-DNA–induced signaling.
Figure 1.
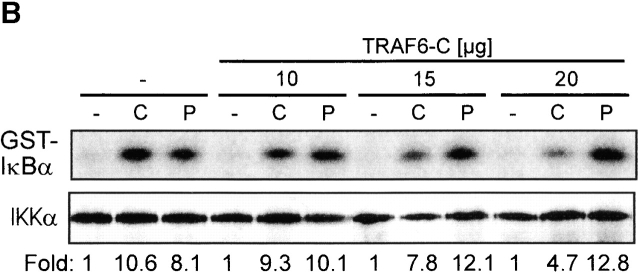
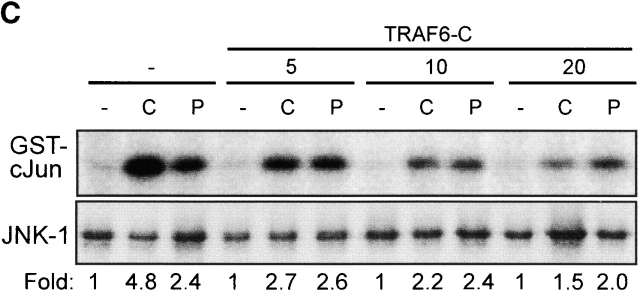
CpG-DNA signals via TRAF6. (A) RAW264.7 cells were transiently transfected with the IL-12 p40-promoter luciferase reporter and increasing amounts of an expression vector for TRAF6-C. Cells were stimulated with 1 μM CpG-ODN (black bars) or 10 μg/ml Poly I:C (white bars) for 8 h and luciferase activity was determined. Luciferase values are represented as percentage of activation for each stimulation relative to cells transfected in the absence of TRAF6-C. In this experiment, CpG-ODN induced 32-fold and Poly I:C 9-fold activation of the reporter gene. (B and C) RAW264.7 cells were transiently transfected with an expression vector for HA-IKKα (B) or HA-JNK-1 (C) either together with a control vector or with increasing amounts of the expression vector for TRAF6-C as indicated. 24 h after transfection cells were left untreated or stimulated with 2 μM CpG-ODN for 20 min or 100 μg/ml Poly I:C for 45 (HA-IKKα) or 90 min (HA-JNK-1). C, CpG-ODN; P, Poly I:C. After stimulation, cells were lysed and kinases were immunoprecipitated with antibodies to the HA tag. In vitro kinase assays were performed with GST-IκBα (B) or GST-cJun(79) (C) as substrate. Western blot analysis was performed with antibodies to IKKα (B) and JNK-1 (C). The radioactivity incorporated into the substrates was quantitated by PhosphorImager analysis (Molecular Dynamics). The obtained values were normalized against the radioactivity obtained in nonstimulated cells and are presented as fold induction. The results shown are representative of three independent experiments.
As detailed above, kinases identified as downstream effectors of TRAF6 are the JNKs and IKKs. Although not shown directly, degradation of IκBα and phosphorylation of IκBα at serine 32/36 after CpG-DNA stimulation (reference 22 and our unpublished observation) suggested that IKKs become activated by CpG-DNA. By transient transfection of RAW264.7 cells with an expression vector for HA-tagged IKKα and subsequent immune complex kinase assays, we found that CpG-DNA does indeed activate the IKK complex (Fig. 1 B). Similar results were obtained when endogenous IKKs (IKKα and IKKβ) were investigated (data not shown). As shown in Fig. 1 B, activation of IKKα by CpG-DNA were reduced when cells were transfected with increasing amounts of TRAF6-C, but activation of IKKα by Poly I:C was not affected by TRAF6-C (Fig. 1 B). These results are in accordance with the IL-12 p40 promoter experiments and with earlier observations that this promoter is activated by transcription factors of the NF-κB family 23. CpG-DNA has been shown to activate JNK1/2 and JNK kinase in APCs, leading to phosphorylation of c-Jun and activation of the transcription factor complex AP-1 12. Receptor-dependent and receptor-independent oligomerization of TRAF6, as well as overexpression of TRAF6, have also been shown to activate JNK activity 19 21. On the other hand, TLR4-induced JNK phosphorylation in 293 cells has been found to be Myd88 dependent, but TRAF6 independent 24. We therefore addressed whether CpG-DNA–induced JNK activation would be TRAF6 dependent. Fig. 1 C shows that TRAF6-C suppresses CpG-DNA–induced JNK-1 activity in a concentration-dependent way. According to the dominant negative activity of TRAF6-C, overexpression of TRAF6 wild-type in RAW264.7 macrophages activates JNK-1 (data not shown). Poly I:C also induced JNK-1; however, this activity was not affected by TRAF6-C.
Taken together, these data point to TRAF6 as an important intermediate of CpG-DNA–induced signal transduction to the IKK complex and stress kinases JNKs. In contrast, Poly I:C activates both the IKK complex and the JNKs in a TRAF6-independent way. These findings are in accordance to previous reports that showed that Poly I:C activates NF-κB in a double-stranded RNA-dependent protein kinase (PKR)- and IKKβ-dependent way 25, probably through direct binding of regulatory domains of PKR 26. Therefore, although it induces similar downstream effector kinases, Poly I:C seems to activate cells by pathways that significantly differ from other stimuli of innate immune cells like LPS or CD40.
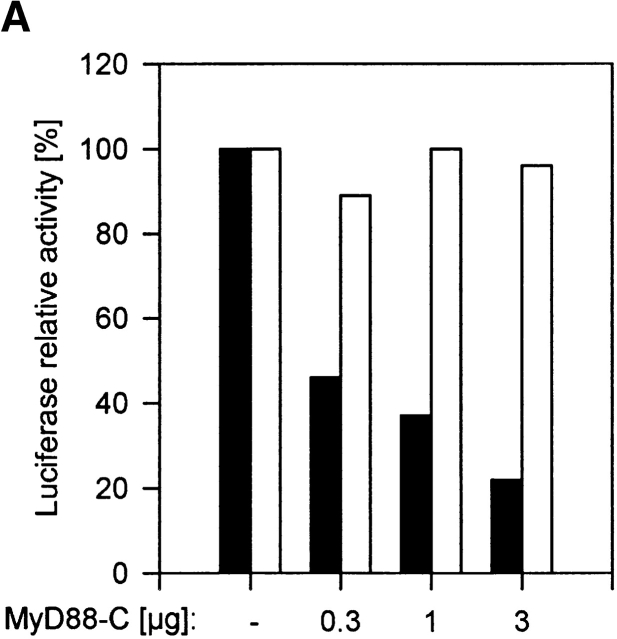
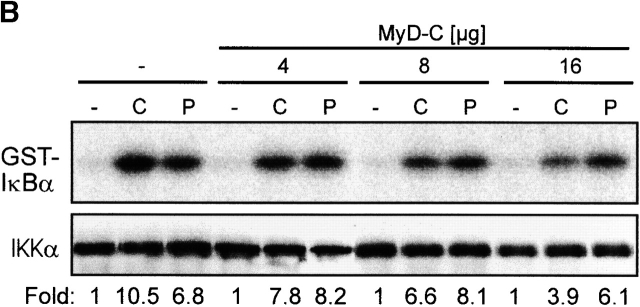
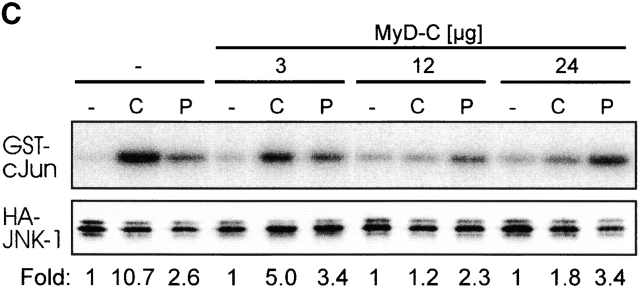
MyD88, originally isolated as IL-6–induced molecule during myeloid differentiation 27, was later defined as essential component of IL-1-, IL-18-, TLR2-, and TLR4-dependent signaling 3 5 16 28. Structurally, it is composed of a COOH-terminal TIR domain, an NH2-terminal death domain, and an intermediate domain. In functional terms, its role as adapter molecule first was characterized at the IL-1R complex 29. The TIR domain permits interaction with the receptor chains, then the IRAKs are recruited to this complex in a transient fashion 7 8 29. Overexpression of the native protein or the NH2 terminus alone is sufficient to induce NF-κB and JNK activation 29 30, whereas the COOH terminus can act as a dominant-negative molecule for TLR/IL-1R dependent signaling 4 5 29. To investigate whether CpG-DNA signals via MyD88, we used this COOH terminus as a dominant-negative inhibitor (MyD-C). In analogy to the experiments with TRAF6 shown above, we transfected RAW264.7 cells with the IL-12 p40 reporter vector, either alone or in the presence of MyD-C. Fig. 2 A shows that increasing amounts of MyD-C inhibit promoter activation, whereas Poly I:C, used as a specificity control, was not affected. Therefore, CpG-DNA uses the TLR/IL-1R signaling pathway to induce gene expression. Accordingly, MyD88-C also inhibited CpG-DNA–induced IKKα activation (Fig. 2 B) as well as JNK activation in a concentration-dependent manner (Fig. 2 C). Again, Poly I:C signaling was not affected. Taken together, the results suggest that CpG-DNA, but not Poly I:C, signals via MyD88 and TRAF6 to activate the IKK complex and JNKs.
Figure 2.
CpG-DNA signals via MyD88. (A) RAW264.7 cells were transiently transfected with the IL-12 p40-promoter luciferase reporter and increasing amounts of an expression vector for MyD88-C. Cells were stimulated with 1 μM CpG-ODN (black bars) or 10 μg/ml Poly I:C (white bars) for 8 h and luciferase activity was then determined. Luciferase values are represented as percentage of activation for each stimulation relative to cells transfected in the absence of MyD88-C. In this experiment, CpG-ODN induced 26-fold and Poly I:C 8-fold activation of the reporter gene. (B and C) RAW264.7 cells were transiently transfected with an expression vector for HA-IKKα (B) or HA-JNK-1 (C) either together with a control vector or with increasing amounts of the expression vector for MyD88-C as indicated. 24 h after transfection cells were left untreated or stimulated with 2 μM of CpG-ODN for 20 min or 100 μg/ml of Poly I:C for 45 min (HA-IKKα) or 90 min (HA-JNK-1). C, CpG-ODN; P, Poly I:C. After stimulation, cells were lysed and kinases were immunoprecipitated with antibodies to the HA tag. In vitro kinase assays were performed with GST-IκBα (B) or GST-cJun(79) (C) as substrate. Western blot analysis was performed with antibodies to IKKα (B) and JNK-1 (C). The radioactivity incorporated into the substrates was quantitated by PhosphorImager analysis. The values obtained were normalized against the radioactivity in nonstimulated cells and are presented as fold induction. The data shown are representative of three independent experiments.
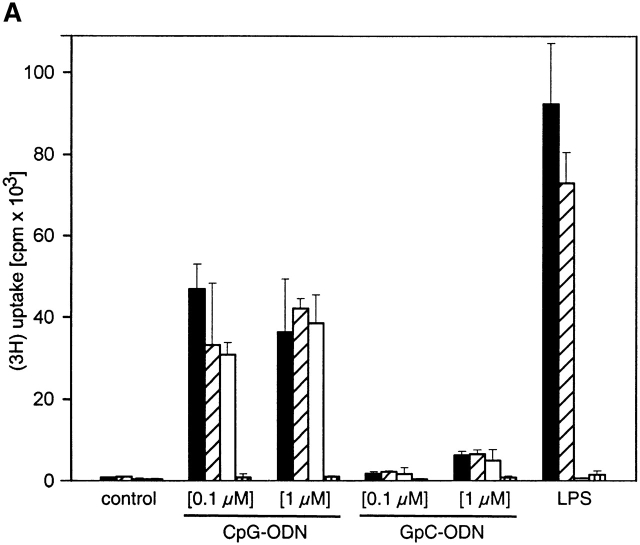
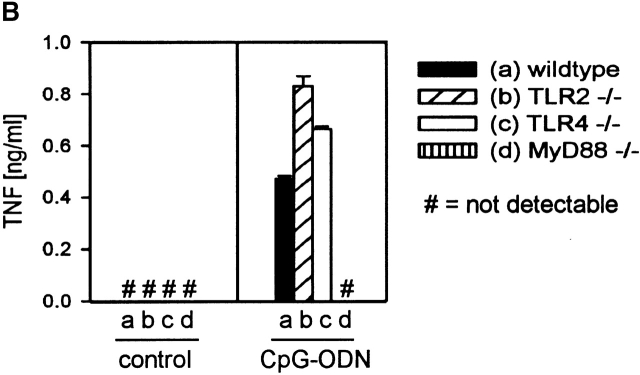
To investigate further whether MyD88 is an essential component for CpG-DNA–driven cell activation, we performed experiments with cells from MyD88-deficient mice. Cells from TLR2- and TLR4-deficient mice were included in these experiments, as TLR2 and TLR4 have been found to be engaged by PRFs and to signal via MyD88 3 4 5. Although spleen cells of wild-type, TLR2-deficient, and TLR4-deficient mice proliferated in response to CpG-DNA, spleen cells from MyD88-deficient mice did not (Fig. 3 A). Destruction of the CpG-motif within CpG-DNA by inversion of the CG dinucleotide to GC (GpC-ODN) abolished its potential to activate spleen cells (Fig. 3 A). As expected, LPS induced proliferation in wild-type and TLR2-deficient cells, but not in TLR4- or MyD88-deficient cells. Moreover, peritoneal macrophages from wild-type, TLR2-deficient, and TLR4-deficient mice produced TNF-α in response to CpG-DNA but peritoneal macrophages of MyD88-deficient mice did not (Fig. 3 B). These data imply that CpG-DNA responses are mediated through a MyD88-dependent but TLR2- and TLR4-independent pathway. Since mice with TLR defects are shown to be hyporesponsive to different PRFs 2, our data are consistent with the view that an unknown CpG-DNA receptor acts upstream of MyD88 and links CpG motif recognition to the TLR/IL-1R signaling pathway. TRAF6 seems to be a pivot in the activation of immature dendritic cells, as it is engaged by CD40 ligation via Th cells, by microbial cell wall components like LPS 11, and by CpG-DNA. All these results are in line with the concept that it is the TLR/IL-1R signaling pathway that translates distinct cell activation signals into a homogeneous biological response. MyD88 appears to be the first intracellular mediator of signaling through the TLRs and the IL-1R. If the situation is analogous to CpG-DNA signaling, MyD88 may be a handle to the molecular identification of the postulated immune receptor for bacterial DNA.
Figure 3.
MyD88 is essential for CpG-DNA–induced immune cell activation. (A) Splenocytes (105) of wild-type mice and TLR2-ng, TLR4-, and MyD88-deficient mice were cultured for 48 h in the presence of CpG-ODN or GpC-ODN (0.1 and 1.0 μM as indicated) or LPS (10 ng/ml). Cells were pulsed with 1 μCi of [3H]thymidine for the last 8 h and 3H uptake was measured in a β-scintillation counter. Each column represents the mean of three independent stimulations; error bars are SD. (B) Peritoneal macrophages (5 × 104) of wild-type mice and TLR4- and MyD88-deficient mice were stimulated with 1 μM CpG-ODN for 24 h and TNF concentrations were determined by ELISA. Each value shown represents the mean and SD of two independent stimulations. #, not detected. This figure represents the results of one out of two similar, independent experiments.
Acknowledgments
We thank Dr. Georg Häcker for critical and helpful discussion of the manuscript, and Sema Eser for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft SFB 391, Teilprojekt B-4, and BMBF grant 0311675/CpG Immunopharmaceuticals GmbH.
References
- Poltorak A., He X., Smirnova I., Liu M.Y., Huffel C.V., Du X., Birdwell D., Alejos E., Silva M., Galanos C. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr micemutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. Cutting edgeToll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Kirschning C.J., Wesche H., Merrill Ayres T., Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C.A., Jr. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- Raabe T., Bukrinsky M., Currie R.A. Relative contribution of transcription and translation to the induction of tumor necrosis factor-alpha by lipopolysaccharide. J. Biol. Chem. 1998;273:974–980. doi: 10.1074/jbc.273.2.974. [DOI] [PubMed] [Google Scholar]
- Muzio M., Ni J., Feng P., Dixit V.M. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- Wesche H., Gao X., Li X., Kirschning C.J., Stark G.R., Cao Z. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J. Biol. Chem. 1999;274:19403–19410. doi: 10.1074/jbc.274.27.19403. [DOI] [PubMed] [Google Scholar]
- Ishida T., Mizushima S., Azuma S., Kobayashi N., Tojo T., Suzuki K., Aizawa S., Watanabe T., Mosialos G., Kieff E. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J. Biol. Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- Lomaga M.A., Yeh W.C., Sarosi I., Duncan G.S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparwasser T., Miethke T., Lipford G., Erdmann A., Hacker H., Heeg K., Wagner H. Macrophages sense pathogens via DNA motifsinduction of tumor necrosis factor-alpha-mediated shock. Eur. J. Immunol. 1997;27:1671–1679. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- Hacker H., Mischak H., Hacker G., Eser S., Prenzel N., Ullrich A., Wagner H. Cell type-specific activation of mitogen-activated protein kinases by CpG-DNA controls interleukin-12 release from antigen-presenting cells. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:6973–6982. doi: 10.1093/emboj/18.24.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv. Immunol. 1999;73:329–368. doi: 10.1016/s0065-2776(08)60790-7. [DOI] [PubMed] [Google Scholar]
- Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., Koretzky G.A., Klinman D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Murphy T.L., Cleveland M.G., Kulesza P., Magram J., Murphy K.M. Regulation of interleukin 12 p40 expression through an NF-kappa B half-site. Mol. Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Minden A., Martinetto H., Claret F.X., Lange-Carter C., Mercurio F., Johnson G.L., Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- Baud V., Liu Z.G., Bennett B., Suzuki N., Xia Y., Karin M. Signaling by proinflammatory cytokinesoligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M., Shirakata Y., Inoue J.I., Nakano H., Okazaki K., Okumura K., Yamamoto T., Nagaoka H., Takemori T. Tumor necrosis factor receptor–associated factor 6 (TRAF6) stimulates extracellular signal–regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J. Exp. Med. 1998;187:237–244. doi: 10.1084/jem.187.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D.V. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- Yi A.K., Tuetken R., Redford T., Waldschmidt M., Kirsch J., Krieg A.M. CpG motifs in bacterial DNA activate leukocytes through the pH-dependent generation of reactive oxygen species. J. Immunol. 1998;160:4755–4761. [PubMed] [Google Scholar]
- Chapman R.E., Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M., Natoli G., Saccani S., Levrero M., Mantovani A. The human Toll signaling pathwaydivergence of nuclear factor κB and JNK/SAPK activation upstream of tumor necrosis factor receptor–associated factor 6 (TRAF6) J. Exp. Med. 1998;187:2097–2101. doi: 10.1084/jem.187.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.M., Ostertag D., Li Z.W., Chang L., Chen Y., Hu Y., Williams B., Perrault J., Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- Clemens M.J., Elia A. The double-stranded RNA-dependent protein kinase PKRstructure and function. J. Interferon. Cytokine. Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- Lord K.A., Hoffman-Liebermann B., Liebermann D.A. Nucleotide sequence and expression of a cDNA encoding MyD88, a novel myeloid differentiation primary response gene induced by IL6. Oncogene. 1990;5:1095–1097. [PubMed] [Google Scholar]
- Xu L., Sanchez A., Yang Z., Zaki S.R., Nabel E.G., Nichol S.T., Nabel G.J. Immunization for Ebola virus infection. Nat. Med. 1998;4:37–42. doi: 10.1038/nm0198-037. [DOI] [PubMed] [Google Scholar]
- Wesche H., Henzel W.J., Shillinglaw W., Li S., Cao Z. MyD88an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J.L., Di Marco F., French L., Tschopp J. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]