Pag3/Papα/Kiaa0400, a Gtpase-Activating Protein for Adp-Ribosylation Factor (Arf), Regulates Arf6 in Fcγ Receptor–Mediated Phagocytosis of Macrophages (original) (raw)
Abstract
The Fcγ receptor (FcγR)-mediated phagocytosis of macrophages is a complex process where remodeling of both the actin-based cytoskeleton and plasma membrane occur coordinately. Several different families of small GTPases are involved. We have isolated a GTPase-activating protein (GAP) for ADP-ribosylation factor (ARF), paxillin-associated protein with ARFGAP activity (PAG)3/Papα/KIAA0400, from mature monocytes and macrophage-like cells. Mammalian ARFs fall into three classes, and the class III isoform (ARF6) has been shown to be involved in FcγR-mediated phagocytosis. Here we report that PAG3 is enriched together with ARF6 and F-actin at phagocytic cups formed beneath immunoglobulin G–opsonized beads in P388D1 macrophages, in which overexpression of ARF6, but not ARF1 (class I) or ARF5 (class II), inhibits the phagocytosis. Overexpression of PAG3, but not its GAP-inactive mutant, attenuated the focal accumulation of F-actin and blocked phagocytosis, although surface levels of the FcγRs were not affected. Other ubiquitously expressed ARFGAPs, G protein–coupled receptor kinase interactors GIT2 and GIT2-short/KIAA0148, which we have shown to exhibit GAP activity for ARF1 in COS-7 cells, did not accumulate at the phagocytic cups or inhibit phagocytosis. Moreover, cooverexpression of ARF6, but not ARF1 or ARF5, restored the phagocytic activity of PAG3-overexpressing cells. We propose that PAG3 acts as a GAP for ARF6 and is hence involved in FcγR-mediated phagocytosis in mouse macrophages.
Keywords: actin remodeling, ARF family GTPases, ARFGAP, membrane remodeling, Rho family GTPases
Introduction
During Fcγ receptor (FcγR)-mediated phagocytosis, remodeling of actin-based cytoskeletal organization and remodeling of plasma membrane structure should be coordinately regulated near and at the phagocytic cups 1 2. Processes for phagocytosis involve accumulation of F-actin beneath bound particles and their dynamic remodeling conforming to the cytoarchitecture of phagocytic cups during engulfment of the particles 3. Engulfment of even large numbers of particles by macrophages is not accompanied by an apparent reduction in surface area, indicating that membrane components are supplied during phagocytosis, possibly by exocytosis of intracellular vesicles or endosomal membrane 2 4 5. In accordance with the complex processes of FcγR-mediated phagocytosis, several distinct families of Ras superfamily small GTPases have been shown to be involved. Inhibition of Cdc42 and Rac1 has been shown, by expression of dominant negative mutants, to inhibit FcγR-mediated phagocytosis by preventing the accumulation of F-actin at the phagocytic cup, though it does not affect particle binding to FcγRs 6 7 8. Involvement of Rho in FcγR-mediated phagocytosis is controversial. It has been shown that inhibition of RhoA by injection of C3 exotoxin inhibits FcγR-mediated phagocytosis by preventing the accumulation of F-actin beneath bound particles and receptor clustering in J774 macrophages as well as blocking FcγR-mediated phagocytosis in FcγRIIA-transfected COS cells 9. On the other hand, recent studies suggest that FcγR-mediated phagocytosis in J774 macrophages may be independent of RhoA function 7 8.
ADP-ribosylation factor (ARF) 6 , an isoform of the ARF family GTPases that also belong to Ras superfamily small GTPases, has also been shown to be involved in the phagocytosis 10. ARFs were originally identified and named for their ability to serve as cofactors in the cholera toxin–catalyzed ADP-ribosylation of the α subunit of Gs 11 and have been shown to function in various membrane trafficking events and in the maintenance of organelle structure 12 13. Members of the family include six isoforms of ARF, and the ARF-like proteins in mammalian cells 14 15. The six ARF isoforms are highly homologous to one another and are classified as class I, II, or III based on sequence similarity 14. Class I includes ARF1, 2, and 3, class II includes ARF4 and 5, and class III includes ARF6. Among them, ARF1 has been most thoroughly studied and has been shown to regulate membrane traffic at multiple sites within the cell, such as endoplasmic reticulum–to-Golgi transport, and to activate phospholipase D 16. The GTP-bound form of ARF1 recruits protein coats, including the clathrin-associated adaptor proteins and the nonclathrin coatomers, to the donor membrane and initiates budding of the membrane to form vesicles; subsequent hydrolysis of the bound GTP to GDP at the target membrane appears to trigger disassembly of the coat from the vesicle, which is necessary for the vesicle to fuse to the target membrane 13 16. On the other hand, ARF6, the ARF which is most distantly related to ARF1, shows a rather wide distribution in the cytoplasm and localizes to an endosomal compartment and membrane ruffling regions. It has been shown to regulate endosomal trafficking and receptor-mediated endocytosis at the cell periphery, recycling of endosomal vesicles to the plasma membrane, actin rearrangements beneath the plasma membrane, and cell spreading 13 17 18 19 20 21 22 23 24 25. ARF6 appears to cycle between the plasma membrane and the recycling endosomes, depending on its nucleotide status; it has been shown that the GTP hydrolysis-defective mutant of ARF6 accumulates at the plasma membrane, and that the GTP binding defective mutant of ARF6 localizes to the recycling endosomes 20. The function of the class II ARFs and the ARF-like proteins is not yet clear. It is noteworthy that, unlike other small GTPase family proteins such as Ras family and Rho family proteins, the intrinsic GTPase activity of ARF proteins is almost undetectable in vitro 11; thus, GTPase-activating proteins (GAPs) appear to be essential for ARFs in their transition from the GTP-bound form to the GDP-bound form.
The involvement of ARF6 in FcγR-mediated macrophage phagocytosis has been demonstrated 10. In this case, both types of ARF6 mutants, one defective in GTP hydrolysis (Q67L) and the other defective in GTP binding (T27N), have been shown to inhibit phagocytosis of IgG-coated erythrocytes and to attenuate the focal accumulation of F-actin beneath the test particles in mouse RAW 264.7 macrophages (hereafter called RAW). Moreover, overexpression of wild-type ARF6 can also exert inhibitory effects on FcγR-mediated phagocytosis 10. On the other hand, it has been also shown that brief treatment (30-min incubation) of macrophage cells with brefeldin A (BFA) does not affect phagocytosis, although the organization and function of the Golgi apparatus in the BFA-treated cells was disrupted 10. BFA is known to inhibit activities of several nucleotide exchanging factors for ARFs and hence inhibits the function of ARFs, with the exception of ARF6 12 18 26 27. Thus, BFA-sensitive ARF isoforms appeared to be nonessential for the phagocytosis 10.
It has recently been shown in several types of cells, such as HeLa, Swiss 3T3, and NRK cells, that actin cytoskeletal structure and membrane structure are regulated as a consequence of intercommunication between Rho family GTPases and ARF family GTPases 22 28 29. In RAW macrophage cells, Rac1-mediated membrane ruffling and actin polymerization have been shown to require the participation of ARF6 23. Therefore, it is likely that intercommunication between Rho and ARF family GTPases also plays an essential role during phagocytosis, for the coordinated regulation of remodeling of actin-based cytoskeletal organization, remodeling of plasma membrane structure, and receptor trafficking as well 1 2. Moreover, it should be also noted that both Rac1 and ARF6 also participate in the activation of phagocyte NADPH oxidase 30 31.
We have isolated a GAP for ARF, paxillin-associated protein with ARFGAP activity (PAG)3, from mature monocytes and macrophage-like cells, as a binding protein of paxillin 32. Paxillin is one of the integrin assembly focal adhesion proteins and also exists at FcγR-mediated phagocytic cups in macrophages 33 34. PAG3 is induced to be expressed on monocyte maturation and localizes to the cell periphery, together with paxillin 32 35. PAG3 is identical to KIAA0400 36 and Papα, which was identified as a Pyk2-binding protein 37. Our previous study using COS-7 cells revealed that PAG3 exhibits a GAP activity for ARF6 rather than ARF1 32, although others have shown that it exhibits phosphatidylinositol 4,5-bisphosphate–dependent GAP activity on ARF1 and ARF5 and less activity on ARF6 in vitro 37. We report here that PAG3 is colocalized with ARF6 and F-actin at phagocytic cups formed beneath IgG-opsonized beads in mouse P388D1 macrophage cells. We also found that ARF6, but not ARF1 or ARF5, is involved in FcγR-mediated phagocytosis in P388D1 macrophage cells, consistent with a previous report with RAW macrophage cells 10. These results prompted us to investigate whether PAG3 acts as a GAP for ARF6 during FcγR-mediated phagocytosis. To do this, we also employed other ARFGAPs, G protein–coupled receptor kinase interactor GIT2 and its spliced variant, GIT2-short/KIAA0148 38 39 40 41, as controls. GIT2 and GIT2-short also bind to paxillin, but with different affinities: GIT2 binds paxillin with a strong affinity, whereas GIT2-short binds paxillin with a weak affinity 38. We demonstrate that overexpression of PAG3, but not its GAP-inactive mutant or GIT2s, inhibits phagocytosis by attenuating focal accumulation of F-actin and provides evidence that PAG3 acts as a GAP for ARF6 during the FcγR-mediated phagocytosis of mouse P388D1 cells. Possible roles of PAG3 and its binding to paxillin and other proteins in linking regulation between ARF and Rho family GTPases during FcγR-mediated phagocytosis are also discussed.
Materials and Methods
Materials.
RPMI 1640, FCS, and β-mercaptoethanol were purchased from Life Technologies. Anti–influenza hemagglutinin (HA) mAb was purchased from BAbCO, anti-ARF6 mAb was from Santa Cruz Biotechnology, Inc., and Cy2-, Cy3-, Cy5-, or horseradish peroxidase–conjugated secondary Abs were from Jackson ImmunoResearch Laboratories. Unless indicated otherwise, all other chemicals were obtained from Sigma-Aldrich or Wako.
Cell Culture.
Mouse macrophage cell line P388D1 was a gift from Y. Ito (Institute for Virus Research, Kyoto University). Cells were maintained in RPMI 1640 supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM β-mercaptoethanol at 37°C under 5% CO2 in a humidified incubator.
Plasmids.
Wild-type PAG3 and its ARFGAP activity–deficient mutant, C436A PAG3, cDNAs were subcloned into pEGFP-C1 vector (CLONTECH Laboratories, Inc.) under the control of the CMV promoter, as described previously 32. cDNA of enhanced green fluorescent protein (EGFP)-tagged PAG3 from pEGFP-PAG3 was subcloned into pBabePuro vector 42 under the control of the murine retrovirus LTR promoter. The HA epitope–tagged GIT2 and GIT2-short expression vectors (pcDNA3/HA-GIT2 and pcDNA3/HA-GIT2-short) and their ARFGAP activity–deficient C11A mutants (pcDNA3/HA-GIT2CA and pcDNA3/HA-GIT2-shortCA) have been described previously 38. cDNAs in pcDNA3 vectors, each encoding ARF1-HA, ARF5-HA, ARF6-HA, and ARF6 Q67L-HA (GTP hydrolysis–deficient mutant), were gifts from K. Nakayama (Tsukuba University, Ibaraki, Japan), and all have the HA tag at their COOH terminals. pcDNA3 encoding ARF6 T27N-HA (GTP binding–deficient mutant) was constructed as described previously 12. Nucleotide sequences were confirmed for all the plasmids after construction.
Transfection.
P388D1 cells (106) were plated onto 35-mm culture dishes (Becton Dickinson) and transfected 24 h later using FuGENE6 (Boehringer) according to the manufacturer's instructions. After transfection, cells were incubated overnight for 16–20 h and then replated in 8-well glass chamber slides (Becton Dickinson) at a density of 104 cells/well, and subjected to each assay after further incubation for 16 h. For moderate expression, pBabe vector, in which cDNA was transcribed from the murine retrovirus LTR promoter, was used. For overexpression, pcDNA3 vector, pEGFP-C1 vector, or its derivatives were used, which contain the CMV promoter. 2 μg of each plasmid was used per 35-mm culture dish. For cotransfection, 1.5 μg of pEGFP-PAG3s was used together with 0.5 μg of pcDNA3/ARFs-HA, as determined by preliminary titration experiments, unless otherwise indicated.
Preparation of IgG-opsonized Beads.
Opsonized microspheres were prepared as described previously 43. In brief, polystyrene beads (3.0-μm diameter; Polysciences, Inc.) were washed twice in PBS and incubated in 8% glutaraldehyde for 16 h. After incubation with 0.1% BSA for 5 h, beads were washed in PBS and incubated for 16 h in 0.5 M ethanolamine. Beads were incubated with 0.2 mg/ml of rabbit polyclonal or mouse monoclonal anti-BSA IgGs for 4 h. All incubations were performed at 25°C. Opsonized beads were washed twice in ice-cold PBS, resuspended in RPMI 1640, and counted in a hemacytometer. Opsonization of the IgG-coated beads was confirmed by positive and negative counterstaining using Cy2-conjugated anti–rabbit IgG and Cy2-conjugated anti–mouse IgG, respectively. Mouse IgG–opsonized beads were used for the simultaneous detection of endogenous PAG3, and rabbit IgG–opsonized beads were used for the other experiments.
Phagocytosis Assay.
P388D1 cells transiently transfected with the indicated constructs were placed in ice-cold medium containing IgG-opsonized beads at a concentration of 50 beads per target cell. Cells were then incubated at 4°C for 20 min to allow attachment of particles to cell surfaces, followed by incubation at 37°C for 30 min, unless otherwise described, for particle internalization. Cells were then washed in ice-cold HBSS to remove unbound beads. For staining attached but not internalized particles, cells were incubated with Cy5-conjugated anti–rabbit IgG before being permeabilized 10. The transfection-positive cells were identified by staining the exogenous proteins (see below). Internalized particles in the transfection-positive cells were counted by phase–contrast microscope. Attached but not internalized particles were identified as the Cy5+ staining. 50 cells expressing each indicated construct and each of the 50 cells of the nonexpressing control were scored for the attached and internalized beads. The association index indicates the number of attached and internalized beads per single cell. The phagocytosis index indicates the number of internalized beads per single cell. The attachment index indicates the number of attached beads per single cell, after incubation only at 4°C for 20 min. Data were expressed as an average (± SE) of three independent experiments (a total of 150 transfection-positive cells were examined in each assay). The difference between transfectants and control cells was compared with a Student's t test. An asterisk denotes significant differences between control cells (P < 0.001).
Immunofluorescence Characterization.
P388D1 cells and cells transiently transfected with the indicated constructs were subjected to phagocytosis assay as described above, then fixed in 4% paraformaldehyde for 10 min at room temperature. After permeabilization with 0.1% Triton X-100 in PBS for 10 min at room temperature, expressions of PAG3s, GIT2s, and ARFs were visualized by immunostaining with appropriate Abs or detecting the autofluorescence from the EGFP tag, as described previously 32 38. Confocal images were acquired using a confocal laser scanning microscope (model 510; Carl Zeiss, Inc.). Focus was adjusted across the center of the majority of phagocytic beads (∼2–4 μm above the surface of the glass chamber plates). Expression levels of exogenous proteins were analyzed for each cell by quantifying the intensities of the fluorescent signals using the computer software associated with the confocal laser scanning microscope (LSM 510 version 2.5; Carl Zeiss, Inc.). Nonspecific background fluorescence levels were then determined by staining the cells with irrelevant Abs coupled with the appropriate dye-conjugated secondary Abs. Also, we confirmed beforehand that the exogenous expression of proteins transcribed from the CMV promoter (in pcDNA3 and pEGFP vectors) was at least 5–10-fold higher than that from the LTR promoter (in pBabe vector) in most cells, and that the exogenous expression of proteins such as PAG3 from the LTR was much the same or only slightly higher than the endogenous expression seen in P388D1 cells. Each figure of microscopic analysis showed representative results that were observed in a majority (>50–80%) of the transfected cells from three independent experiments (>200 cells).
Immunoblotting.
P388D1 cells and cells transfected with the indicated constructs were lysed on ice with 1% NP-40 buffer (1% NP-40, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 5 mM EDTA, 1 mM Na3VO4, 10 μM Na2MoO4, 1 mM PMSF, 1% aprotinin, 2 μg/ml leupeptin, 3 μg/ml pepstatin A) as described previously 44. Protein concentrations were determined using a Dc protein assay kit (Bio-Rad Laboratories) with BSA as a standard. Each 20 μg of protein was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were then incubated with mouse anti-HA mAb or mouse anti-GFP mAb (CLONTECH Laboratories, Inc.), followed by horseradish peroxidase–conjugated anti–mouse IgG. Specific binding was detected using enzyme-linked chemiluminescence, according to the manufacturer's instructions (Amersham Pharmacia Biotech). Each figure shows representative results from at least two independent experiments.
Cell Surface FcγR Staining.
P388D1 cells were transfected using FuGENE6 with 2 μg of pEGFP-PAG3s or 2 μg of pEGFP-C1 empty vector as a control. For HA-tagged proteins, each 1.8 μg of pcDNA3 plasmid was cotransfected with 0.2 μg of pEGFP-C1 empty vector as a marker. Only transfection-positive cells, as detected by autofluorescence of the EGFPs 45 46, were then subjected to analysis of their surface expression of FcγRII/III. For analyzing FcγRII/III expressions, transfected cells (106) were incubated with 5 μg/ml of PE-conjugated 2.4G2 mAb (BD PharMingen) or isotype-matched irrelevant control Abs, in PBS supplemented with 2% FCS and 0.01% sodium azide for 30 min at 4°C. Cells were then washed three times in PBS, and fluorescence intensities were measured by a FACScan™ flow cytometer (Becton Dickinson).
Results
PAG3 Accumulates in Phagocytic Cups Together with F-Actin and ARF6.
We have shown that an ARFGAP, PAG3, is expressed in human mature monocyte and macrophage-like cells, is localized at the cell periphery, and acts as a GAP for ARF6 in COS-7 cells 32. ARF6 has been shown to be involved in the FcγR-mediated phagocytosis in macrophages 10. Therefore, we examined a possible role of PAG3 in the FcγR-mediated phagocytosis in macrophages in this study.
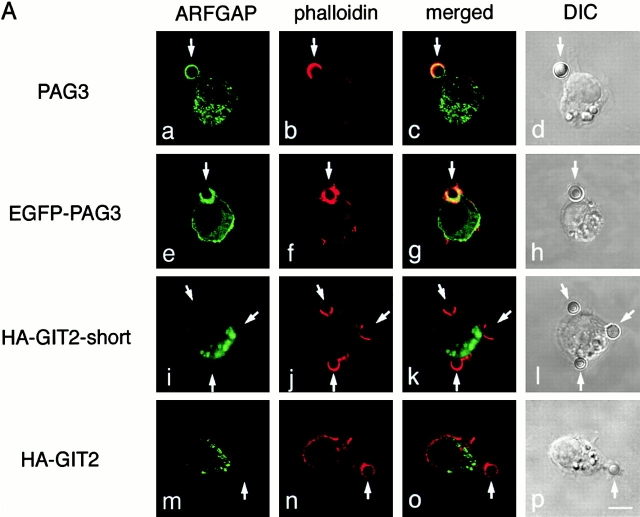
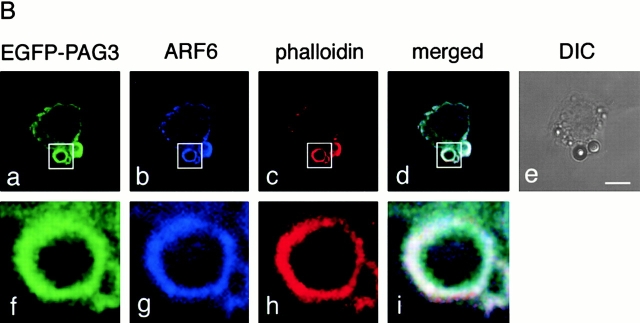
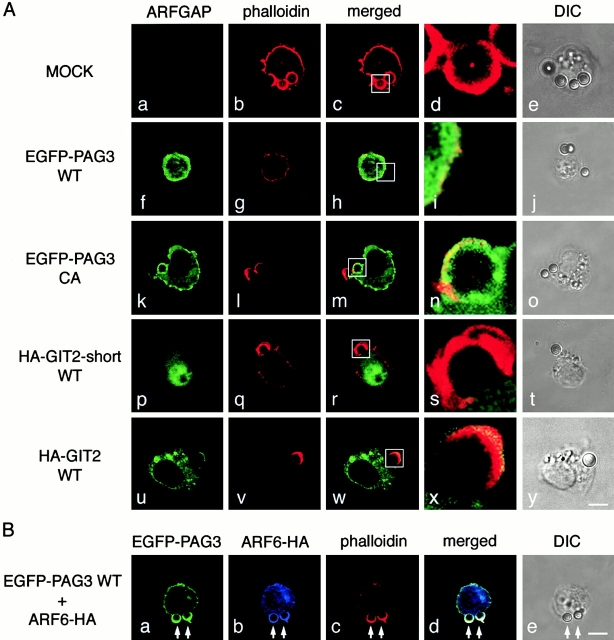
First, we investigated the subcellular localization of PAG3 in phagocytic macrophage cells. P388D1 mouse macrophages were briefly incubated at 37°C with IgG-opsonized beads, fixed, and stained for endogenous PAG3 and actin filaments. As shown in Fig. 1 A, PAG3 was found to accumulate at the phagocytic cups formed beneath the IgG-opsonized beads, and was seen to largely, but not completely, overlap with F-actin. Next we examined whether PAG3 colocalized with ARF6 at the phagocytic cups. For this purpose, an EGFP-tagged PAG3 was used, since the opsonized beads used for this assay had already been coated with rabbit or mouse IgG, and we only have access to rabbit IgG for PAG3 and mouse IgG for ARF6. EGFP-PAG3 was expressed from the pBabe vector to ensure only moderate levels of expression to avoid possible effects attributable to its overexpression (see Materials and Methods). We have previously confirmed that PAG3 can be tagged with EGFP without significant biological alteration, retaining its subcellular localization 32. Accumulation of EGFP-PAG3 was then clearly detected at the phagocytic cups, together with ARF6 and F-actin (Fig. 1 B).
Figure 1.
PAG3, but not GIT2s, accumulates to phagocytic cups, together with F-actin and ARF6. P388D1 cells and cells transfected with pBabe/EGFP-PAG3, pBabe/HA-GIT2-short, or pBabe/HA-GIT2 were incubated with IgG-opsonized beads for 20 min at 4°C, and fixed after incubation at 37°C for 5 min. (A) Endogenous PAG3 (panel a) was visualized using an anti-PAG3 Ab coupled with Cy2-anti–rabbit IgG, and EGFP-PAG3 (panel e), HA-GIT2-short (panel i), and HA-GIT2 (panel m) were visualized by detecting the auto-fluorescence from the EGFP, or by staining cells with an anti-HA mAb coupled with Cy2-anti–mouse IgG. F-actin was visualized with Texas red X–conjugated phalloidin (panels b, f, j, and n). The third panels represent the merging of the left two panels. The right panels show the differential interference contrast (DIC) images of the same fields of the three preceding panels. Arrows indicate positions of phagocytic beads. The expression levels of EGFP-PAG3 in each cell were almost equal or only slightly higher compared with the levels of endogenous PAG3, as assessed by staining EGFP-PAG3–expressing cells with an anti-PAG3 Ab, coupled with Cy3-anti–rabbit IgG, and comparing their fluorescent signals with those of untransfected cells (the transfected cells gave 2.4 ± 0.1–fold higher signals than the untransfected cells; see Materials and Methods). (B) EGFP-PAG3 (panel a) and F-actin (panel c) were visualized as above; endogenous ARF6 was detected using anti-ARF6 mAb coupled with Cy5-anti–mouse IgG (panel b). Panel d represents the merging of panels a–c, and panel e shows the DIC images of the same fields of panels a–d. Higher magnifications of images of panels a–d (indicated by rectangles) are each shown below (panels f–i). Bars, 5 μm.
To assess the specificity of PAG3 localization to the phagocytic cups, we employed two other ubiquitously expressed ARFGAPs, GIT2 and its short isoform, GIT2-short/KIAA0148 38 39 40 41. Like PAG3, both GIT2 and GIT2-short are able to bind to paxillin, although their affinities differ significantly 38. Because Abs raised against the GIT2 proteins were weak for immunostaining, we expressed the HA-tagged cDNAs that were also constructed in the pBabe vector to ensure moderate levels of expression. As shown in Fig. 1 A, HA-GIT2 and HA-GIT2-short did not accumulate at the phagocytic cups, unlike in the case of PAG3.
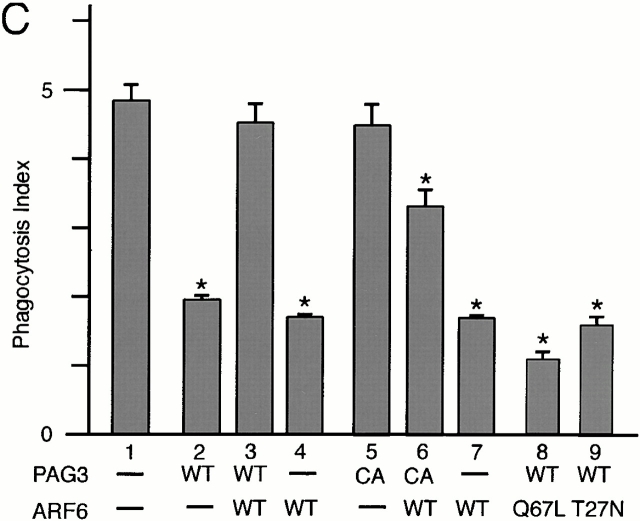
It has been shown that simple overexpression of wild-type ARF6 blocks FcγR-mediated phagocytosis in RAW cells 10. It has also been demonstrated that ARF isoforms whose activities are influenced by BFA may not be essential for the phagocytosis 10. Therefore, we examined whether or not each class of ARF isoforms participates in the phagocytic activities in our experimental system. The HA-tagged ARF isoforms were overexpressed using the CMV promoter of the pcDNA3 vector, and expression levels of the HA-tagged ARFs in each cell were examined using confocal laser scanning microscopy and its attached computer software, before data acquisition (see Materials and Methods). ARF1 was used to represent class I and ARF5 was used to represent class II. As shown in Fig. 2 B, transient overexpression of wild-type ARF6-HA reduced the number of IgG-opsonized beads ingested by the transfected cells, as has been shown with RAW cells 10. In contrast, transient overexpression of ARF1-HA and ARF5-HA, which were expressed at similar levels to ARF6-HA, did not exert such inhibitory effects on phagocytosis (Fig. 2 B).
Figure 2.
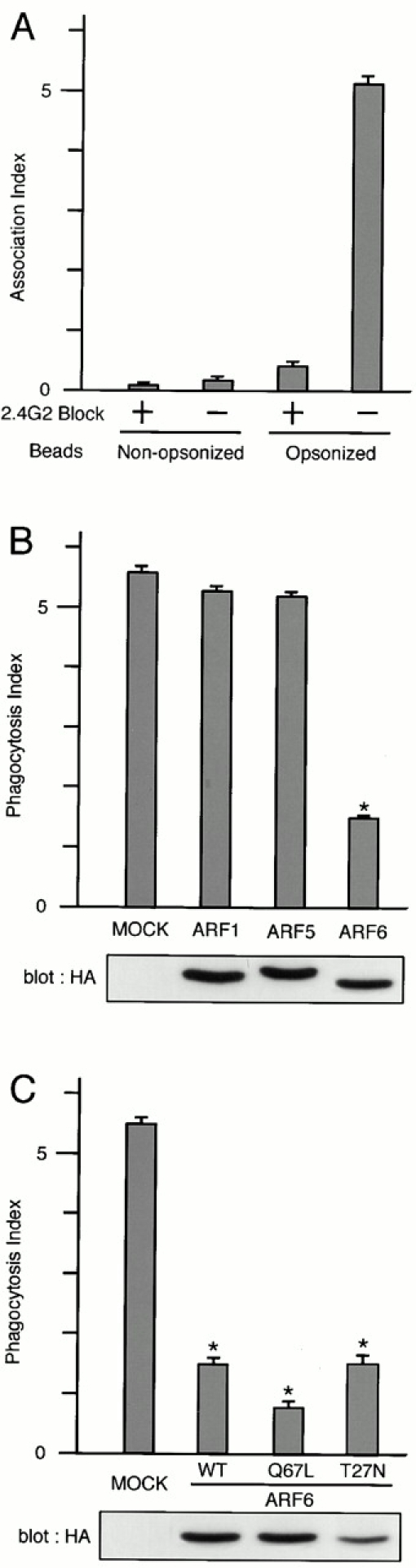
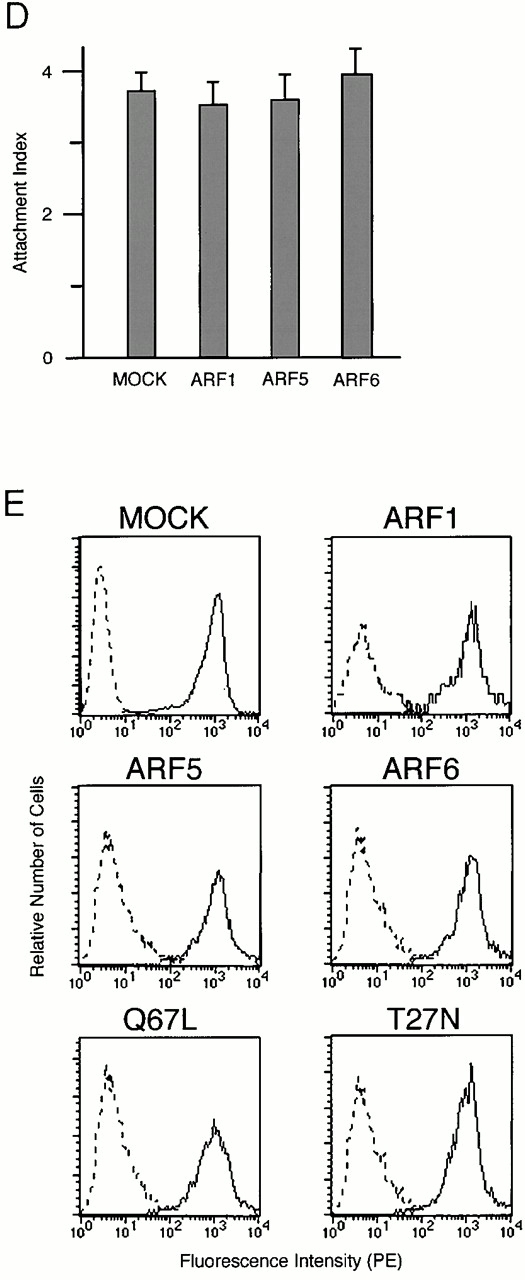
Effects of overexpression of ARF isoforms on FcγR-mediated phagocytosis. (A) Phagocytic activities of P388D1 cells against IgG-opsonized and nonopsonized polystyrene beads. P388D1 cells, preincubated with (+) or without (−) anti–mouse FcγRII/III mAb 2.4G2 for 30 min at 4°C, were incubated with polystyrene beads nonopsonized or opsonized with rabbit IgG for 20 min at 4°C, followed by 30 min at 37°C. After removal of unbound beads, the number of beads attached and ingested per single cell (the association index) was counted as described in Materials and Methods. (B and C) Inhibition of FcγR-mediated phagocytosis by overexpression of the class III ARF (ARF6), but not other classes of ARFs. P388D1 cells transiently transfected with the indicated constructs in pcDNA3 vectors were incubated with IgG-opsonized beads as above. Each ARF cDNA was tagged with HA tag. The number of beads ingested per single cell (the phagocytosis index) was determined for cells expressing the indicated constructs and for nonexpressing controls (MOCK), as described in Materials and Methods. Wild-type cDNAs of ARF1, ARF5, and ARF6 were used in B, and wild-type and Q67L and T27N mutants of ARF6 were used in C. The exogenous expression of HA-tagged ARFs, as detected by immunoblotting using anti-HA mAb, is shown below. The expression levels of exogenous wild-type ARF6-HA were 8.1 ± 0.5–fold higher than endogenous ARF6. This estimation was obtained by immunostaining the mock-transfected cells and cells expressing ARF6-HA using an anti-ARF6 mAb coupled with Cy3-anti–mouse IgG, and differences in fluorescent intensities of single cells in these two samples were compared as described in Materials and Methods. Abs that specifically recognize ARF1 or ARF5 were not available. (D and E) Overexpression of ARF isoforms does not affect the binding to IgG-opsonized beads and surface expression of FcγRs. Binding to IgG-opsonized beads was assessed by incubating cells with the beads for 20 min at 4°C. Unbound beads were then removed by washing, and numbers of beads attached to the transfected cells are shown (the attachment index) (D). Other procedures are as described above. (E) Surface levels of FcγRs on cells transiently transfected with the indicated ARF cDNA constructs and, hence, expressing the exogenous proteins were assessed by staining the nonpermeabilized cells with 2.4G2 mAb (solid line; see Materials and Methods). Control uses irrelevant rat IgG (broken line). In A–D, data are expressed as the mean ± SE of three independent experiments. *P < 0.001.
Unlike other small GTPases such as the Rho and Ras family members, it has been well documented that both the GTP hydrolysis–deficient and the GTP binding–deficient mutants of ARF proteins are able to exhibit inhibitory effects on the trafficking and recycling of cell surface receptors 10 17. Similarly, blockage of FcγR-mediated phagocytosis was also observed in the transient overexpression of both the GTP hydrolysis–deficient (Q67L) and the GTP binding–deficient (T27N) mutants of ARF6 in our system (Fig. 2 C), again similar to those reported previously with RAW cells 10.
On the other hand, the binding ability of FcγRs to the IgG-opsonized beads, as assessed by incubating macrophages with the test beads only at 4°C, was not significantly affected by overexpression of these proteins, including ARF6 mutants (Q67L and T27N) (Fig. 2 D; data not shown). Surface expression of FcγRs, as assessed by staining cells with anti–mouse FcγRII/III mAb 2.4G2 was also almost unaffected by these proteins (Fig. 2 E). We also confirmed that P388D1 cells did not exhibit significant phagocytic activity towards nonopsonized polystyrene beads, and that phagocytosis of the IgG-coated beads in P388D1 cells was substantially inhibited by preincubation of the cells with anti-FcγRII/III mAb 2.4G2 (Fig. 2 A), as has been shown previously with other macrophage cells 47.
Overexpression of PAG3, but Not Its GAP-inactive Mutant or GIT2s, Inhibits FcγR-mediated Phagocytosis.
Next we examined whether PAG3 is involved in FcγR-mediated phagocytosis. Although our previous study indicates that PAG3 can act as a GAP for ARF6 in COS-7 cells 32, it has been proposed that different GAPs may be involved in the recruitment of different coatomer proteins to the same ARF 48. Therefore, we investigated whether PAG3 acts on ARF6 during FcγR-mediated phagocytosis in macrophages.
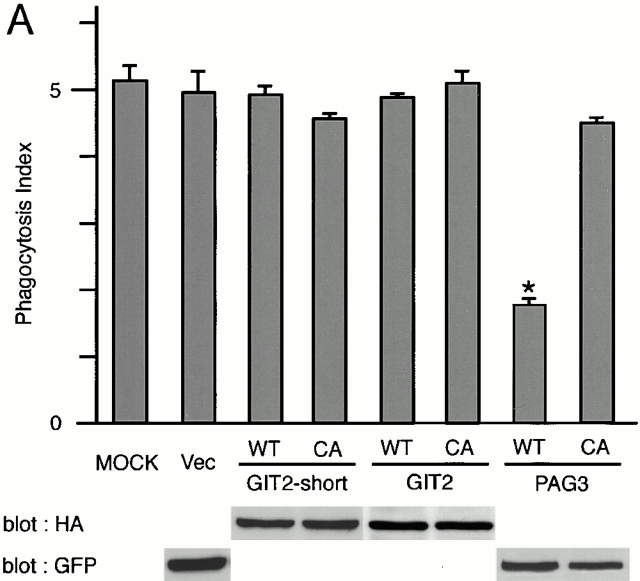
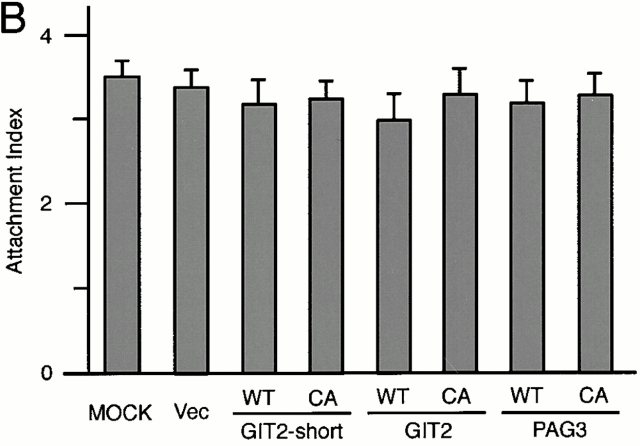
It has been shown previously that the overexpression of ARFGAPs, but not their GAP-inactive mutants, are able to act in an inhibitory manner towards ARF functions in several assay systems 32 38. Therefore, PAG3 and GIT2s were overexpressed at high levels using the CMV promoter in P388D1 cells. As shown in Fig. 3 A, overexpression of EGFP-PAG3 inhibited phagocytosis of IgG-opsonized beads. In contrast, such inhibition was not observed by the GAP-inactive CA mutant of EGFP-PAG3, nor by HA-GIT2, HA-GIT2-short, and their GAP-inactive CA mutants, although they were all overexpressed at similar levels (Fig. 3 A). Again, surface expression of the FcγRII/III and the ability to bind opsonized beads were not significantly altered by the overexpression of these proteins, including wild-type EGFP-PAG3 (Fig. 3b and Fig. c).
Figure 3.
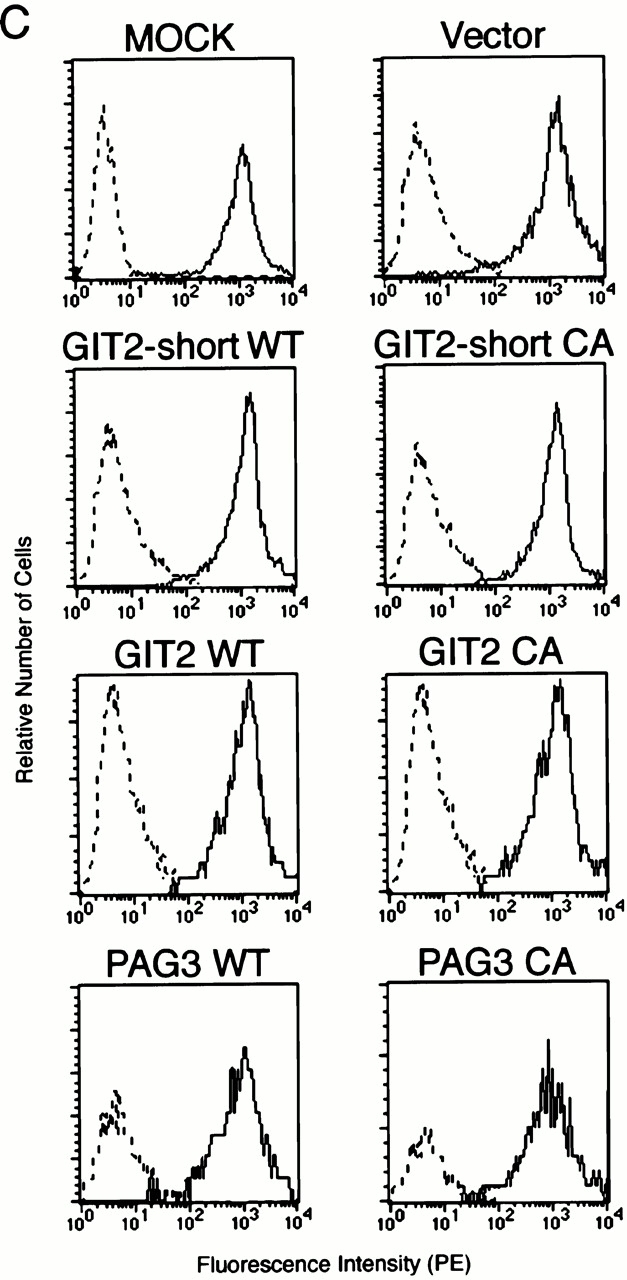
Overexpression of PAG3, but not its GAP-inactive mutant or GIT2s, inhibits FcγR-mediated phagocytosis. (A and B) P388D1 cells (MOCK) and cells transiently transfected with the empty pEGFP-C1 vector (Vec), or the plasmids containing cDNAs for wild-types (WT) or the GAP-inactive mutants (CA) of EGFP-PAG3, HA-GIT2, and HA-GIT2-short, were incubated with IgG-opsonized beads for 20 min at 4°C, followed by 30 min at 37°C to define the phagocytosis index (A); or only for 20 min at 4°C to define the attachment index (B). Cells expressing the exogenous proteins were examined as described in Materials and Methods. Data are expressed as the mean ± SE of three independent experiments. *P < 0.001. The exogenous expression of HA-tagged GIT2s, EGFP-tagged PAG3s and EGFP, detected by immunoblotting using anti-HA mAb and anti-GFP mAb, is shown in A. (C) Equivalent expression of FcγRs on cells transiently transfected with the indicated constructs was demonstrated by flow cytometry analysis. Surface levels of FcγRs of only transfection-positive cells were assessed as in the legend to Fig. 2. 2.4G2 staining (solid line) and rat IgG control (broken line) are shown. Mock-transfected cells without plasmid DNA were also included as a control. Overexpression of EGFP-PAG3 was confirmed by immunostaining of the mock-transfected cells and cells expressing EGFP-PAG3 with polyclonal anti-PAG3 Abs coupled with Cy3-anti–rabbit IgG, and differences in fluorescent intensities of single cells from these two samples were compared as in the legend to Fig. 2. The level of overexpression of EGFP-PAG3 was estimated to be 23.7 ± 1.1–fold higher than that of endogenous PAG3.
Overexpression of PAG3 Affects Actin–Cytoskeletal Remodeling.
ARF6 has been shown to be involved in actin rearrangements in HeLa and Chinese hamster ovary cells 18 21 22 23 24 25, and also in the phagocytosis of particles in RAW cells 10. Consistent with the notion that PAG3 acts as a GAP for ARF6 in the phagocytosis, we found that overexpression of EGFP-PAG3, but not its GAP-inactive mutant, attenuated the focal accumulation of F-actin beneath IgG-opsonized particles and inhibited phagocytic cup formation (Fig. 4 A), though cells overexpressing EGFP-PAG3 could bind to the test beads almost normally, as already described in Fig. 3 B (see also Fig. 4 A). On the other hand, overexpression of HA-GIT2 and HA-GIT2-short did not affect the focal accumulation of F-actin and phagocytic cup formation (Fig. 4 A), again supporting the distinct and specific function of PAG3 in the FcγR-mediated phagocytosis. The GAP-inactive mutants of GIT2s were also ineffective (data not shown). In these experiments, a significant proportion of the EGFP-PAG3 CA mutant, and also a small proportion of HA-GIT2, could be detected at the phagocytic cups. Since these proteins were overexpressed at high levels in the cytosol, we do not know whether these localizations imply some sort of biological significance.
Figure 4.
Overexpression of PAG3, but not its GAP-inactive mutant or GIT2s, attenuates the focal accumulation of F-actin beneath IgG-opsonized beads and its restoration by cooverexpression of ARF6. P388D1 cells and cells transiently transfected with the indicated plasmid constructs were incubated with IgG-opsonized beads and processed for laser confocal microscopic analysis, as described in the legend to Fig. 1, except that HA-tagged ARF6 was visualized with anti-HA mAb coupled with Cy5-anti–mouse IgG. All plasmid constructs were derived from pcDNA3 or pEGFP, and the overexpression of exogenous proteins was confirmed as in the legends to Fig. 2 and Fig. 3. (A) Cells were mock-transfected (MOCK; panels a–e), or transfected with plasmid DNAs for wild-type EGFP-PAG3 (panels f–j), its GAP-inactive mutant (panels k–o), wild-type HA-GIT2-short (panels p–t), and wild-type HA-GIT2 (panels u–y). Exogenous ARFGAP proteins (panels a, f, k, p, and u) and F-actin (panels b, g, l, q, and v) were visualized as described in the legend to Fig. 1. The third column of panels (c, h, m, r, and w) represent the merging of the first two columns of panels, and their higher magnifications are shown in the fourth column of panels (indicated by rectangles; panels d, i, n, s, and x). The last column of panels (e, j, o, t, and y) show the DIC images of the same fields of the left four panels. (B) Cells were cotransfected with 1.5 μg of plasmid DNA for wild-type EGFP-PAG3 together with 0.5 μg of plasmid DNA for ARF6-HA; and EGFP-PAG3 (panel a), ARF6-HA (panel b), and F-actin (panel c) were visualized as above (see also the legend to Fig. 5 C, third column). Panel d represents the merging of the three left panels. Panel e shows the DIC image of the same field of the first four panels. Arrows indicate the position of phagocytic beads. Bars, 5 μm.
Overexpression of ARF6, but Not ARF1 or ARF5, Can Restore the PAG3-induced Inhibition of FcγR-mediated Phagocytosis.
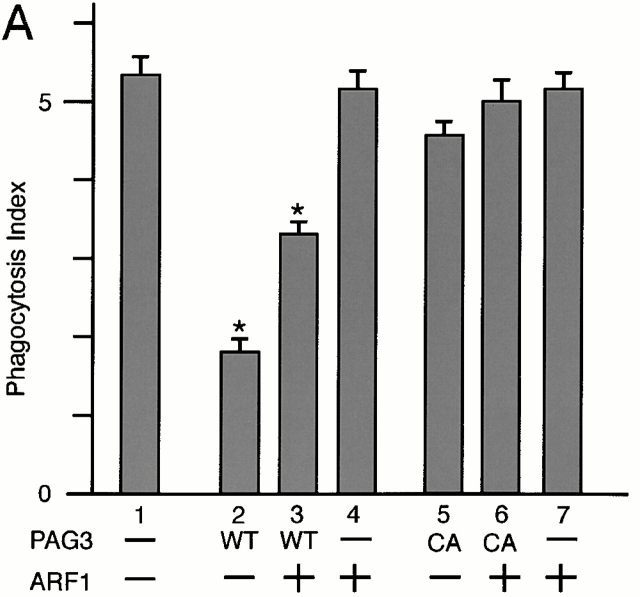
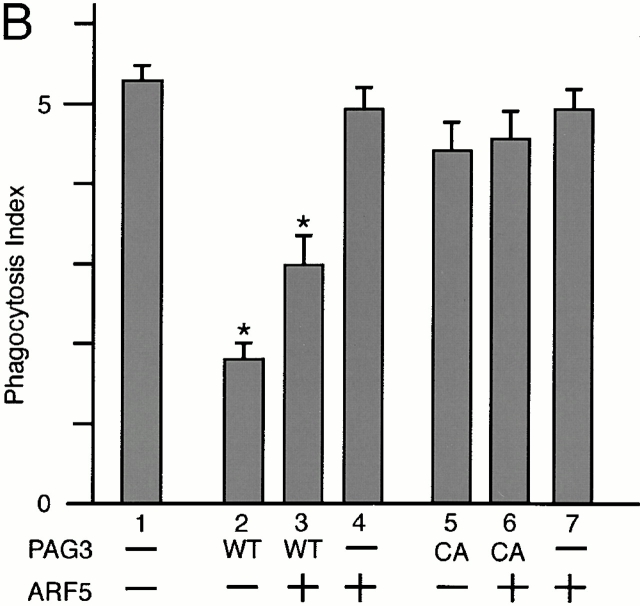
Finally, to verify the functional relationship between PAG3 and ARF6, we examined whether overexpression of ARF6 could restore the inhibition of FcγR-mediated phagocytosis induced by the overexpression of PAG3, and vice versa. Unlike other Ras superfamily small GTPases, ARFs do not exhibit detectable intrinsic GTPase activities 11. Therefore, ARFs appear to primarily depend on ARFGAPs for hydrolyzing bound GTP, and the molecular ratio between ARFs and their GAPs may be important for biological functions of ARF molecules.
Plasmid DNA for PAG3 was cotransfected with plasmid DNA for ARF isoforms, where all plasmids were designed for the overexpression. As shown in Fig. 5 C, we found that increasing amounts of plasmid for EGFP-PAG3 together with decreasing amounts of plasmid for ARF6-HA gave bell-shaped dose responses of the phagocytosis index, and a condition was found where both EGFP-PAG3 and ARF6-HA were overexpressed but the phagocytic activity of the transfected cells was restored to almost normal. In contrast, a control experiment using the GAP-inactive mutant of EGFP-PAG3, instead of wild-type EGFP-PAG3, did not give such bell-shaped dose responses, but gave responses declining linearly in accordance with amounts of ARF6-HA plasmid DNA transfected (Fig. 5 C). Cooverexpression of EGFP-PAG3 with the Q67L or the T27N mutant of ARF6-HA was also unable to restore phagocytic activity (Fig. 5 C). Moreover, such conditions for the apparent restoration of the phagocytosis index were found neither with cooverexpression of EGFP-PAG3 and ARF1-HA nor with cooverexpression of EGFP-PAG3 and ARF5-HA (Fig. 5a and Fig. b), although we examined with several different ratios between PAG3 and these ARF isoforms in addition to those shown in Fig. 5 (data not shown). These results again are consistent with a notion that PAG3 acts as a GAP for the class III ARF, ARF6, but not other classes of ARFs, and is thereby involved in the FcγR-mediated phagocytosis in macrophages. Accumulation of F-actin beneath the IgG-opsonized beads was also observed in cells overexpressing both EGFP-PAG3 and ARF6-HA, where their phagocytic activities were restored to almost normal (Fig. 4 B).
Figure 5.
Cooverexpression of ARF6, but not other classes of ARF isoforms, restores the PAG3-induced inhibition of FcγR-mediated phagocytosis. P388D1 cells and cells transiently transfected with the indicated plasmid constructs in pEGFP and pcDNA3 vectors for overexpression were incubated with IgG-opsonized beads, and the phagocytosis index was determined, as described in the legend to Fig. 2. Plasmid DNAs for EGFP-PAG3 (WT) or its GAP-inactive mutant (CA) were cotransfected with plasmid DNAs for ARF1-HA (A), ARF5-HA (B), and ARF6-HA (C). In A–C, 2.0 μg of plasmid DNA for EGFP-PAG3 (bar 2), 1.5 μg of plasmid DNA for EGFP-PAG3 plus 0.5 μg of plasmid DNA for each ARF-HA (bar 3), and 2.0 μg of plasmid DNA for each ARF-HA (bar 4) were used; the GAP-inactive mutant of EGFP-PAG3 was used instead of wild-type EGFP-PAG3 in bars 5–7, as indicated. In bars 8 and 9 in C, 1.5 μg of plasmid DNA for EGFP-PAG3 plus 0.5 μg of plasmid DNA for the Q67L mutant (bar 8) or the T27N mutant (bar 9) of ARF6-HA were used. Mock-transfected controls were also included (bar 1 in A–C). The overexpression of exogenous proteins were confirmed as described in the legend to Fig. 2 and Fig. 3, with each cell examined for the phagocytic activities. In bar 3 in C, these cells expressed EGFP-PAG3 at levels 25–30-fold higher than endogenous PAG3, and expressed ARF6-HA at levels 7–9-fold higher than endogenous ARF6 (also see Fig. 4 B). In bars 2 and 4 in C, the expression levels of EGFP-PAG3 and ARF6-HA were essentially the same as those described in the legend to Fig. 2 and Fig. 3. Expression of ARF1-HA and ARF5-HA in A and B was also similar to those in Fig. 2. Data are expressed as the mean ± SE of three independent experiments. *P < 0.001.
Discussion
In this study, we have demonstrated that PAG3 is involved in FcγR-mediated phagocytosis in macrophages and provided evidence for the first time that PAG3 acts as a GAP for ARF6 during phagocytosis. We have also shown that endogenous PAG3 accumulates at phagocytic cups together with ARF6 and F-actin, and on the other hand, its overexpression attenuates the focal accumulation of F-actin beneath IgG-opsonized beads and blocks phagocytosis. FcγR-mediated phagocytic activity of macrophages is essential for the immune response and for inflammation, and also participates in development and tissue remodeling 1 2. It has also been implicated in several noninfectious diseases such as rheumatoid arthritis and atherosclerosis 49 50 51. Moreover, there are striking similarities in the processes for the remodeling of cytoskeletal architecture and membrane structure between the extension of pseudopods during phagocytosis and the formation of leading edges during cell migration 52; the latter has been also our inquiry with PAG3 (32; also see below).
One of our major concerns in this study is the specificity of PAG3 for ARF isoforms. We have previously shown that PAG3 acts as a GAP for ARF6, rather than ARF1, in COS-7 cells 32, where each class of ARF isoforms was overexpressed and subjected to activation by treating the cells briefly with AlF (a mixture of 30 mM NaF and 50 μM AlCl3; reference 18), an activator of heterotrimeric G proteins 24 25. However, results obtained from in vitro assays regarding the specificity of PAG3 for different ARF isoforms 37 were contradictory to our in vivo results, as mentioned above. Moreover, it has been suggested that different GAPs may be involved in the recruitment of different coatomer proteins to the same ARF 48. We therefore tested whether PAG3 acts as a GAP for ARF6 during FcγR-mediated phagocytosis in macrophages, in which ARF6 has been shown to play a role 10. We showed that PAG3 localizes and accumulates at the phagocytic cups formed beneath IgG-opsonized beads, and that overexpression of PAG3, but not its GAP-inactive mutant, is inhibitory for the phagocytic activity. Thus, we examined which class(es) of ARF isoforms is the target of PAG3 during macrophage phagocytosis. Our results indicated that PAG3 acts on the class III ARF, ARF6, but not on the class I ARF, ARF1, or on the class II ARF, ARF5, during FcγR-mediated phagocytosis. We also employed other ubiquitously expressed ARFGAPs, GIT2 and its isoform GIT2-short 38 39 40 41, and showed that these proteins neither accumulate at the phagocytic cups when expressed at moderate levels nor inhibit the phagocytic activity even when overexpressed. Again, although our in vivo assays in COS-7 cells indicated that both GIT2 (unpublished results) and GIT2-short 38 exhibit GAP activities for ARF1, but not for ARF6, these proteins have been shown to exhibit similar GAP activities toward different classes of ARF isoforms in vitro, including ARF1, 2, 4, and 6, which was conducted in the absence of coatomers or effectors 40 41. Our results here are again consistent with our previous results, and GIT2 and GIT2-short do not appear to exhibit efficient GAP activities for ARF6 during FcγR-mediated phagocytosis in macrophages. A possible reason for the discrepancy in the specificities of ARFGAPs measured in vivo and in vitro has already been discussed 32 38.
We showed here that overexpression of PAG3 attenuates the focal accumulation of F-actin. Phagocytosis proceeds in discrete but coordinated stages, in which both the actin-based cytoskeletal remodeling and the membrane remodeling take place 1 2. Actin-based cytoskeletal remodeling is primarily regulated by Rho family GTPases 7 8 9, and several downstream effectors of these GTPases have been identified 8. Some of the effector proteins, such as the Arp2/3 complex, have been shown to function in macrophages to regulate phagocytosis 8. On the other hand, membrane remodeling is the major role for ARF family GTPases 12 13 16. Although several ARF family GTPases, including ARF6, have been shown to participate also in actin cytoskeletal remodeling (18, 22–25, 28, and this study), molecular mechanisms by which ARFs regulate actin cytoskeleton are still largely unknown. In this regard, it has been recently shown that Rho and ARF family GTPases intercommunicate with each other to determine actin–cytoskeletal architecture 22 23 24 25 28 29. Hence, some mechanism must exist that links the regulation of these two different families of GTPases, temporally and spatially. This putative linkage may also be important for coordination of the actin-based cytoskeletal remodeling and the membrane remodeling. Lipid modification of the plasma membrane is likely to be involved. For example, Rho family GTPases each contain different lipid modification 53, and several proteins regulating Rho and ARF family GTPases also contain different pleckstrin homology domains, which could interact differently with the plasma membrane, depending on the lipid composition 54. Moreover, several enzymes that modify membrane lipids act downstream of these GTPases 55 56. On the other hand, it is also possible that some proteins physically bridge Rho and ARF family GTPases and/or proteins regulating these GTPases. POR1, partner of Rac1, exhibits a potential for interacting with both Rac1 and ARF6 in vitro 57 58, though its physiological relevance has not yet been clarified 23. PAG3 contains several different protein–protein interaction modules including a zinc finger motif, ankyrin repeats, proline-rich sequences, and a src homology 3 domain, and also contains a pleckstrin homology domain 32 37. PAG3 binds to paxillin 32, which is localized to FcγR-mediated phagocytic cups 33 34. It has been shown that paxillin can associate with a p21-activated kinase (PAK)-interacting guanine nucleotide exchanger β called βPIX 59 60, a guanine nucleotide exchanging factor for Rac1 61. We also found that paxillin can directly bind to a Rac/Cdc42 downstream effector kinase PAK3 60, though these two proteins have been previously proposed to be indirectly associated 59. Moreover, we found that PAG3 binds directly to amphiphysin IIm (unpublished results), which is a macrophage-specific amphiphysin II isoform and required for FcγR-mediated phagocytosis in murine resident peritoneal macrophages by recruiting a GTPase, dynamin, to the nascent phagosome 46. Furthermore, it should be noted that PAG3 was identified also as a binding protein to a protein tyrosine kinase, Pyk2 37, which has been shown to participate in FcγRIIA/IIIB-mediated phagocytosis in human neutrophils 62. Therefore, PAG3 has the potential to serve to coordinate the regulation of different families of GTPases in FcγR-mediated phagocytosis, including Rho and ARF family GTPases and dynamin, and may also be involved in the coordination of activities of these GTPases with signal transduction events of protein tyrosine phosphorylation. However, the biological relevance of these hypotheses largely remains to be examined.
Macrophage phagocytosis provides an excellent model system for understanding the mechanism for the coordination of actin-based motility, membrane/vesicular trafficking, and signal transduction 1 2. Identification of PAG3, which exhibits a multiple protein–protein interaction property, as a GAP for ARF6 in macrophage FcγR-mediated phagocytosis will thus provide a way for the further understanding of the fine mechanisms for this complicated and fundamental model system.
Acknowledgments
We are grateful for Manami Hiraishi and Yumiko Shibata for their technical assistance and Mayumi Yoneda for her secretarial work. We also thank Drs. Yoshiaki Ito (Kyoto University) for P388D1 cells, Keiichi Nakayama (Tsukuba University) for ARF cDNAs, Takahiro Nagase (Kazusa DNA Research Institute, Chiba, Japan) for ARFGAP cDNAs, Hermut Land (National Cancer Institute, Frederick, MD) for pBabe vector, Tomohiro Kurosaki (Kansai Medical College, Osaka, Japan) for FACScan™ analysis and useful comments, and Kathy Barker and Helena Akiko Popiel for their critical reading of the manuscript and useful comments.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan; grants from Takeda Medical Foundation, Mitsubishi Foundation, Ciba-Geigy Foundation (Japan) for the Promotion of Science, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Novartis Foundation for the Promotion of Science. Osaka Bioscience Institute was founded as a commemoration of the 100th anniversary of the municipal government of Osaka City and is supported by Osaka City.
Footnotes
Abbreviations used in this paper: ARF, ADP-ribosylation factor; BFA, brefeldin A; DIC, differential interference contrast; EGFP, enhanced green fluorescent protein; GAP, GTPase-activating protein; HA, hemagglutinin; PAG, paxillin-associated protein with ARFGAP activity.
References
- Aderem A., Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Greenberg S. Modular components of phagocytosis. J. Leukoc. Biol. 1999;66:712–717. doi: 10.1002/jlb.66.5.712. [DOI] [PubMed] [Google Scholar]
- Swanson J.A., Baer S.C. Phagocytosis by zippers and triggers. Trends Cell Biol. 1995;5:89–93. doi: 10.1016/s0962-8924(00)88956-4. [DOI] [PubMed] [Google Scholar]
- Cox D., Tseng C.C., Bjekic B., Greenberg S. A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 1999;274:1240–1247. doi: 10.1074/jbc.274.3.1240. [DOI] [PubMed] [Google Scholar]
- Bajno L., Peng X.R., Schreiber A.D., Moore H.P., Trimble W.S., Grinstein S. Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J. Cell Biol. 2000;149:697–705. doi: 10.1083/jcb.149.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Chang P., Zhang Q., Reddy P.G., Bokoch G.M., Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J. Exp. Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E., Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- May R.C., Caron E., Hall A., Machesky L.M. Involvement of the Arp2/3 complex in phagocytosis mediated by FcγR or CR3. Nat. Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- Hackam D.J., Rotstein O.D., Schreiber A., Zhang W., Grinstein S. Rho is required for the initiation of calcium signaling and phagocytosis by Fcγ receptors in macrophages. J. Exp. Med. 1997;186:955–966. doi: 10.1084/jem.186.6.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cox D., Tseng C.C., Donaldson J.G., Greenberg S. A requirement for ARF6 in Fcγ receptor-mediated phagocytosis in macrophages. J. Biol. Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- Kahn R.A., Gilman A.G. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- Peters P.J., Hsu V.W., Ooi C.E., Finazzi D., Teal S.B., Oorschot V., Donaldson J.G., Klausner R.D. Overexpression of wild-type and mutant ARF1 and ARF6distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Goud B. The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M., Price S.R., Tsai S.-C., Moss J., Vaughan M. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J. Biol. Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- Clark J., Moore L., Krasinskas A.L., Way J., Battey J., Tamkun J., Kahn R.A. Selective amplification of additional members of the ADP-ribosylation factor (ARF) familycloning of additional human and Drosophila ARF-like genes. Proc. Natl. Acad. Sci. USA. 1993;90:8952–8956. doi: 10.1073/pnas.90.19.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Vaughan M. Molecules in the ARF orbit. J. Biol. Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Li G., Colombo M.I., Stahl P.D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H., Klausner R.D., Donaldson J.G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H., Donaldson J.G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C., van Donselaar E., Hsu V.W., Yang C., Stahl P.D., Peters P.J. ARF6 targets recycling vesicles to the plasma membraneinsights from an ultrastructural investigation. J. Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Khachikian Z., Radhakrishna H., Donaldson J.G. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H., Al-Awar O., Khachikian Z., Donaldson J.G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangement. J. Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Calafat J., Janssen H., Greenberg S. ARF6 is required growth factor- and Rac-mediated membrane ruffling in macrophages at a stage distal to Rac membrane targeting. Mol. Cell. Biol. 1999;19:8158–8168. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awar O., Radhakrishna H., Powell N.N., Donaldson J.G. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol. Cell. Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans R.L., Szanto S., van Aelst L., D'Souza-Schorey C. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 2000;20:3685–3694. doi: 10.1128/mcb.20.10.3685-3694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh M.M., Whitney J.A., Carrol K., Zhang C.-J., Boman A.L., Rosenwald A.G., Mellman I., Kahn R.A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Chardin P., McCormick F. Brefeldin Athe advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- Norman J.C., Jones D., Barry S.T., Holt M.R., Cockcroft S., Critchley D.R. ARF1 mediates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fiber formation in intact and permeabilized Swiss 3T3 fibroblasts. J. Cell Biol. 1998;143:1981–1995. doi: 10.1083/jcb.143.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare A., Paris S., Albertinazzi C., Dariozzi S., Andersen J., Mann M., Longhi R., de Curtis I. p95-APP1 links membrane transport to Rac-mediated reorganization of actin. Nat. Cell Biol. 2000;2:521–530. doi: 10.1038/35019561. [DOI] [PubMed] [Google Scholar]
- Abo A., Pick E., Hall A., Totty N., Teahan C.G., Segal A.W. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- Dana R.R., Eigsti C., Holmes K.L., Leto T.L. A regulatory role for ADP-ribosylation factor 6 (ARF6) in activation of the phagocyte NADPH oxidase. J. Biol. Chem. 2000;275:32566–32571. doi: 10.1074/jbc.M005406200. [DOI] [PubMed] [Google Scholar]
- Kondo A., Hashimoto S., Yano H., Nagayama K., Mazaki Y., Sabe H. A new paxillin-binding protein, PAG3/Papα/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration. Mol. Biol. Cell. 2000;11:1315–1327. doi: 10.1091/mbc.11.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S., Chang P., Silverstein S.C. Tyrosine phosphorylation of the γ subunit of Fcγ receptors, p72syk and paxillin during Fc receptor-mediated phagocytosis in macrophages. J. Biol. Chem. 1994;269:3897–3902. [PubMed] [Google Scholar]
- Allen L.H., Aderem A. Molecular definition of distinct cytoskeletal structures involved in complement- and Fc receptor–mediated phagocytosis in macrophages. J. Exp. Med. 1996;184:627–637. doi: 10.1084/jem.184.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaki Y., Uchida H., Hino O., Hashimoto S., Sabe H. Paxillin isoforms in mouse. Lack of the γ isoform and developmentally specific β isoform expression. J. Biol. Chem. 1998;273:22435–22441. doi: 10.1074/jbc.273.35.22435. [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Nagase T., Nakajima D., Seki N., Ohira M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. VIII. 78 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1997;4:307–313. doi: 10.1093/dnares/4.5.307. [DOI] [PubMed] [Google Scholar]
- Andreev J., Simon J.P., Sabatini D.D., Kam J., Plowman G., Randazzo P.A., Schlessinger J. Identification of a new Pyk2 target protein with Arf-GAP activity. Mol. Cell. Biol. 1999;19:2338–2350. doi: 10.1128/mcb.19.3.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaki Y., Hashimoto S., Okawa K., Nakamura K., Yagi R., Yano H., Kondo A., Iwamatsu A., Mizoguchi A., Sabe H. An ARFGAP protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin, and actin cytoskeletal organization. Mol. Biol. Cell. 2001;12:645–662. doi: 10.1091/mbc.12.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T., Seki N., Tanaka A., Ishikawa K., Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- Vitale N., Patton W.A., Moss J., Vaughan M., Lefkowitz R.J., Premont R.T. GIT proteins, a novel family of phosphatidylinositol 3,4,5-triphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 2000;275:13901–13906. doi: 10.1074/jbc.275.18.13901. [DOI] [PubMed] [Google Scholar]
- Premont R.T., Claing A., Vitale N., Perry S.J., Lefkowitz R.J. The GIT family of ADP-ribosylation factor GTPase-activating proteins. J. Biol. Chem. 2000;275:22373–22380. doi: 10.1074/jbc.275.29.22373. [DOI] [PubMed] [Google Scholar]
- Morgenstern J.P., Land H. Advanced mammalian gene transferhigh titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner D.J., Hall C.F., Jackson S. Fcγ receptor-mediated activation of phospholipase D regulates macrophage phagocytosis of IgG-opsonized particles. J. Immunol. 1999;162:2266–2274. [PubMed] [Google Scholar]
- Sabe H., Hata A., Okada M., Nakagawa H., Hanafusa H. Analysis of the binding of the Src homology 2 domain of Csk to tyrosine-phosphorylated proteins in the suppression and mitotic activation of c-Src. Proc. Natl. Acad. Sci. USA. 1994;91:3984–3988. doi: 10.1073/pnas.91.9.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold E.S., Underhill D.M., Morrissette N.S., Guo J., McNiven M.A., Aderem A. Dynamin 2 is required for phagocytosis in macrophages. J. Exp. Med. 2000;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold E.S., Morrissette N.S., Underhill D.M., Guo J., Bassetti M., Aderem A. Amphiphysin IIm, a novel amphiphysin II isoform, is required for macrophage phagocytosis. Immunity. 2000;12:285–292. doi: 10.1016/s1074-7613(00)80181-8. [DOI] [PubMed] [Google Scholar]
- Unkeless J.C. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J. Exp. Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S., Spang A., Schekman R. A primer on vesicle budding. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- Blackburn W.D., Jr., Heck L.W., Koopman W.J., Gresham H.D. A low molecular weight, heat-labile factor enhances neutrophil Fc receptor-mediated lysosomal enzyme release and phagocytosis. Arthritis Rheum. 1987;30:1006–1014. doi: 10.1002/art.1780300907. [DOI] [PubMed] [Google Scholar]
- Torsteinsdottir I., Arvidson N.G., Hallgren R., Hakansson L. Monocyte activation in rheumatoid arthritis (RA)increased integrin, Fcγ and complement receptor expression and the effect of glucocorticoids. Clin. Exp. Immunol. 1999;115:554–560. doi: 10.1046/j.1365-2249.1999.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S., Meng X.P., Ramasamy S., Harrison D.G., Galis Z.S. Relative oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix metalloproteinases in vitroimplications for atherosclerotic plaque stability. J. Clin. Invest. 1996;98:2572–2579. doi: 10.1172/JCI119076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggeler J., Werb Z. Initial events during phagocytosis by macrophages viewed from outside and inside the cellmembrane–particle interactions and clathrin. J. Cell Biol. 1982;94:613–623. doi: 10.1083/jcb.94.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski M.S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Ma A.D., Abrams C.S. Pleckstrin homology domains and phospholipid-induced cytoskeletal reorganization. Thromb. Haemost. 1999;82:399–406. [PubMed] [Google Scholar]
- Van Aelst L., D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Honda A., Nogami M., Yokozeki T., Yamazaki M., Nakamura H., Watanabe H., Kawamoto K., Nakayama K., Morris A.J., Frohman M.A., Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Boshans R.L., McDonough M., Stahl P.D., van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh H., Williger B.T., Exton J.H. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J. Biol. Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- Turner C.E., Brown M.C., Perrotta J.A., Riedy M.C., Nikolopoulos S.N., McDonald A.R., Bagrodia S., Thomas S., Leventhal P.S. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP proteina role in cytoskeletal remodeling. J. Cell Biol. 1999;145:851–863. doi: 10.1083/jcb.145.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Tsubouchi A., Mazaki Y., Sabe H. Interaction of paxillin with p21-activated kinase (PAK)association of paxillin α with the kinase-inactive and the Cdc42-activated forms of PAK3. J. Biol. Chem. 2001;276:6037–6045. doi: 10.1074/jbc.M005854200. [DOI] [PubMed] [Google Scholar]
- Manser E., Loo T.H., Koh C.G., Zhao Z.S., Chen X.Q., Tan L., Tan I., Leung T., Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Hazan-Halevy I., Seger R., Levy R. The requirement of both extracellular regulated kinase and p38 mitogen-activated protein kinase for stimulation of cytosolic phospholipase A2 activity by either FcγRIIA or FcγRIIIB in human neutrophils. J. Biol. Chem. 2000;275:12416–12423. doi: 10.1074/jbc.275.17.12416. [DOI] [PubMed] [Google Scholar]