DNA Double-Strand Breaks: Prior to but not Sufficient in Targeting Hypermutation (original) (raw)
Abstract
The activation-induced cytidine deaminase (AID) is required for somatic hypermutation (SHM) and class-switch recombination (CSR) of immunoglobulin (Ig) genes, both of which are associated with DNA double-strand breaks (DSBs). As AID is capable of deaminating deoxy-cytidine (dC) to deoxy-uracil (dU), it might induce nicks (single strand DNA breaks) and also DNA DSBs via a U-DNA glycosylase-mediated base excision repair pathway (‘DNA-substrate model’). Alternatively, AID functions like its closest homologue Apobec1 as a catalytic subunit of a RNA editing holoenzyme (‘RNA-substrate model’). Although rearranged Vλ genes are preferred targets of SHM we found that germinal center (GC) B cells of AID-proficient and -deficient Vλ1-expressing GC B cells display a similar frequency, distribution, and sequence preference of DSBs in rearranged and also in germline Vλ1 genes. The possible roles of DSBs in relation to AID function and SHM are discussed.
Keywords: AID, class-switch recombination, DSB, Igλ, somatic hypermutation
Introduction
Somatic hypermutation (SHM) of B cell Ig variable region genes contributes to the diversification of the antibody repertoire. Antigen-activated B cells migrate into B cell follicles of secondary lymphoid tissues, where they undergo rapid expansion and establish oligoclonal germinal centers (GCs). The GC is a specialized microenvironment where B cells receive specific signals enabling them to further diversify their V(D)J rearranged Ig heavy chain (IgHC) and light chain (IgLC) genes through SHM and alter their effector/homing capacity through class-switch recombination (CSR) in the IgH constant region (1). The majority (>90%) of somatic mutations in Ig genes are point mutations, the rest are small deletions and duplications. Tetramers with a RGYW and its inverse complement WRCY (R is A or G, Y is C or T, and W is A or T) are intrinsic mutational hot spots where ∼50–60% of all mutations are found (2). Mutations are restricted to the hypermutation domain a region of ∼2 kb downstream of the Ig promoter encompassing the rearranged V(D)J gene segment and, in the case of the VJ rearranged λ LC locus, reaches into the 5′ end of the constant region (3). The preferential targeting of mutations to this domain points to the existence of cis-acting element(s) capable of targeting the ‘hypermutator’ to V gene regions. The Ig promoter, the Ig enhancers, but not the rearranged V(D)J region itself (4) have been identified as critical cis acting elements in controling SHM (2, 5, 6). The mutation load of rearranged Ig genes correlates with transcription, and can be tuned experimentally by transcription (7, 8).
Activation-induced cytidine deaminase (AID) has been identified as a key protein in controlling SHM and CSR (9–11), two central events of late B cell development. In humans, mutations in AID are causative for the development of the autosomal recessive form of the hyper-IgM syndrome 2 (HIGM2) (11). AID, a 24-kD protein, is similar to the cytidine deaminase Apobec-1 and both are encoded in close proximity on human chromosome 12p13. Apobec-1 serves as the catalytic subunit of the apolipoprotein B (apoB) mRNA-editing complex and associates with the Apobec-1 Complementation Factor (ACF), a 65-kD protein that contains three RNA-binding motifs. Within this complex, Apobec-1 deaminates specifically the C6666 of the apo-B mRNA into a U6666, converting codon 2153 from a glutamine (CAA) into a premature stop codon (UAA) (12, 13). AID might function like other members belonging to this gene family as a catalytic subunit of a RNA editing holoenzyme, editing for example the mRNA of a protein involved in either nicking, repairing or synthesizing DNA. Alternatively, as AID can deaminate deoxycytidine (9), AID itself might be part of the ‘hypermutator’ and ‘class-switcher’ to function more directly as a nick or single nucleotide gap-inducing enzyme. Together with ACF-like factor(s) AID might specifically deaminate dC in secondary structures of transcribed DNA. Hydrolysis of dC to dU are frequent spontaneous lesions that are efficiently repaired by base excision repair (BER) and if occurring on both strands can lead to double-strand breaks (DSBs; reference 14).
Indirect and direct evidence for DNA DSBs as intermediates in hypermutating Ig genes has accumulated (15–19). The generation of DSBs is favored by transcription and like for somatic hot spot mutations these DSBs occur preferentially at RGYW motifs within the hypermutation domain (16, 20). These DSBs likely occur as a result of two single strand DNA breaks which are introduced by an unknown nuclease and cause staggered DSBs. Based on these data, models related to the one of Brenner and Milstein (21) have been proposed, in which mutations are introduced by an error prone fill-in reaction of staggered DNA breaks. In accordance to these models the DNA polymerases η and ζ, both capable of translesion synthesis, i.e., bypassing and continuing DNA synthesis in the presence of a lesion, have recently been linked to SHM (22–24).
Materials and Methods
Mice.
The generation and genotyping of AID- and Igκ-deficient mice have been described elsewhere (10, 25).
Immunization.
Igκ knockout mice were immunized with 0.2 ml of a 10% sheep red blood cells solution in PBS (16). AID knockout mice were immunized with NP-CG. For the immunization with NP-CG, NP(28)-CGG® (Biosearch Technologies, Inc.) is resuspended at 1 mg/ml in PBS, an equal volume of Alu-Gel-S® (Serva) is added, mixed, incubated overnight at 4°C, and 0.2 ml of this suspension (100 μg NP(28)-CGG) is injected intraperitoneally. For the analysis of DNA DSBs mice were killed 7 d after immunization, for the analysis of SHM mice were killed 10 d after immunization.
Cell Sorting and DNA Isolation.
Sorting of GC and non-GC B cells was done using a combination of magnet activated cell sorting (MACS®) (Miltenyi Biotec) and fluorescence activated cell sorting (FACS®) using a FACStar™ (Becton Dickinson). The isolation of high molecular weight DNA from these fractions has been described elsewhere (16). Sorting of Vλ1&2-expressing GC and non-GC B cell subsets was achieved with a Vλ1&2-specific mAb.
Analysis of SHM in Germline Vλ1 Gene Segments.
For the amplification of germline Vλ1 gene segments a seminested PCR assay was applied using the reverse Vλ1 intron primer in combination with the Vλ1&2 external primer for the first round and the Vλ1&2 internal primer for the second round of PCR amplification. The PCR amplification was performed as described previously (26).
Amplification, Cloning, and Sequencing of Splinkerette-ligated Vλ Genes.
The ligation of the splinkerettes has been described elsewhere (26). The quantity of the DNA used is based on a defined number of sorted cells. Based on previous experiments and as determined by semiquantitative PCR reactions with Ku70 specific primers (see below) this method of DNA quantification is reproducible. Specific amplification of adaptor-ligated Vλ1&2 genes from genomic DNA was achieved by using a nested PCR strategy. In the first round the external splinkerette primer was used in combination with the external Vλ primer. For the second round of amplification the internal splinkerette primer was used in combination with the internal Vλ primer. To detect any Vλ/adaptor hybrids, we used the same PCR conditions as described for the amplification of the Vλ1 genes from single cells (26). PCR products were resolved on a 2% (wt/vol) agarose gel, visualized with ethidium bromide under UV-light and isolated from agarose gel slices using a QiaQuick® matrix (QIAGEN). After isolation the PCR products were cloned into the TOPO pCRII® vector from the TOPO TA Cloning® kit (Invitrogen) and sequenced using the DyeDeoxy Terminator Cycle Sequencing® kit (Applied Biosystems).
Oligonucleotides.
Vλ1&2 external primer: 5′-GGGTATGCAACAATGCGCATCTTGTC-3′; Vλ1&2 internal primer: 5′-GCGAAGAGAAGCTTGTGACTCAGGAATCTGCA-3′; and Vλ1 intron primer: 5′-AATGATTCTATGTTCTGCCAAGTC-3′. External splinkerette primer: 5′-CGAAGAGTAACCGTTGCTAGGAGAGACC-3′. Internal splinkerette primer: 5′-GTGGCTGAATGAGACTGGTGTCGAC-3′. Splinkerette oligomers: 5′-CGAAGAGTAACCGTTGCTAGGAGAGACCGTGGCTGAATGAGACTGGTGTCGACACTAGTGG-3′ (long strand, 61-mer); 5′-CCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA-3′ (short strand, 44-mer); 5′-ACACGGCTTCCTTAATGTGA-3′ (KU70 forward primer); and 5′-GGCTGGCTTTAGCACTGTCA (KU70 reverse primer).
Online Supplemental Material.
The exact location of all break sites shown in Figs. 2 and 4 are indicated in five separate pdf files. Online supplemental material can be found at http://www. jem.org/cgi/content/full/jem.20011749/DC1.
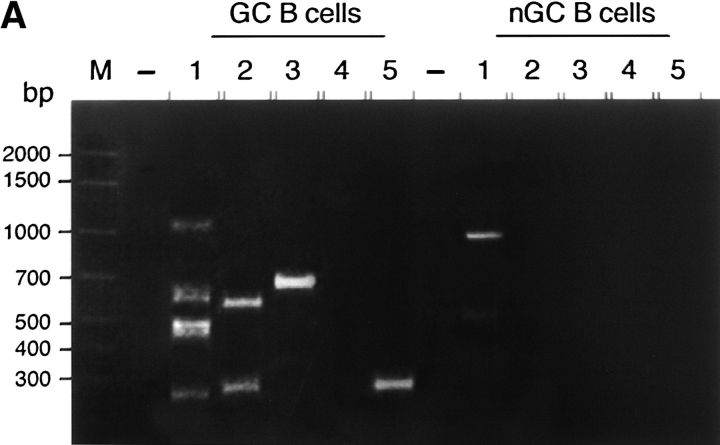
Figure 2.
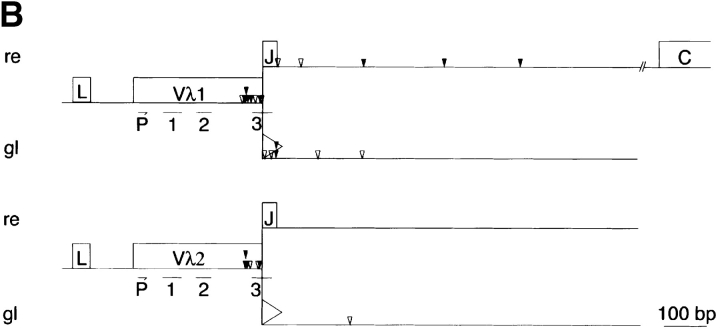
DSBs in the Igλ locus of GC B cells from Igκ knockout mice. (A) Detection of DSBs. LM-PCR products of the 5′ break sites from 10,000 (1), 5,000 (2), 2,500 (3), 1,250 (4), and 625 (5) cell-equivalents of GC B cells (CD19+, PI−, PNAhigh) and non-GC B cells (CD19+, PI−, PNAlow) from Igκ knockout mice. Numbers at the side indicate the location of the DNA size markers (basepairs). (B) DSBs in rearranged (re) and germline (gl) Vλ gene segments of hypermutating Igκ-deficient GC B cells. The distribution and location of the DSBs found are depicted as arrows. Vertically aligned arrows indicate DSBs obtained from independent LM-PCR reactions. Closed arrows indicate DNA-DSBs at an RGYW motif, open arrows indicate DNA-DSBs outside an RGYW motif. P, location of the Vλ internal primer. 1, 2, 3, location of the complementary determining regions (CDR1, 2, and 3, respectively). Asterisks above arrows indicate DSBs in Vλ1Jλ3 rearranged λ LC genes. Triangles indicate the recognition signal sequence.
Figure 4.
DSBs in the Igλ locus of AID-deficient GC B cells and non-GC B cells. (A) AID does not control the generation of DSBs in hypermutating Ig genes. LM-PCR products derived from the amplification of the 5′ break sites in Vλ1&2 gene segments from 10,000 (1), 5,000 (2), 2,500 (3), 1,250 (4), and 625 (5) cell equivalents of GC B cells (Vλ1, CD19+, PI−, PNAhigh) from AID+/+ and AID−/− mice as well as non-GC B cells (Vλ1, CD19+, PI−, PNAlow) from AID-deficient mice were separated on a 2% (wt/vol) TAE agarose gel and visualized with ethidium bromide under UV-light. The DNA size markers are indicated in basepairs. As revealed by Southern blotting and sequencing most PCR products are Vλ1&2 specific. (B) DSBs in rearranged and nonrearranged Vλ genes of GC B cells from AID deficient mice (see text). For abbreviations, see legend to Fig. 2.
Results and Discussion
To determine whether AID functions as a catalytic subunit of an RNA-editing holoenzyme or more directly upstream of DSBs as a nick-inducing enzyme, i.e., by deaminating dCs within the hypermutation domain (9, 27, 28) a ligation-mediated (LM)-PCR protocol (16) was applied. This protocol allows us to identify ex vivo the frequency, distribution and sequence preference of DSBs in Igλ LC genes of hypermutating B cells. The Igλ locus arose by gene duplication (Fig. 1) . Compared with Igκ, Igλ-LC–expressing B cells normally represent <5% of all B cells (25). Therefore, the occurrence of DSBs in the Igλ locus is first analyzed in GC (CD19+, PNAhigh) and non-GC (CD19+, PNAlow) B cells of Igκ-deficient mice where only λLC-expressing B cells develop (25). High molecular weight DNA was carefully isolated from these B cell fractions and aliquots corresponding to a defined number of sorted B cells were ligated to blunt-ended DNA adapters. The Vλ/adaptor hybrids were PCR amplified in two rounds. The two primer sets used hybridize specifically within the 5′ region of Vλ1&2 genes and to the complement of the long strand of the splinkerette, respectively (16, 28). If DSBs exist in the Vλ1&2 region of hypermutating B cells, the PCR products should occur in a size range of the hypermutation domain. As previously demonstrated for the targeted VHB1–8 gene (16) distinct PCR products were found in hypermutation competent GC B cells and only infrequently in small, non-GC B cells (Fig. 2 A and B). Southern blot analysis with a radiolabeled Vλ1 probe and sequencing revealed the specificity of nearly all PCR products (data not shown). After sequencing the PCR products, the identity, location, and site preference of DSBs in the Igλ locus were determined. DSBs are found in a region of 100–2,000 basepairs downstream of rearranged Vλ1&2 genes and interestingly also in nonrearranged, i.e., germline configured Vλ1&2 segments of the λ LC locus (Fig. 2 B). Considering the 19 tetramers with a RGYW/WRCY consensus in the Vλ1&2 segments, 38% of randomly distributed DSBs are expected to occur at these sites. However, 57% (43/76) of all DSBs locate at these sites, indicating a preference of DSBs to occur in RGYW/WRCY motifs. A hot spot of DSBs composed of repetitive Gs locates 5′ of the complementary determining region 3 (CDR3) and is also a hot spot for mutations in Vλ1 as was also shown for IgH genes (19, 26, 29).
Figure 1.
Genomic organization of the mouse Igλ locus (see text).
The presence of a Jλ element or a recognition signal sequence, which is used for the rearrangement of the V to J element in the Ig LC loci, at the 3′ end of Vλ segments distinguishes between DSBs in rearranged and germline Vλ genes, respectively. Excluding the DSBs within Vλ segments, the relative frequency of DSBs in rearranged versus germline Igλ gene segments can be determined. Of the remaining 29 DSBs, 48% (14/29) of the DSBs were found downstream of rearranged and 52% (15/29) downstream of germline Vλ1 gene segments. As transcription favors the generation of DSBs (16, 17) and each of the two autonomous Igλ enhancers, Eλ2–4 and Eλ3–1 can independently activate transcription of rearranged and nonrearranged Vλ genes (30), a high frequency of DSBs in germline Vλ gene segments is expected. Only 14% (2/14) of the DSBs in Vλ2 gene segments are found downstream of rearranged and 86% (12/14) downstream of germline Vλ2 gene segments. This finding likely relates to the fact that VJ rearrangements at the Igλ locus of B cell precursors preferentially (90%) make use of Vλ1 segments, leaving most Vλ2 alleles in germline configuration (25). In this context it should be mentioned, that according to the enhancer flip-flop model (30) a single enhancer suffices to activate sequentially several promoters. Therefore, DSBs in germline VH or Vλ gene segments are also expected to be introduced, albeit taking the cooperation between the intronic and 3′ enhancers at the IgHC and Igλ LC locus and the distance to upstream V gene promoters into account at lower frequency.
To finally determine whether AID functions upstream or downstream of DSBs, Vλ1+ expressing GC (CD19+, Vλ1+, PNAhigh) and non-GC B cells (CD19+, Vλ1+, PNAlow) were sorted 7 d after the immunization with 100 μg NP-CG from the spleen of AID-deficient and AID-proficient mice (Fig. 3) . Genomic DNA isolated from a defined number of sorted B cells was used to determine the frequency and distribution of DSBs in Vλ1&2 genes. The DNA content of the different DNA samples used was equal as verified by semiquantitative PCR (data not shown). The adaptor-ligated DNA samples were titrated and the Vλ1&2 specific 5′ break sites amplified. The titration step allows a better estimation of the DSB frequency. Apparently, AID-deficiency does not affect significantly the frequency (number of PCR products) and distribution of DSBs (size range of PCR products) along the rearranged and germline Vλ1&2 genes of GC B cells (Fig. 4 A). To determine, whether these breaks differ qualitatively, the PCR products were cloned and sequenced. Considering the 19 tetramers with a RGYW consensus in Vλ1&2 segments, 38% of randomly distributed DSBs are expected to occur at these sites. However, 56% (15/27) of all DSB found in AID-deficient GC B cells locate at a RGYW consensus motif. As summarized in Fig. 4 B, the generation, distribution as well as the preference of DSBs to occur in RGYW motifs appear not to be controlled by AID. If rearranged and germline-configured Vλ genes are equal substrates of an unknown nuclease, are they also equally targeted by the SHM system? To (re)-address this question in our system the mutation frequencies of rearranged and nonrearranged Vλ1 genes were determined by amplifying and sequencing these regions from single class-switched Vλ1+, CD19+, Igμ-δ- memory B cells isolated from the spleen of C57Bl/6 mice (26). Despite the fact that DSBs are found at an equal frequency in rearranged and germline Vλ1 genes, the frequency of mutations differs. While 57% (42/74) of rearranged Vλ1 genes were mutated at a frequency of 0.61% (144 mutations in 23,680 basepairs sequenced), only 21% (7/34) of germline Vλ1 genes sequenced were mutated at a frequency of 0.11% (12 mutations in 10,880 base pairs sequenced). Taking into account that SHM has been active in 57% of the cells, the actual mutation frequency is 1.07% for rearranged and 0.19% for germline Vλ1 genes. In line with previous studies, ∼60% of class switched Vλ1+, CD19+, Igμ-δ- memory B cells have a mutated, rearranged Vλ1 gene (8, 26) and mutations in germline configured Vλ segments occur, but at a significantly lower frequency (3, 31). Therefore, although DSBs are introduced at similar frequencies in germline and rearranged Vλ1&2 genes, SHM is preferentially targeted to the rearranged Vλ1&2 genes, indicating that a DSB itself does not suffice for optimal targeting of the hypermutation machinery. We propose that DSBs usually are efficiently repaired in a nonmutagenic manner and in general do not lead to cell death.
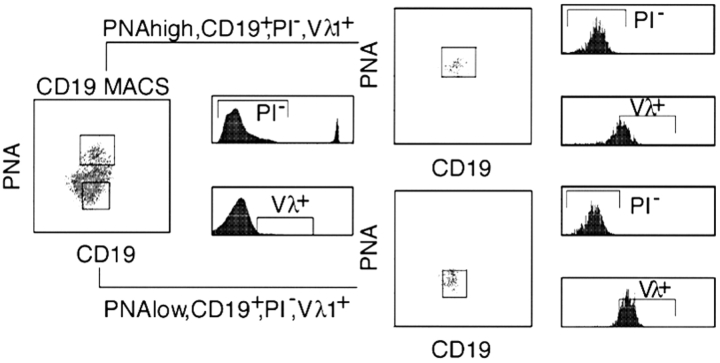
Figure 3.
Splenic B cells were positively enriched by magnet activated cell sorting (MACS®) using CD19-specific beads. AID-deficient B cells are shown as an example. Hereafter cells were stained with a FITC-conjugated CD19 specific mAb (clone 1D3), a PE-conjugated Vλ1/2-specific mAb (clone LS136), and biotinylated peanut agglutinin (PNA). Biotinylated PNA was revealed indirectly with streptavidin-allophycocyanin (SA-APC; Molecular Probes). GC-B cells (PNAhigh) in which SHM is known to be ongoing have compared with non-GC B cells (PNAlow) more binding sites for PNA. Dead cells were excluded by propidium iodine staining (PI+). Viable (PI−) B cells (CD19+) were sorted into a PNAlow, Vλ1&2+, and a PNAhigh, Vλ1&2+ fraction (MoFlo® Cytomation).
The frequency of SHM in AID proficient and AID-deficient GC (PNAhigh, Vλ1+, CD19+) and non-GC (PNAlow, Vλ1+, CD19+) B cells was determined by single cell PCR of rearranged Vλ1 genes (26). While the mutation frequency of rearranged Vλ1 genes in AID-proficient GC B cells is 0.64%, the frequency of mutations in AID-deficient GC B cells is similar to the Taq-error frequency determined for our assay, 0.018 and 0.025%, respectively.
In conclusion, SHM is preferentially targeted to rearranged Vλ gene segments. As the generation of DSBs is dependent on transcription, or at least on the initiation of transcription (16, 17), the occurrence of DSBs in germline and rearranged Vλ genes at a similar frequency likely relates to the presence of two independent λ enhancers. Although DSBs are introduced at a similar frequency in VJ rearranged and nonrearranged Vλ1 genes, the data clearly show that most of the DSBs are AID independent. Therefore, we can formally raise three possibilities: (i) none of the DSBs are involved in SHM; (ii) only a small fraction of DSBs are involved in SHM; and (iii) AID functions downstream of DSBs. The first possibility is in view of previous studies unlikely because several independent groups have provided direct and indirect evidence for DSBs being associated with SHM (for a review, see reference 28). Like the first possibility, the second possibility places AID upstream of DSBs. This suggests that the DSBs in AID-deficient GC B cells are not related to SHM, however leaving the possibility that a minority of DSBs found in AID proficient GC B cells are indeed related to the process of SHM, either as an intermediate or as a byproduct. The last possibility that AID functions downstream of the DSBs is also possible but appears to be unlikely because of a recent publication by Petersen et al. (32) indicating that AID is required to initiate the focus formation of Nijmegen breakage syndrome protein and phophorylated H2A histone family member X protein at sites of CSR. Present studies aim on the identification and characterization of the RNA substrate, RNA-binding protein within the putative RNA-editing complex, the identification of the nuclease, and the DNA repair pathway involved.
Acknowledgments
The authors would like to thank Sue Cooper and Roy Allenspach for their expert technical assistance, Tracy Hayden and Hubertus Kohler for fluorescence-activated cell sortings, Erwin Schilliger for preparing figures, the BII animal caretaker team for their biotechnical help, and F. McBlane and G. Kline for critically reading the manuscript. Many thanks to Drs. Sigfried Weiss and Holger Engel at the GBF in Braunschweig, Germany for providing valuable sequence information on the mouse Igλ locus.
Special thanks to F. Hoffmann La Roche, Ltd. (Basel, Switzerland) which founded and supported the Basel Institute for Immunology.
The online version of this article contains supplemental material.
References
- 1.Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. [DOI] [PubMed] [Google Scholar]
- 2.Neuberger, M.S., and C. Milstein. 1995. Somatic hypermutation. Curr. Opin. Immunol. 7:248–254. [DOI] [PubMed] [Google Scholar]
- 3.Azuma, T. 1998. Somatic hypermutation in mouse λ chains. Immunol. Rev. 162:97–105. [DOI] [PubMed] [Google Scholar]
- 4.Yelamos, J., N. Klix, B. Goyenechea, F. Lozano, Y.L. Chui, A. Gonzalez Fernandez, R. Pannell, M.S. Neuberger, and C. Milstein. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 376:225–229. [DOI] [PubMed] [Google Scholar]
- 5.Storb, U., A. Peters, E. Klotz, N. Kim, H.M. Shen, K. Kage, B. Rogerson, and T.E. Martin. 1998. Somatic hypermutation of immunoglobulin genes is linked to transcription. Curr. Top. Microbiol. Immunol. 229:11–19. [DOI] [PubMed] [Google Scholar]
- 6.Winter, D.B., and P.J. Gearhart. 1998. Dual enigma of somatic hypermutation of immunoglobulin variable genes: targeting and mechanism. Immunol. Rev. 162:89–96. [DOI] [PubMed] [Google Scholar]
- 7.Fukita, Y., H. Jacobs, and K. Rajewsky. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 9:105–114. [DOI] [PubMed] [Google Scholar]
- 8.Bachl, J., and C. Olsson. 1999. Hypermutation targets a green fluorescent protein-encoding transgene in the presence of immunoglobulin enhancers. Eur. J. Immunol. 29:1383–1389. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu, M., V.S. Sankaranland, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 10.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 11.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 12.Mehta, A., M.T. Kinter, N.E. Sherman, and D.M. Driscoll. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA- binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20:1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lellek, H., R. Kirsten, I. Diehl, F. Apostel, F. Buck, and J. Greeve. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275:19848–19856. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg, E.C., G.C. Walker, and W. Siede. 1995. DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington. 698 pp.
- 15.Sale, J.E., and M.S. Neuberger. 1998. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 9:859–869. [DOI] [PubMed] [Google Scholar]
- 16.Bross, L., Y. Fukita, F. McBlane, C. Demolliere, K. Rajewsky, and H. Jacobs. 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 13:589–597. [DOI] [PubMed] [Google Scholar]
- 17.Papavasiliou, F.N., and D.G. Schatz. 2000. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 408:216–221. [DOI] [PubMed] [Google Scholar]
- 18.Lo, A.K., A.K. Ching, P.L. Lim, and Y.L. Chui. 1997. Strand breaks in immunoglobulin gene hypermutation. Annu. NY Acad. Sci. 815:432–435. [DOI] [PubMed] [Google Scholar]
- 19.Kong, Q., and N. Maizels. 2001. DNA breaks in hypermutating immunoglobulin genes: evidence for a break-and-repair pathway of somatic hypermutation. Genetics. 158:369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sale, J.E., M. Bemark, G.T. Williams, C.J. Jolly, M.R. Ehrenstein, C. Rada, C. Milstein, and M.S. Neuberger. 2001. In vivo and in vitro studies of immunoglobulin gene somatic hypermutation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner, S., and C. Milstein. 1966. Origin of antibody variation. Nature. 211:242–243. [PubMed] [Google Scholar]
- 22.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 23.Rogozin, I.B., Y.I. Pavlov, K. Bebenek, T. Matsuda, and T.A. Kunkel. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536. [DOI] [PubMed] [Google Scholar]
- 24.Zan, H., A. Komori, Z. Li, A. Cerutti, A. Schaffer, M.F. Flajnik, M. Diaz, and P. Casali. 2001. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 14:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou, Y.R., H. Gu, and K. Rajewsky. 1993. Generation of a mouse strain that produces immunoglobulin κ chains with human constant regions. Science. 262:1271–1274. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs, H., Y. Fukita, G.T. van der Horst, J. de Boer, G. Weeda, J. Essers, N. de Wind, B.P. Engelward, L. Samson, S. Verbeek, et al. 1998. Hypermutation of immunoglobulin genes in memory B cells of DNA repair-deficient mice. J. Exp. Med. 187:1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poltoratsky, V., M.F. Goodman, and M.D. Scharff. 2000. Error-prone candidates vie for somatic mutation. J. Exp. Med. 192:F27–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs, H., and L. Bross. 2001. Towards an understanding of somatic hypermutation. Curr. Opin. Immunol. 13:208–218. [DOI] [PubMed] [Google Scholar]
- 29.Reynaud, C.A., S. Frey, S. Aoufouchi, A. Faili, B. Bertocci, A. Dahan, E. Flatter, F. Delbos, S. Storck, C. Zober, and J.C. Weill. 2001. Transcription, β-like DNA polymerases and hypermutation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijgerde, M., F. Grosveld, and P. Fraser. 1995. Transcription complex stability and chromatin dynamics in vivo. Nature. 377:209–213. [DOI] [PubMed] [Google Scholar]
- 31.Weiss, S., and G.E. Wu. 1987. Somatic point mutations in unrearranged immunoglobulin gene segments encoding the variable region of λ light chains. EMBO J. 6:927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen, S., R. Casellas, B. Reina-San-Martin, H.T. Chen, M.J. Difillippantonio, P.C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D.R. Pilch, et al. 2001. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]