AID Mediates Hypermutation by Deaminating Single Stranded DNA (original) (raw)
Abstract
Activation-induced deaminase (AID) is a protein indispensable for the diversification of immunoglobulin (Ig) genes by somatic hypermutation (SHM), class switch recombination (CSR), and gene conversion. To date, the precise role of AID in these processes has not been determined. Here we demonstrate that purified, tetrameric AID can deaminate cytidine residues in DNA, but not in RNA. Furthermore, we show that AID will bind and deaminate only single-stranded DNA, which implies a direct, functional link between hypermutation and transcription. Finally, AID does not target mutational hotspots, thus mutational targeting to specific residues must be attributed to different factors.
Keywords: B lymphocytes, immunoglobulin gene, activation-induced deaminase, somatic hypermutation
Introduction
Secondary antibody diversification is achieved by somatic hypermutation (SHM), gene conversion, and class switch recombination (CSR)* of the Ig genes during the adaptive immune response. In B cells, SHM and gene conversion effectively mutate the Ig variable (V) regions, thereby changing the affinity of antibody for antigen. CSR replaces the Igμ heavy chain constant region with a downstream constant region, thereby altering the physical properties of the resulting antibodies. All three events depend on activation-induced cytidine deaminase (AID), but the exact role of this protein has remained elusive.
It has been proposed that AID directly deaminates DNA, as ectopic overexpression of this protein has been shown to result in dramatically increased levels of mutation in nongerminal center B cells (1), non-B cells (2), and even Escherichia coli (3). However, mutation could arise through multiple mechanisms, and the available data could fit any of the following theoretical possibilities. First, as initially conceived, AID could be an RNA editing enzyme whose overexpression would lead to promiscuous RNA editing, as is the case for overexpressed APOBEC-1 (4). Second, AID could be deaminating cytidine, dCTP or CTP, thereby potentially altering the composition of the cellular nucleotide pools, and it is well understood that even subtle pool alterations can lead to mutagenesis in bacteria (5). Finally, DNA itself could be the substrate for AID. Indeed, parallel work has persuasively pointed to uracil DNA glycosylase (ung) as a factor in the somatic hypermutation pathway (6, 7): ung would remove the uridine product of cytidine deamination from DNA. Here, we demonstrate that purified AID is a DNA deaminase and that its preferred, in vitro substrate is ssDNA.
Materials and Methods
Purification of strepAID.
Starting with human AID cDNA, we cloned strepAID into the pet3d vector (Novagen) and used the construct to transform Rosetta (Novagen). Protein expression was induced by treating bacterial cultures at OD 0.8 with 2 mM IPTG for 3 h.
To purify strepAID, bacteria were lysed with B-PER (Pierce Chemical Co.). Lysates were precleared on Q-sepharose FF (Amersham Biosciences) and then loaded onto freshly prepared phosphocellulose (P-11; Whatman). StrepAID was eluted from P-11 with 0.4 M KCl. The eluate was diluted and loaded onto monoQ (Amersham Biosciences). Purified strepAID tetramer was gradient-eluted from monoQ at 500 mM salt. A detailed protocol can be furnished upon request.
DT40 aid−/− Complementation Assays.
We cloned human AID cDNA, strepAID, or GST-AID into the pMSCV-gfp retroviral vector (CLONTECH Laboratories, Inc.). Pantropic virus produced in 293GP cells (CLONTECH Laboratories, Inc.) was used to infect DT40 aid−/− cells (8). Gfp+ cells were sorted four days after infection, and plated in bulk. To assess gene conversion, cells were stained with anti-chicken IgM-PE (clone M1; Southern Biotechnology Associates, Inc.) and visualized on a FACSVantage™.
Electromobility Shift Assay and UV-Cross-Linking.
Oligonucleotide substrate (100 ng) was T4 polynucleotide kinase labeled with 0.1 mCi γ-P32ATP. 1 ng substrate was combined with 100–200 ng strepAID in 10 μl binding reactions (containing 10 mM Hepes pH7.6, 10% glycerol, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 0.5 mM DTT) at room temperature for 40 min.
For electromobility shift assay (EMSA) analysis, the entire reaction was loaded on 4–20% acrylamide gels (BMA or Bio-Rad Laboratories) and electrophoresed in tris-glycine running buffer at 100 V on a Bio-Rad Laboratories Mini-Protean apparatus until the bromophenol blue dye front had reached the bottom of the gel.
For UV cross-linking, reactions (in 0.5 ml microfuge tubes) were placed on ice, 1.5 cm from the light source and exposed for 10 min at 254 nm in a Stratagene UV Stratalinker. These samples were heated to 95°C for 5 min in 1% SDS before loading on the same 4–20% acrylamide gels electrophoresed at 200 V with 0.1% SDS in the running buffer.
Gels were dried and images were stored on phosphor screens and analyzed on STORM or Typhoon Phosphorimagers with ImageQuant software.
Deamination Assays.
The standard buffer used in the deamination assay (Figs. 3 b, 4, and online supplemental Fig. S2 b) contained 100 mM KCl, 6 mM Hepes, 5 mM EDTA, at pH 7.6. at a temperature of 30°C. However, AID was able to deaminate DNA in a range of conditions including 50–150 mM salt, 7.6–9.0 pH, and 23–37°C.
Figure 3.

Recombinant strepAID can deaminate ssDNA. (a) Schematic of the SNuPE reaction used to detect dC-to-dU conversion on oligonucleotide substrates. (b) The SNuPE reaction is specific: given a template with a uridine at a known position, Bst polymerase will extend a primer with the addition of the complementary base (α-P32dATP), but not with any other deoxynucleotide (α-P32dTTP, α-P32dGTP, or α-P32dCTP). (c) StrepAID-treatment of a template containing a single dC can lead to its deamination to dU. This conversion event requires active strepAID (duplicate lanes 5 and 6), and does not happen when the template is incubated with BSA (duplicate lanes 1 and 2) or with a strepAID active site mutant (E58Q/C87A/C90A, duplicate lanes 3 and 4). In addition, dsDNA is not deaminated by strepAID (lanes 7 and 8). Right panel: SNuPE of U-containing oligos serves as a calibration curve (lane 12 = signal from 100% U-containing template, lane 11 = signal from 10% U template and 90% C template, lane 10 = signal from 1% U template-99% C template, lane 9 = C-template alone).
10 ng of gel-purified oligonucleotide substrates (for sequences see below) were incubated with 100–200 ng AID for 30 min. A tenth of each reaction (1 ng) was used for primer extension with Bst polymerase (a thermophilic Klenow fragment (New England Biolabs, Inc.; Figs. 3 b, 4, and online supplemental Fig. S2 b) or Taq (unpublished data). Briefly, a 25 μl reaction containing 1 ng template, 1 ng primer, and Bst buffer was incubated at 70°C before the addition of 0.5 units Bst. The reaction temperature was first lowered to the appropriate primer annealing temperature for 10 s, and then raised to 65°C (the optimal temperature for Bst). After 1 min extension at 65°C the reaction was terminated by the addition of sample loading buffer, heated to 95°C, and loaded on a 14% denaturing gel (Sequagel; National Diagnostics). It is important to note that these reaction conditions were calibrated specifically for the template-primer pairs reported here and should be faithfully followed: longer annealing or extension times, or higher amounts of polymerase will increase background signal.
After drying, gels were exposed to phosphor storage screens (Amersham Biosciences) and those were scanned on a Typhoon imager using ImageQuant software (Molecular Dynamics).
SNuPE Oligonucleotide Substrates.
RNA oligos were custom synthesized by Dharmacon, Inc., and DNA oligos were made by FisherOligos.
Templates.
ACCint: agtaaatgaaACCgaatgtatgagaatagaagagataatga.
ACCend: aACCgaatgtatgagaatagaagagataatgaataatagaa.
TCGint: agtaaatgaaTCGgaatgtatgagaatagaagagataatga.
TCGend: aTCGgaatgtatgagaatagaagagataatgaataatagaa.
Primers.
PrExACC: atctcttctattctcatacattcG (annealing T = 58°C).
PrExTCG: atctcttctattctcatacattcC (annealing T = 57°C).
Oligonucleotide Substrates for EMSA and uv-Cross-Linking.
ssRNA: aUCGgaauguaugagaauagaagagauaaugaauaauagaa.
ssDNA: aTCGgaatgtatgagaatagaagagataatgaataatagaa.
dsDNA (hp): multiple sequences, including: (a) aTCGgaatgtatgagaatagaagagataatgaataatagaattctattattcattatctcttctattctcatacattcCGAt; (b) alternating dCdG 66mer.
TLC Oligonucleotide Substrates.
End labeled RNA: Cgaauguaugagaauagaagagauaaugaauaauagaa.
Body-labeled RNA was created by transcribing pBluescript II SK- (linearized with XhoI) with T7 RNA polymerase.
Online Supplemental Material.
We use TLC, EMSA, and SNuPE to demonstrate (a) that AID does not deaminate RNA and (b) that APOBEC1 binds and deaminates ssDNA in vitro. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20030481/DC1.
Results and Discussion
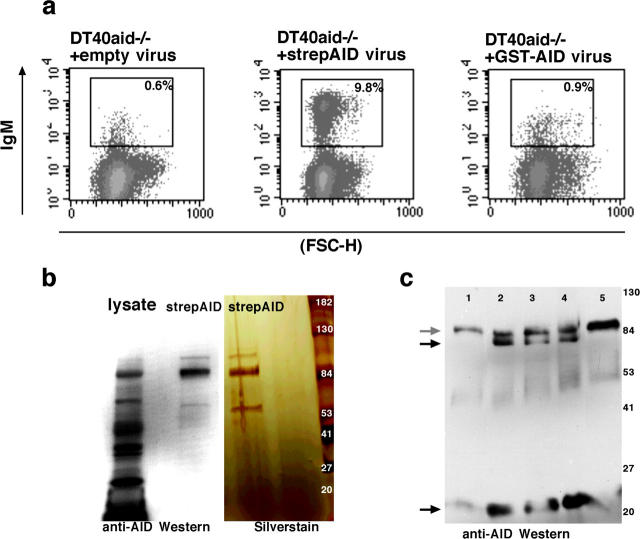
We sought to determine which of a number of possible substrates (RNA, CTP, dCTP, cytidine, or DNA) was deaminated by purified AID protein in vitro. Before overexpressing tagged protein in bacteria for purification purposes, we wanted to know that the tag did not interfere with the function of the protein in vivo. Thus we attempted to complement DT40 aid−/− cells (8) with (a) empty virus, (b) minimally tagged AID (N-terminal strep-Tag, henceforth strepAID), and (c) GST-AID. DT40 aid−/− cells infected with untagged AID and strepAID were able to recover gene conversion within 4 wk after infection (Fig. 1 a, and unpublished data), whereas GST-AID was unable to complement AID deficiency (Fig. 1 a). We conclude that GST-AID is inactive in vivo, possibly because the bulky tag interferes with the proper folding of the protein.
Figure 1.
Minimally tagged, strepAID is active in vivo and assembles into a tetramer in vitro. (a) DT40aid−/− cells can recover gene conversion activity after infection with strepAID virus, but not after infection with GST-AID virus (assay as per reference 8). (b) Phosphocellulose-purified recombinant strepAID runs with the predicted size of a tetramer. Left: Western with anti-AID antibody (reference 10). Right: silver stained gel. (c) Western blot which shows that AID tetramer (gray arrow) can be dissociated into trimer and monomer (black arrows) upon boiling in buffer containing denaturants and EDTA (lanes 2–4) but not in standard sample buffer (lane 1) or in EDTA alone (20 mM EDTA and 3% β-mercaptoethanol, lane 5). The molecular weight assignments for trimers and monomers is based on their relative position on the denaturing gel compared with protein standards. Lanes 2–4 show samples boiled in: 50% formamide and 50 mM EDTA (lane 2), 0.8 M urea and 50 mM EDTA (lane 3), 25 mM Tris (pH 8), 10 mM EDTA, and 2% SLS (lane 4).
We decided to overexpress minimally tagged, strepAID protein in bacteria and attempted to purify it to homogeneity. Even though the protein was soluble, we noticed that at most 5% of the tagged protein would bind Strep-Tactin resin, possibly because the tag was buried in the protein. We then decided to purify strepAID through standard biochemical methods. One of the preferred resins for binding DNA deaminases is phosphocellulose (9). We found that strepAID did bind P-11 resin and eluted from it with a peak around 0.4M KCl. We have thus purified strepAID and putative active site mutants of AID such as a E58Q/C87A/C90A mutant (henceforth referred to as mutant strepAID) to relative purity (Fig. 1 b).
To verify the identity of the phosphocellulose-bound product, putative strepAID protein was boiled in SDS loading buffer, run on a denaturing gel, and Western blotted. Recombinant protein reacted both with anti-AID antibody and with streptavidin (10; Fig. 1 b and unpublished data). We also noticed that phosphocellulose-bound recombinant protein would invariably run with the predicted size of an AID tetramer. The tetramer could be broken up only when the protein was boiled in strong denaturants (such as urea, guanidine) and EDTA (Fig. 1 c). These experiments suggest that in solution, strepAID is a tetramer. Indeed, many deaminases are multimers (mostly dimers or tetramers), and many retain that configuration even on denaturing acrylamide gels (examples include APOBEC-1, itself a dimer and BSD, a tetramer; references 11 and 12).
We tested the cytidine deaminase activity of the purified strepAID tetramer in a standard TLC assay. Incubation of CTP (or dCTP) with strepAID tetramer for 1hr did not result in deamination to UTP (or dUTP). In contrast, cdd, a Bacillus subtilis cytidine deaminase (9), converted over 90% of CTP (or dCTP) to UTP (or dUTP) within minutes (online supplemental Fig. S1, lane 2 vs. lane 4, and unpublished data). We also tested the ability of the purified strepAID tetramer to deaminate cytidine, in a spectrophotometric conversion assay (13). Incubation of cytidine with strepAID tetramer for 1 h did not result in conversion to uridine although cdd converted 50 to 75% of cytidine to uridine within minutes (unpublished data). We conclude that neither cytidine nor CTP (or dCTP) are good substrates for AID activity in vitro.
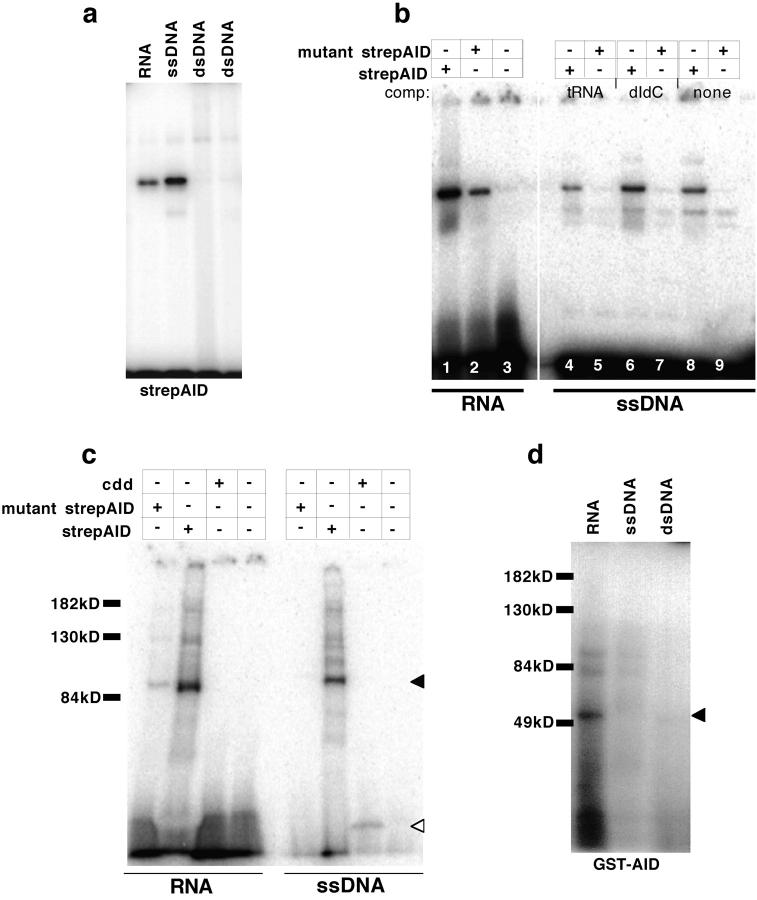
If DNA or RNA were good substrates, AID binding to oligonucleotides would be expected. To determine whether strepAID would bind nucleic acids under physiologic conditions we performed standard EMSA. Indeed, strepAID bound single stranded RNA and DNA, but not dsDNA (Fig. 2 a). Binding was robust, as it could withstand a 25-fold excess of either double stranded DNA competitor (polydIdC; Fig. 2 b, lane 6), or tRNA competitor (Fig. 2 b, lane 4) and was observed with or without cytidine in the substrate (unpublished data). Mutant strepAID did not bind to ssDNA at all (Fig. 2 b, lanes 5, 7, and 9), though it did retain some binding to RNA (Fig. 2 b, lane 2).
Figure 2.
Recombinant strepAID preferentially binds ssDNA. (a) EMSA: strepAID can bind single stranded DNA and RNA but does not bind double stranded DNA. (b) EMSA: an active site mutant of AID (E58Q/C87A/C90A) retains some ability to bind RNA (compare lanes 1 and 2) but cannot bind single stranded DNA (lanes 5, 7, and 9). Binding of strepAID is resistant to a 25-fold excess of tRNA (lane 4) or polydIdC (lane 6). (c) AID can be uv-cross-linked to RNA and ssDNA, and the cross-linked species run with the predicted size of an AID tetramer (closed triangle). Mutant AID can be uv-cross-linked to RNA, and the bacterial cytidine deaminase cdd can be cross-linked weakly to DNA (open triangle). (d) GST-AID can be uv-cross-linked to RNA but not to ssDNA or dsDNA (closed triangle).
When the binding reaction was UV-irradiated and analyzed by SDS-PAGE, the P32-labeled protein-DNA complex migrated at the same position as the strepAID tetramer (relative to the protein standards). StrepAID cross-linked to ssDNA but did not cross-link to double stranded DNA, or to RNA-DNA substrates (online supplemental Fig. S2 and unpublished data); neither mutant strepAID nor inactive, GST-AID showed any cross-linking to ssDNA but did cross-link to RNA (Fig. 2, c and d). Mutants of better-studied deaminases, such as APOBEC-1, lose substrate binding as well as activity when the cysteine residues of the active site are lost (14). Hence, it is possible that the preferred substrate for AID in vitro is DNA, but that AID, which is highly basic, nonspecifically interacts with RNA, perhaps on the basis of charge.
To directly assess whether AID would deaminate RNA, we compared its activity to that of recombinant GST-APOBEC-1 (henceforth APOBEC-1) in a TLC assay (15). Incubation of APOBEC-1 with an artificial RNA oligomer containing a single cytidine at the 5′end, led to the deamination of the cytidine to uridine (online supplemental Fig. S1, lane 7). In contrast, incubation with AID did not lead to cytidine deamination (online supplemental Fig. S1, lane 8). It is possible that AID is structurally limited to catalysis on internal cytidines, so we also incubated these proteins with internally labeled RNA substrates. Again, though APOBEC-1 was able to catalyze deamination, AID was not (online supplemental Fig. S1, lanes 10 and 11). While we cannot exclude the possibility that AID acts on a specific RNA (of unknown sequence), our data thus far suggests that RNA is not a good substrate for AID in vitro.
To ascertain that strepAID would not only bind but also deaminate DNA we developed a single nucleotide primer extension assay (SNuPE). In this assay, a primer that ends just short of the potential deoxyuridine is annealed to the DNA. A polymerase (Taq or Bst, a thermophilic Klenow fragment) is then added to the reaction, along with a single radioactive nucleotide. Under stringent annealing conditions, the polymerase will extend the primer by one (radioactive) nucleotide only if the nucleotide precisely complements the template (16; Fig. 3 a). The primer extension reaction can then be run on a sequencing gel and visualized by autoradiography. In control reactions, we found that Taq or Bst would add P32-dATP to a primer only if the template had a deoxyuridine at the appropriate position, but not if the template had a different nucleotide (e.g., deoxycytidine, deoxyguanine or deoxyadenosine; Fig. 3 b). We did not detect the incorrect addition of any of the other three P32-dNTPs to the primer under these reaction conditions (Fig. 3 b).
To determine the sensitivity of this assay, we diluted template containing a deoxyuridine at a known position with the identical template but containing deoxycytidine in that position. We found that the assay would consistently detect the uridine-containing template when it comprised as little as 1% of the template pool (Fig. 3 c, lane 10). Hence, using single nucleotide primer extension we can specifically detect uridine in DNA.
To measure the conversion of deoxycytidine to deoxy-uridine in DNA, and hence, the DNA deaminating capacity of strepAID, we created an artificial oligonucleotide substrate containing a single deoxycytidine close to the 5′end. We then treated this substrate with strepAID and assayed the conversion of the lone deoxycytidine to deoxyuridine by SNuPE. We could indeed detect deoxyuridine in DNA after strepAID treatment (Fig. 3 c, lanes 5 and 6 shown in duplicate). The level of deamination, in comparison to standards containing uridine, ranged from 1% to a maximum of 10% of the input molecules (Fig. 3 c, lanes 5 and 6, and unpublished data). This reaction was rapid (occurred within 10 min though it peaked at around 30 min, unpublished data), and did not occur when the substrate was incubated with BSA, mutant strepAID, or cdd (Fig. 3 c, lanes 1–4, and unpublished data). In addition, strepAID did not deaminate double stranded DNA (Fig. 3 c, lanes 7 and 8). Deamination by strepAID was observed within a range of deamination conditions: (a) salt levels were important (they were kept within a 50–150 mM range), (b) the reaction could tolerate moderate levels of EDTA (5–10 mM [17]), (c) it would work at a wide range of pH (from 7.6 to 9.0 were tested), (d) and it would work with varying efficiencies from room temperature to 37°C. In addition, like other deaminases (11) the protein was still active after being heated at 65°C for 30 min before the addition of substrate. Our data suggests that, under a range of conditions, strepAID is able to rapidly deaminate single stranded DNA in vitro.
We also tested whether APOBEC-1, could deaminate DNA. APOBEC-1, a known RNA deaminase, has been shown to confer a mutator phenotype on E. coli (3_)_. RNA, ssDNA, and dsDNA oligonucleotide substrates of random sequence could all be cross-linked to APOBEC-1 (online supplemental Fig. S2 a, gray arrow, pairs denote duplicate experiments). In contrast to strepAID, APOBEC-1 deaminated RNA of random sequence (online supplemental Fig. S1, lanes 7 and 10). Furthermore, like strepAID, APOBEC-1 deaminated ssDNA, but not dsDNA (online supplemental Fig. S2 b, duplicate lanes 2 and 3 vs. 7 and 8). Thus, the mutator phenotype conferred on both bacterial and mammalian cells by APOBEC-1 overexpression (3, 4) is most likely due to the ability of this enzyme to target ssDNA under conditions where the enzyme is uncoupled from the confines of the larger RNA editing complex within which it normally functions. It thus seems that APOBEC-1 may have dual specificities whereas AID appears to be active only on ssDNA.
Point mutations are not introduced randomly in the variable regions of Ig genes. Rather, mutations preferentially accumulate over “hotspot” motifs, and investigators have mined the database of known mutations to create tables assigning mutability probabilities to nucleotide triplets (18). Overexpression of AID appears to be sufficient for hypermutation, so we asked whether AID itself could impart hotspot preference to the hypermutation machinery. To approach this question, we created DNA substrates that contained deoxycytidine in the context of known mutational hotspots (GCT, AGC, and its reverse complement TCG) and in the context of coldspots (ACC, TCT) (18). StrepAID deaminated deoxycytidine within the ACC coldspot and within the TCG hotspot equally well (Fig. 4 , lanes 4 and 5 vs. lanes 10 and 11 all in duplicate), but deamination of deoxycytidine in the context of AGC, GCT, or TCT, was at background levels (unpublished data). It is theoretically possible that the sequence context of the trinucleotides is important and that the “inactive” trinucleotides (like AGC) were not couched in the optimal context, though the contribution of neighboring bases to the targeting of a particular trinucleotide has not been studied. We have tested ACC, TCG, and AGC in several sequence contexts and have found that ACC and TCG were deaminated whereas AGC was not (unpublished data). We conclude that although AID prefers to deaminate deoxycytidines within certain sequences, this selectivity does not mirror the preference of the hypermutation machinery, and hence, that hotspot preference may be attributed to a different factor. This unknown protein may act early in the reaction, marking specific deoxycytidines for deamination. Alternatively, hotspot preference may reflect the preference of the repair machinery for error correction (19).
Figure 4.
Recombinant strepAID does not display hotspot preference. StrepAID deaminates a single C in the context of a TCG hotspot or an ACC coldspot. In these reactions, the C-to-U deamination efficiency is equivalent (∼1% to 5% of templates are deaminated). The experiments shown are done in duplicate using independent protein preparations. Left panel: SNuPE of U-containing oligos as a calibration curve (lane 1 = signal from 10% U-containing template and 90% C-containing template, lane 2 = signal from 1% U template-99% C template, lane 3 = C-template alone).
Taken together, our data suggest that the role of AID in hypermutation is to bind and deaminate single stranded DNA. In vivo, single stranded DNA may arise transiently during transcription, in which case our data would strongly predict that AID activity must be tightly coupled to the transcriptional apparatus, as previously envisioned (20). Alternatively, more stable single stranded DNA structures might arise through the formation of stalled transcription forks, R-loops (as has been described for CSR [21]), or through the action of a single or double stranded nuclease. If AID functions as a single stranded DNA deaminase in vivo, it will be interesting to see what processes generate its target DNA.
Acknowledgments
We wish to thank Eva Johansson and Sine Larsen, who contributed purified cdd protein, Jobst Greeve, who contributed purified GST-APOBEC-1, C.A. Reynaud and J.-C. Weill who provided anti-AID antibody, and H. Arakawa and J.-M. Buerstedde who provided us with DT40 aid−/− cells. We also thank S.D. Fugmann for critical comments on the manuscript, and N.O. Davidson and V. Blanc for advice with the TLC protocol and the Nussenzweig lab for sharing of unpublished data.
F.N. Papavasiliou is an Irene Diamond Assistant Professor and S.K. Dickerson is a National Science Foundation fellow. This work has been partially supported by grants from the Achelis and Bodman Foundations, the New York Community Trust, and the New York Academy of Medicine.
The online version of this paper contains supplemental material.
S.K. Dickerson and E. Market contributed equally to this work.
Footnotes
*
Abbreviations used in this paper: AID, activation-induced cytidine deaminase; CSR, class switch recombination; EMSA, electromobility shift assay; SNuPE, single nucleotide primer extension assay.
References
- 1.Martin, A., P.D. Bardwell, C.J. Woo, M. Fan, M.J. Shulman, and M.D. Scharff. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 415:802–806. [DOI] [PubMed] [Google Scholar]
- 2.Yoshikawa, K., I.M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036. [DOI] [PubMed] [Google Scholar]
- 3.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka, S., M.E. Balestra, L.D. Ferrell, J. Fan, K.S. Arnold, S. Taylor, J.M. Taylor, and T.L. Innerarity. 1995. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. USA. 92:8483–8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunz, B.A. 1988. Mutagenesis and deoxyribonucleotide pool imbalance. Mutat. Res. 200:133–147. [DOI] [PubMed] [Google Scholar]
- 6.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 7.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa, H., J. Hauschild, and J.M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 9.Mejlhede, N., and J. Neuhard. 2000. The role of zinc in Bacillus subtilis cytidine deaminase. Biochemistry. 39:7984–7989. [DOI] [PubMed] [Google Scholar]
- 10.Faili, A., S. Aoufouchi, Q. Gueranger, C. Zober, A. Leon, B. Bertocci, J.C. Weill, and C.A. Reynaud. 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 3:815–821. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M., S. Sekido, Y. Isogai, and I. Yamaguchi. 2000. Expression, purification, and characterization of blasticidin S deaminase (BSD) from Aspergillus terreus: the role of catalytic zinc in enzyme structure. J. Biochem. (Tokyo). 127:955–963. [DOI] [PubMed] [Google Scholar]
- 12.Oka, K., K. Kobayashi, M. Sullivan, J. Martinez, B.B. Teng, K. Ishimura-Oka, and L. Chan. 1997. Tissue-specific inhibition of apolipoprotein B mRNA editing in the liver by adenovirus-mediated transfer of a dominant negative mutant APOBEC-1 leads to increased low density lipoprotein in mice. J. Biol. Chem. 272:1456–1460. [DOI] [PubMed] [Google Scholar]
- 13.Neuhard, J. 1968. Pyrimidine nucleotide metabolism and pathways of thymidine triphosphate biosynthesis in Salmonella typhimurium. J. Bacteriol. 96:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navaratnam, N., S. Bhattacharya, T. Fujino, D. Patel, A.L. Jarmuz, and J. Scott. 1995. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 81:187–195. [DOI] [PubMed] [Google Scholar]
- 15.Blanc, V., S. Litvak, and A. Araya. 1995. RNA editing in wheat mitochondria proceeds by a deamination mechanism. FEBS Lett. 373:56–60. [DOI] [PubMed] [Google Scholar]
- 16.Whitehurst, C.E., M.S. Schlissel, and J. Chen. 2000. Deletion of germline promoter PD beta 1 from the TCR beta locus causes hypermethylation that impairs D beta 1 recombination by multiple mechanisms. Immunity. 13:703–714. [DOI] [PubMed] [Google Scholar]
- 17.Driscoll, D.M., J.K. Wynne, S.C. Wallis, and J. Scott. 1989. An in vitro system for the editing of apolipoprotein B mRNA. Cell. 58:519–525. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro, G.S., K. Aviszus, D. Ikle, and L.J. Wysocki. 1999. Predicting regional mutability in antibody V genes based solely on di- and trinucleotide sequence composition. J. Immunol. 163:259–268. [PubMed] [Google Scholar]
- 19.Kunkel, T.A., and M. Diaz. 2002. Enzymatic cytosine deamination: friend and foe. Mol. Cell. 10:962–963. [DOI] [PubMed] [Google Scholar]
- 20.Peters, A., and U. Storb. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 4:57–65. [DOI] [PubMed] [Google Scholar]
- 21.Tian, M., and F.W. Alt. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275:24163–24172. [DOI] [PubMed] [Google Scholar]