Overlapping Roles of CXCL13, Interleukin 7 Receptor α, and CCR7 Ligands in Lymph Node Development (original) (raw)
Abstract
Lymphoid tissue development is associated with local accumulation of CD4+ CD3− IL-7Rαhi hematopoietic cells that deliver lymphotoxin (LT)α1β2 signals to resident stromal cells. Previous studies have established an important role for CXCL13 (BLC) in the development of Peyer's patches (PP) and some peripheral lymph nodes (LNs), but the chemokine requirements for several LN types, including mesenteric LNs, remain undefined. Using CXCL13−/− mice that additionally carry the paucity of LN T cell mutation (plt/plt), we discovered that CCR7 ligands function in peripheral LN development. We also tested for a genetic interaction during LN development between CXCL13 and a cytokine receptor required in PP development, IL-7Rα. Mice deficient for both CXCL13 and IL-7Rα displayed a striking absence of LNs, including mesenteric LNs. These data extend the role of CXCL13 to the development of all LNs and establish a previously unappreciated role for IL-7Rα in this process. Both circulating and LN CD4+ CD3− IL-7Rαhi cells are shown to express LTα1β2 in an IL-7Rα–dependent manner. Furthermore, CXCL13 was found to be sufficient to mediate CD4+ CD3− IL-7Rαhi cell recruitment in vivo to an ectopic site. These findings indicate that CXCL13 and CCR7 ligands promote accumulation of CD4+ CD3− IL-7Rαhi cells, delivering IL-7Rα–dependent LTα1β2 signals critical for LN development.
Keywords: organogenesis, CXCR5, SLC, ELC, lymphotoxin
Introduction
Genetic studies have identified a key requirement for membrane-bound lymphotoxin (LT)α1β2 and the LTβ receptor (LTβR) in the development of LNs and Peyer's patches (PPs; 1). Evidence from a series of experiments has implicated a population of hematopoietic cells, commonly termed CD4+ CD3− IL-7Rαhi cells, as the critical source of LTα1β2 during lymphoid tissue development (2–5). These cells are found to colonize sites of future PP and LN development and in addition to LTα1β2, they express CD45, the integrin α4β7, and the CXCL13 (BLC) receptor, CXCR5 (2, 6). When these cells are identified by their expression of IL-7Rα or α4β7, a related population of CD4− LTα1β2+ cells is identified in similar numbers (6). IL-7Rα is critical for the development of PPs, and this is thought to be due at least in part to its role in inducing LTα1β2 on CD4+ CD3− IL-7Rαhi cells in the PP anlagen (6, 7). LN development has not yet been reported to require IL-7Rα (8, 9).
The requirement for a circulating cell population to induce LN and PP development indicated a need for chemoattractant molecules to mediate recruitment and clustering of the cells at sites of lymphoid tissue genesis. Consistent with this, CXCL13 and CXCR5 have been identified as important players in development of peripheral LNs and PPs (10, 11). CD4+ IL-7Rαhi cells chemotax to CXCL13 in vitro and a recent study demonstrated that the transfer of wild-type CD4+ CD3− cells to CXCR5-deficient mice was sufficient to restore some aspects of PP development (5, 7). However, it is not clear whether CXCL13 is sufficient to mediate CD4+ CD3− IL-7Rαhi cell recruitment to sites of lymphoid tissue induction. Furthermore, mesenteric LNs and some other LN types continue to develop in CXCR5- and CXCL13-deficient mice and the chemokine requirements for development of these LNs have not been defined.
Here, we demonstrate that CCL19 (ELC) and/or CCL21 (SLC) ligands for CCR7 contribute to the induction of several LN types and we reveal that CXCL13 and IL-7Rα make overlapping contributions that are essential for the development of all LNs.
Materials and Methods
Mice.
C57BL/6 (B6) mice were purchased from Charles River Laboratories and IL-7Rα−/−, γc−/−, and RAG-1−/− mice on a B6 background were purchased from Jackson Laboratories. Rat insulin promoter (RIP)-CXCL13 transgenic mice were on a B6 background (12). CXCR5−/− (10) and CXCL13−/− mice (11) were backcrossed seven to nine times onto a B6 background. plt/plt mice on a BALB/c background (13) were used to cross with CXCL13−/− on a mixed B6/129 background. CXCL13−/− mice were screened by PCR on tail DNA as previously described (11). PCR primers used to type CXCR5+/− versus CXCR5−/− mice were: TGCAAAACTGTGATCGCTCTGC (forward, wild-type), CTTGACGAGTTCTTCTGAGGGGA (forward, mutant), and AACCTTGGCAAAGAGGAGTTCC (reverse). PCR primers used to type +/+, plt/+, and plt/plt were: ATGGGGTCTAGGAAAACATGG (forward), and AAATTATGAGTATTTCACCTGAGTGTG (reverse). These primers amplify a sequence polymorphism adjacent to the plt locus with a product of 76 bp for BALB/c mice and 96 bp for B6 mice. The plt genotype correlated with the presence of a 76-bp band and was occasionally verified by histology or Western blotting for absence of CCL21 protein in LNs. Screening of neonatal IL-7Rα1/− and IL-7Rα−/− mice was performed on thymus by counting cell numbers and staining with anti–IL-7Rα antibody. All experiments were performed in compliance with institutional guidelines for animal care.
LN Enumeration.
Adult mice were injected intraperitoneally with 300 μl of 1% Chicago sky blue 6B dye (also called Pontamine Blue; Sigma-Aldrich) in PBS 7–14 d before they were killed. The dye becomes concentrated within lymphoid tissues and facilitates the macroscopic detection of even very small LNs. LN nomenclature was performed according to Tilney (14). The term “cervical LN” refers to LNs that were termed superficial cervical LNs by Ansel et al. (11) and mandibular LNs by other authors (15). Periaortic LNs are also called iliac LNs. The chain of mesenteric LNs was counted as a single LN. All other LN sets were considered to consist of maximally two nodes, one on each side of the body axis, even though superficial cervical nodes and others can consist of pairs of adjacent LNs. “Percentages of LNs present” were calculated for each LN type using the total number of LNs found in mice from a given strain relative to the maximal number of LNs of that type found in the same number of wild-type B6 mice.
Flow Cytometric Analysis and Immunohistochemical Analysis.
Embryonic or neonatal mice were killed by decapitation, blood outflow was collected in Alsever's solution, and RBCs were lysed in Tris-ammonium chloride for 1 min. Mesenteric LNs were identified and removed using a stereomicroscope. LN suspensions were generated in complete RPMI medium using microscopy slides and lymphocyte size cells were counted using a hematocytometer. To detect surface expression of LTα1β2, cells were pretreated with FcR blocking antibody (2.4.G2; BD Biosciences), 0.5% normal mouse and rat serum, and if indicated, anti-LTβ blocking antibody (BB.F6; reference 16). LTβR-Fc (16) was added and detected using a biotinylated goat anti–human IgG (Jackson ImmunoResearch Laboratories) pretreated for 30 min with 4% normal mouse and rat serum. Finally, streptavidin-allophycocyanin (Molecular Probes) was added along with other surface markers. FITC-conjugated antibodies to B220, CD11b (Caltag), CD11c, and CD3 (BD Biosciences) were used to identify lineage-positive cells. PE-labeled antibodies to α4β7 (DATK32) or IL-7Rα (SB/14) along with PerCP-conjugated anti-CD4 (RM4-5; BD Biosciences) was used to stain for CD4+ CD3− cells. To detect CCR7 expression, cells were resensitized for 30 min at 37°C and then stained with CCL19-Fc (17) or hLFA3-Fc (16) as described above for LTβR-Fc. To detect CXCR5 expression, cells were stained with CXCR5 rabbit antiserum (17) in 4% normal mouse and rat serum, followed by biotinylated goat anti–rabbit IgG (BD Biosciences) and streptavidin-allophycocyanin. Stained cells were analyzed using a four-color FACSCalibur® (BD Biosciences) and FlowJo® software (BD Biosciences). Flow cytometric and immunohistochemical analysis of nonperfused embryonic/newborn pancreas was performed as previously described for adult pancreas (12).
Quantitative RT-PCR Analysis.
PCR primers and probes for CCL19, CCL21, and HPRT were as previously described (18). PCR primers were specific for CCL19-atg but did not distinguish between CCL21-ser and CCL21-leu. Additional primer pairs and probes, including their specificity, orientation (forward [F], reverse [R]), and sequence were as follows: CXCL13 (F tggccagctgcctctctc, R ttgaaatcactccagaacacctaca, probe aggccacggtattctggaagcccat). Quantitative RT-PCR was performed on an ABI7700 sequence detection instrument (Taqman; Applied Biosystems) according to the manufacturer's instructions.
Online Supplemental Material.
Fig. S1 shows photographs of inguinal LN regions in wild-type, IL-7Rα−/−, RAG-1−/−, CXCL13−/−, and CXCL13−/− IL-7Rα−/− mice. Fig. S1 is available at http://www.jem.org/cgi/content/full/jem.20021294/DC1.
Results
CCR7 Ligands Function in LN Development.
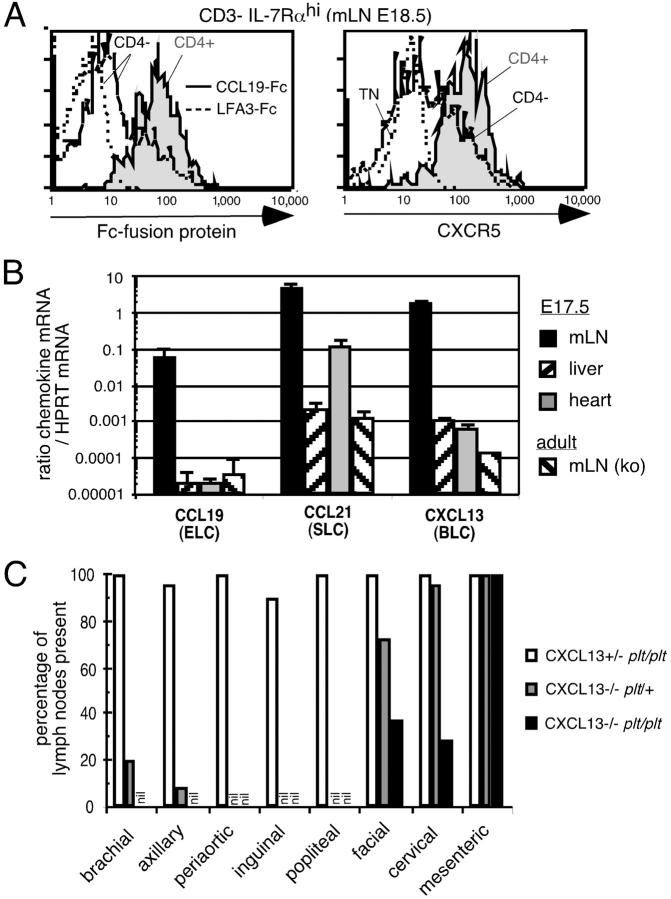
The failure of LN development in CXCL13−/− and CXCR5−/− mice is characterized by variable, incomplete penetrance for affected LNs and consistent development of mesenteric LNs (11, 19). Previous studies have indicated that CD4+ CD3− cells from embryonic intestine express CCR7 mRNA and chemotax to CCL19 (ELC) and CCL21 (SLC) in addition to CXCL13 (7), and we find that CD4+ CD3− IL-7Rαhi cells from embryonic mesenteric LNs are uniformly positive for CCL19-Fc binding as well as for expressing CXCR5 (Fig. 1 A). In previous work, we have established that CCL19-Fc is a faithful reporter of CCR7 expression levels on lymphoid cells (17). CD4− CD3− IL-7Rαhi cells showed weaker staining for both CCR7 and CXCR5 compared with the CD4+ subset (Fig. 1 A). Consistent with CCL19 and CCL21 participating in early events during LN development, mRNA for these chemokines as well as for CXCL13 was found at high levels in the mesenteric region of embryonic day (E) 17.5 mice (Fig. 1 B). CCR7-deficient mice and plt/plt mice that lack the CCL19 gene and the CCL21-ser gene have not previously been found to lack LNs or PPs (13, 20–22). However, we have observed the occasional absence of inguinal LNs in plt/plt and CCR7−/− mice and a single case of axillary LN deficiency in plt/plt mice (Fig. 1 C and not depicted). To test the possibility that the contribution of CCR7 ligands to LN development was partially overlapping with CXCL13, we intercrossed plt/plt mice with mice carrying a targeted deletion of the CXCL13 gene (11). CXCL13−/− plt/plt mice were found to display a markedly greater LN deficiency than observed in CXCL13−/− mice, with frequent absence of facial and cervical LNs (Fig. 1 C). CXCL13−/− plt/plt mice also exhibited a more complete deficiency in brachial and axillary LNs than CXCL13−/− mice. By contrast, mesenteric LNs continued to develop in these multi-chemokine–deficient mice (Fig. 1 C). These observations establish that CCR7 ligands have a broad role in LN development that partially overlaps with the role of CXCL13.
Figure 1.
The chemokines CXCL13 (BLC), CCL19 (ELC), and CCL21 (SLC) play overlapping roles in the development of facial, cervical, brachial, and axillary LNs. (A) Representative histograms showing the staining with CCL19-Fc fusion protein (left) or CXCR5 on the CD4+ (shaded) and CD4− (open) subset of CD3− IL-7Rαhi cells from E 18.5 mesenteric LNs (mLN) of wild-type mice. As a negative control, staining with the fusion protein hLFA3-Fc is shown for embryonic CD4− CD3− IL-7Rαhi cells (left) and CXCR5 staining is shown for CD4− CD3− IL-7Rα− triple negative cells (TN, right) in dotted lines. CD3− indicates low or negative for the lineage markers B220, CD3, CD11c, and CD11b. (B) Quantitative PCR analysis of chemokine expression in the indicated organs from E17.5 B6 mice. As negative controls, mesenteric LN samples from adult CCL19/CCL21-ser (plt/plt) or CXCL13-deficient mice were used. CCL21 expression in fetal heart and the weak expression in fetal liver and in plt/plt mesenteric LNs is likely due to lymphatic expression of the second CCL21 gene, CCL21-leu. Three samples were analyzed per group except for the CXCL13−/− mesenteric LN. ND, not determined. (C) Quantitative assessment of the presence of the indicated LN types detected macroscopically after injection of Chicago sky blue dye (refer to Materials and Methods) in littermate mice deficient for CCL19/CCL21-ser (plt/plt), CXCL13, or CCL19/CCL21-ser and CXCL13 combined. 10–13 mice were investigated per group.
IL-7Rα-dependent LTα1β2 Expression by CD4+ CD3− IL-7Rαhi Cells in LNs and in Blood.
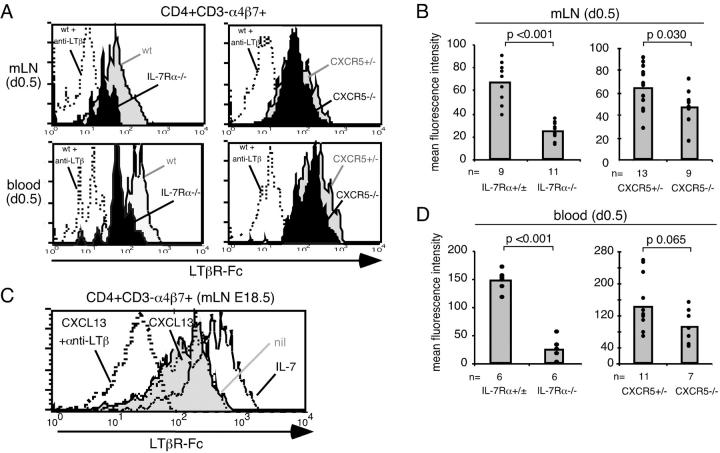
A key function of CD4+ CD3− cells is thought to be the provision of LTα1β2 signals necessary for LN and PP development (1). IL-7Rα functions in PP development by inducing LTα1β2 on CD4+ CD3− cells (6, 7), and we previously observed that CXCL13 up-regulates LTα1β2 on B cells (11). Therefore, we reasoned that IL-7Rα and CXCL13 may both contribute to LTα1β2 induction in LN CD3− IL-7Rαhi cells. LTα1β2 levels were measured on cells from newborn (day 0.5) IL-7Rα−/− and CXCL13−/− or CXCR5−/− mesenteric LNs. To facilitate identification of CD4+ CD3− cells in IL-7Rα−/− mice, α4β7 was used as a marker in place of IL-7Rα (2, 6). CD4+ CD3− α4β7+ cells from mesenteric LNs of IL-7Rα−/− mice showed strongly decreased staining with LTβR-Fc compared with wild-type cells (Fig. 2, A and B) although the staining remained above the background level determined by pretreatment of cells with an LTβ-specific antibody that blocks LTβR-Fc binding to LTα1β2 (Fig. 2 A). CD4− CD3− α4β7+ cells exhibited bimodular staining with LTβR-Fc and their frequency in IL-7Rα−/− mice was too low to allow reliable measurement of LTα1β2 levels (not depicted). Analysis of cells from CXCR5−/− mice showed LTα1β2 expression levels close to the wild-type control (Fig. 2, A and B). In agreement with the in vivo observations, in vitro exposure of CD4+ CD3− α4β7+ and CD4− CD3− α4β7+ cells from embryonic mesenteric LNs to IL-7 was sufficient to promote up-regulation of LTα1β2 (Fig. 2 C and not depicted). By contrast, CXCL13 did not lead to a detectable up-regulation of LTβR-Fc staining although the high constitutive expression of LTα1β2 on CD4+ CD3− α4β7+ cells in the absence of chemokine or cytokine addition may have obscured small effects of CXCL13 (Fig. 2 C). To examine at which step LTα1β2 induction occurs in vivo, we asked whether a difference in LTα1β2 expression could be observed between CD4+ CD3− α4β7+ cells in blood versus mesenteric LNs. Surprisingly, we observed levels of surface LTα1β2 expression on circulating CD4+ CD3− α4β7+ cells that were similar to those on cells in mesenteric LNs and this expression was also partially dependent on IL-7Rα (Fig. 2, A and D) but not CXCR5 (Fig. 2, A and D).
Figure 2.
IL-7Rα but not CXCR5 signals promote induction of LTα1β2 on CD4+ CD3− α4β7+ cells in developing mesenteric LNs and in blood. (A) Representative histograms showing LTβR-Fc staining on CD4+ CD3− α4β7+ cells from day 0.5 mesenteric LNs (mLN) or blood of wild-type (wt) or CXCR5+/− (shaded) and IL-7Rα−/− or CXCR5−/− mice (solid) as indicated. As a negative control, LTβR-Fc staining of wild-type cells pretreated with anti-LTβ blocking antibody is shown (dotted line). (B) Compilation of data as shown in A, showing the mean fluorescence intensity (MFI) of LTβR-Fc staining on CD4+ CD3− α4β7+ cells from mesenteric LNs of newborn mice of the indicated genotypes. (C) LTβR-Fc staining of sorted CD4+ CD3− α4β7+ cells from mesenteric LN cells of E18.5 mice that had been cultured for 6 h in vitro without treatment (nil, shaded), with 2 μg/ml CXCL13 (thin dotted line), or with 20 ng/ml IL-7 (open). As a negative control, LTβR-Fc staining of wild-type cells treated with anti-LTβ blocking antibody is shown (thick dotted line). CD3− indicates low or negative for the lineage markers B220, CD3, CD11c, and CD11b. Data are representative of two experiments. Similar observations were made for CD4− CD3− α4β7+ cells in the same cultures. (D) Compilation of data as in A, showing the MFI of LTβR-Fc staining on CD4+ CD3− α4β7+ cells from blood of newborn mice of the indicated genotypes. In B and D, plotted values represent the MFI after subtraction of the background LTβR-Fc staining on cells treated with LTβ blocking antibody. The data are compiled from two to four independent experiments, with littermate mice used as controls. IL-7Rα−/− mice were either CXCL13+/+ or CXCL13+/−, and data were pooled as no difference was observed between these two groups.
Mice with Combined Deficiency for CXCL13 and IL-7Rα Lack all LNs.
The substantial reduction in LTα1β2 expression on CD4+ CD3− α4β7+ cells of IL-7Rα−/− mice prompted us to investigate the presence of LNs in these mice. Although earlier studies have not reported a LN deficiency in IL-7Rα–deficient mice (8, 9), we observed that about half of the IL-7Rα single–deficient mice we analyzed lacked one or both macroscopically detectable axillary, periaortic, inguinal, and cervical LNs (Fig. 3 A). The LNs that remained in these animals were often very small, typically smaller than those present in lymphocyte-deficient RAG-1−/− mice (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20021294/DC1). Microscopic examination of sites lacking LNs revealed the occasional presence of rudimentary lymphoid nodules (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20021294/DC1, and not depicted). The presence of facial and mesenteric LNs was largely unaffected by IL-7Rα deficiency although their size was clearly reduced (Fig. 3, A and C, and not depicted). Analysis of mice deficient in the IL-2R common γ chain (γc) that is required for signaling by several cytokine receptors, including the IL-7 receptor, revealed a similar deficiency in LNs (Fig. 3 A).
Figure 3.
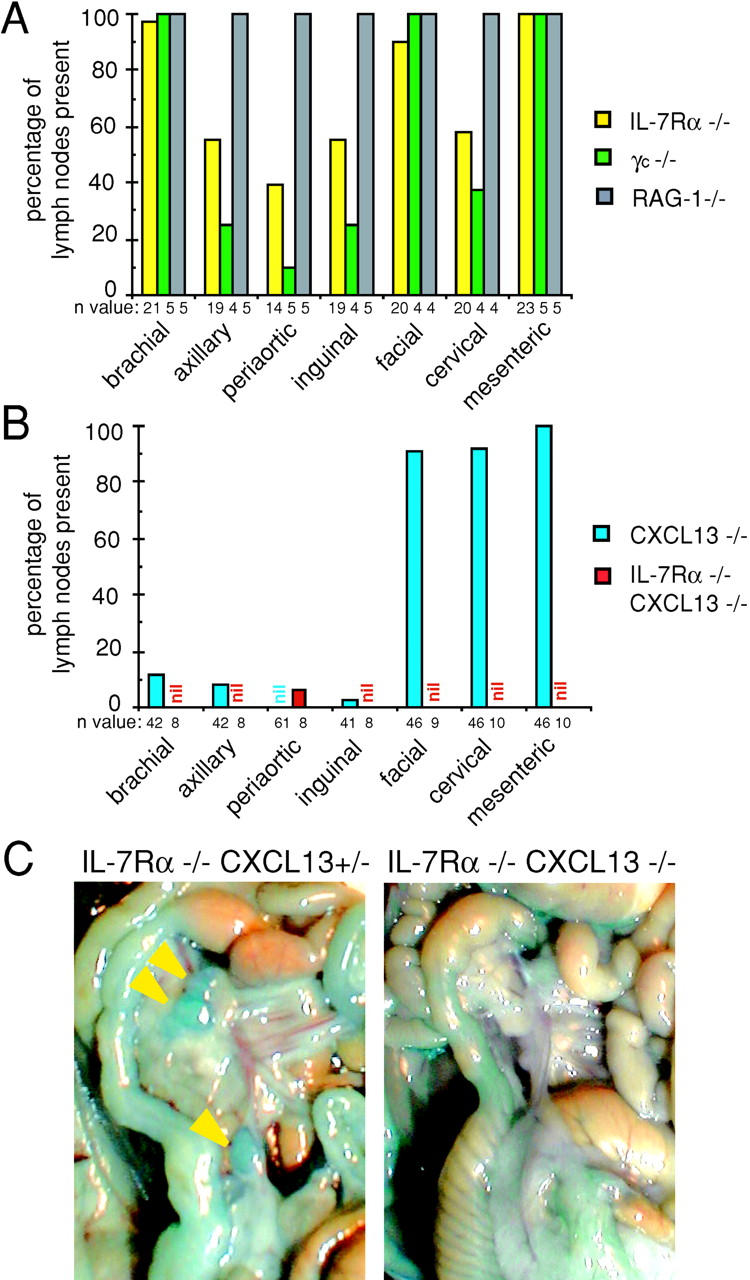
Deficiency of macroscopically detectable LNs in IL-7Rα−/− and IL-7Rα−/− CXCL13−/− mice. (A and B) Quantitative assessment of the presence of the indicated LNs in mice deficient for IL-7Rα, γc chain, RAG-1 (A), CXCL13, or IL-7Rα and CXCL13 combined (B), compared with wild-type levels (100%). Importantly, with the exception of one periaortic LN, all LNs were missing in IL-7Rα−/− CXCL13−/− mice. The LN enumeration data for CXCL13−/− mice is equivalent to that previously reported (reference 11) and is shown here to facilitate the comparison with the other mouse strains. No difference in LN presence was observed between IL-7Rα−/− CXCL13+/+ and IL-7Rα−/− CXCL13+/− mice. 100% of IL-7Rα−/− mice also lacked sacral LNs (not depicted). The n value indicates the number of mice that were investigated using Chicago sky blue dye. (C) Representative photograph of mesentery region in IL-7Rα−/− CXCL13+/− and IL-7Rα−/− CXCL13−/− mice that had been injected with Chicago sky blue dye, showing the presence of several small mesenteric LNs in IL-7Rα−/− CXCL13+/− mice (arrow heads) and the absence of detectable mesenteric LNs in IL-7Rα−/− CXCL13−/− mice.
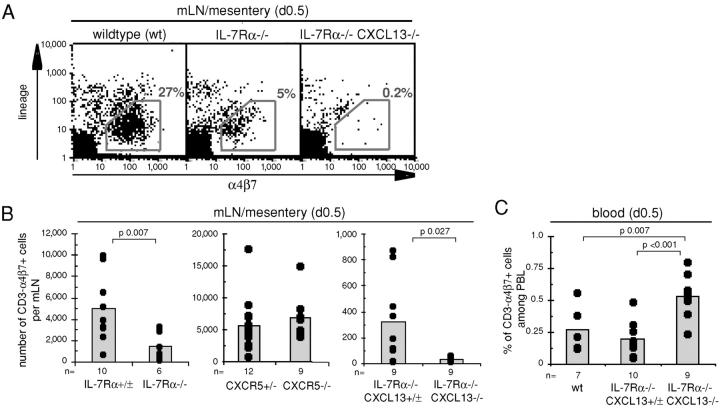
To explore whether IL-7Rα and CXCL13 contribute to the same process in LN development, mice with combined IL-7Rα and CXCL13 deficiency were investigated. Strikingly, all seven LN types investigated were absent in these mice, including facial and mesenteric LNs, with the exception of a single animal that retained a periaortic LN (Fig. 3, B and C, and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20021294/DC1). Enumeration of CD3− α4β7+ cells in day 0.5 mesenteric LNs revealed that they were reduced in IL-7Rα−/− mice but not in CXCR5−/− or CXCL13−/− mice (Fig. 4, A and B , and not depicted). Consistent with a very early block in mesenteric LN development in CXCL13−/− IL-7Rα−/− mice, CD3− α4β7+ cells were almost absent in the mesenteric region of day 0.5 animals (Fig. 4, A and B). Analysis of CD3− α4β7+ cell frequencies in newborn blood showed that wild-type, IL-7Rα−/−, CXCR5−/−, and CXCL13−/− mice had low but comparable frequencies whereas in CXCL13−/− IL-7Rα−/− mice their frequency was slightly increased (Fig. 4 C and not depicted). These findings indicate that CXCL13 and IL-7Rα make overlapping contributions that are critical for appropriate accumulation of CD3− IL-7Rαhi cells during LN development.
Figure 4.
IL-7Rα and CXCR5 signals promote the accumulation of CD3− α4β7+ cells in developing mesenteric LNs. (A) Representative dot plots of day 0.5 mesenteric LNs or mesentery region from wild-type, IL-7Rα−/−, and IL-7Rα−/− CXCL13−/− mice showing reduced numbers and absence of CD3− α4β7+ cells in IL-7Rα−/− and IL-7Rα−/− CXCL13−/− mice, respectively. (B) Histograms depicting the number of CD3− α4β7+ cells per mesenteric LN of newborn mice of the genotype indicated. In each case, 40% of CD3− α4β7+ cells were CD4+. (C) Histogram showing the percentage of CD3− α4β7+ cells in peripheral blood leukocytes of newborn mice of the genotype indicated. n, number of mice investigated. Each set is a compilation of two to three different experiments, with littermate mice used as controls. In B and C, CD3− indicates low or negative for the lineage markers B220, CD3, CD11c, and CD11b except for IL-7Rα+/− mice (B) where only B220, CD3, and CD11c were used as lineage markers.
Accumulation of CD4+ IL-7Rαhi Cells in Pancreatic Islets of RIP-CXCL13 Mice.
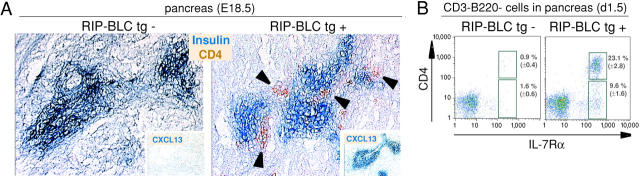
Previous studies have demonstrated that ectopic expression of CXCL13 in the pancreatic islets leads to recruitment of B and T lymphocytes in large numbers (12). To determine whether CXCL13 was sufficient to mediate recruitment of CD4+ CD3− IL-7Rαhi cells, we analyzed the pancreas of RIP-CXCL13 transgenic mice at E 18.5, a time when few T or B lymphocytes are present in circulation. As expected from previous studies of RIP activity, CXCL13 transgene expression was detected in the fetal pancreas of transgenic but not control mice (Fig. 5 A, insets). Immunohistochemical staining for CD4 revealed the presence of numerous CD4+ cells in the pancreas of transgenic but not control mice, with most of the cells situated proximal to the CXCL13-expressing islet cells (Fig. 5 A). No CD3+ cells could be identified in the pancreas at this time (not depicted). Flow cytometric analysis confirmed the presence of CD4+ CD3− IL-7Rαhi cells in the pancreas of transgenic mice and revealed their frequency to be ∼20-fold above the background level detected in littermate controls (Fig. 5 B). CD4− IL-7Rαhi cells were also detected in the transgenic pancreas, although at a lower frequency than the CD4+ subset (Fig. 5 B) consistent with their lower expression of CXCR5 (Fig. 1 A). These findings support the conclusion that CXCL13 plays a direct role in promoting the accumulation of CD4+ CD3− IL-7Rαhi cells in nascent LNs.
Figure 5.
CD4+ IL-7Rαhi cells in the pancreas of RIP-CXCL13 transgenic mice. (A) Immunohistochemical analysis of pancreas from RIP-CXCL13 transgenic (tg+) and littermate control (tg−) E 18.5 embryos to detect insulin-producing β cells (blue) and CD4+ cells (brown). Insets, adjacent sections stained to detect CXCL13 transgene expression (blue). (B) Frequency of IL-7Rαhi CD3− B220− cells in day 1.5 pancreas tissue of transgenic and littermate control mice. Numbers show average percentages of CD4+ and CD4− IL-7Rαhi cells ± standard deviation in seven tg− and five tg+ mice.
Discussion
The above observations establish that CCR7 ligands function in the development of several LN types and that CXCL13 (BLC) and IL-7Rα function in the development of all LNs. In the case of peripheral LNs, CXCL13 and IL-7Rα each play such a strong role that mice lacking either molecule fail to develop some of their LNs. Deficiency in mesenteric, cervical, and facial LNs becomes evident only when both molecules are absent, indicating overlapping contributions by the CXCL13 and IL-7Rα pathways. We also demonstrate that IL-7Rα–dependent signals promote LTα1β2 expression on circulating CD4+ CD3− cells and that CXCL13 mediates in vivo recruitment of these LTα1β2-expressing cells.
The finding that CCL21 (SLC) and/or CCL19 (ELC) function in the development of facial, cervical, brachial, and axillary LNs provides the second example of chemokine receptor involvement in lymphoid tissue organogenesis. An initial indication that CCR7 may function in lymphoid tissue development came from the observation that CD4+ CD3− IL-7Rαhi cells from embryonic intestine can respond to CCL19 and CCL21, and that CCL19 was up-regulated at an early step during PP development in an IL-7Rα–dependent manner (7). As CD4+ CD3− cells express both CXCR5 and CCR7 and respond similarly in vitro to CXCL13, CCL19, and CCL21, we suggest that these molecules may function at a similar early step in LN development, such as in the recruitment and clustering of CD4+ CD3− cells, with CXCL13 playing the dominant role. The continued ability of CXCL13−/− plt/plt mice to develop some LN types, including mesenteric LNs, might reflect the incomplete deficiency of CCL21 in plt/plt mice because this spontaneous mutant mouse strain retains lymphatic CCL21 expression (21). As the plt mutation arose by a spontaneous deletion, it is formally possible that the LN development defect we observed reflects the role of some other gene that was disrupted by the deletion, although the finding of similar peripheral LN deficiencies in plt/plt and CCR7−/− mice makes this unlikely. Analysis of CXCR5/CCR7 double deficient mice should help to further address this issue. Our studies leave open the possibility that additional chemokine receptors, such as CXCR4 (5), participate in inducing recruitment and adhesion of CD3− IL-7Rαhi cells, particularly during mesenteric LN development.
Previous experiments have established a critical role for the IL-7Rα chain in PP but not LN development (8, 9, 23). The finding that IL-7Rα−/− mice lack a fraction of their axillary, inguinal, periaortic, and cervical LNs demonstrates that this receptor has an important nonredundant role in LN organogenesis. Mice deficient in the IL2Rγc chain, which partners with IL-7Rα in the IL-7 receptor but not the TSLP receptor (24), have been suggested to have a partial deficiency in some peripheral LNs (25) and our findings confirm and extend this observation. Similarly, mice deficient in the tyrosine kinase jak-3 that transmits signals by γc chain–containing receptors have been shown to lack some peripheral but not mesenteric LNs (26). By showing that lymphocyte-deficient RAG-1−/− mice retained all seven LN types analyzed, we have excluded that the LN deficiencies in the IL-7Rα and γc mutants is due to the role of IL-7 receptor signaling in lymphocyte development. Although the extent of LN deficiency in IL-7−/− mice has not yet been established (27), the present findings suggest that the IL-7Rα requirement in LN development is due to its involvement in IL-7Rα/γc heterodimers that transmit signals in response to IL-7.
The consistently smaller LN size in IL-7Rα−/− compared with RAG-1−/− mice (see Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20021294/DC1) indicates that IL-7Rα signaling may have a more general role in LN development than indicated by the measurement of LN presence or absence. In this regard it is important to note that IL-7Rα deficiency, but not CXCR5 or CXCL13 deficiency, led to decreased numbers of CD3− α4β7+ cells in mesenteric LNs at birth. IL-7Rα deficiency did not have a significant effect on the frequency of these cells in newborn blood, suggesting that this pathway was required for the cells to accumulate within the developing mesenteric LN. As the IL-7Rα signal was important for promoting maximal expression of LTα1β2 on CD4+ CD3− α4β7+ cells, a likely explanation is that the accumulation of CD3− α4β7+ cells is the result of ongoing recruitment, and the lower LTα1β2 expression on CD4+ CD3− α4β7+ cells in the absence of IL-7Rα results in weaker induction of chemokines and adhesion molecules needed for cell recruitment or retention (6, 7). Consistent with this possibility, the accumulation of CD4+ CD3− cells is reduced to a similar extent in the mesenteric region of newborn IL-7Rα−/− mice (Fig. 4) as in LTα−/− mice (28). IL-7Rα signals could also be involved in proliferation or survival of CD3− α4β7+ cells, similar to the role of this pathway in lymphocyte development. It remains to be determined whether the reduced LTα1β2 expression or reduced numbers of CD4+ CD3− cells are sufficient to explain the small LN size in IL-7Rα−/− mice, or whether IL-7Rα signaling has additional functions necessary for LN development.
In experiments to determine whether a local source of IL-7Rα ligand is responsible for the induction of LTα1β2 in CD4+ CD3− α4β7+ cells during LN development, we observed similar levels of LTα1β2 expression on CD4+ CD3− α4β7+ cells from blood compared with mesenteric LNs. This finding suggests that CD4+ CD3− α4β7+ cells are induced to express LTα1β2 before entry into early LNs. This possibility is consistent with the lack of evidence for focal expression of IL-7Rα ligands in developing PPs (6, 7) and with the observation that transgenic expression of IL-7 diffusely within intestinal enterocytes is sufficient to partially restore defective PP development in IL-7−/− mice (29). In further support of a systemic action for IL-7Rα ligands, a recent study showed rescue of LN development in TRAF6−/− mice by intraembryonic IL-7 injection (30).
Although IL-7Rα signals play a major role in LTα1β2 induction on mesenteric CD4+ CD3− IL-7Rαhi cells, a considerable level of LTα1β2 remains on these cells in the absence of IL-7Rα (Fig. 2, A and B), implicating the involvement of other LTα1β2-inducing signals. Our studies fail to establish a role for CXCL13 in mediating LTα1β2 induction in these cells, although a small contribution by CXCL13 is not excluded. TRANCE and TRANCE receptor, which are both expressed on CD4+ CD3− IL-7Rαhi cells, play a critical role in the development of LNs but not PPs (28), and a recent study demonstrated in vitro that TRANCE receptor signaling via TRAF6 induces LTα1β2 expression CD4+ CD3− cells (30). Although this study establishes an important role for TRAF6-dependent signals in induction of LTα1β2, TRAF6−/− CD4+ CD3− cells retained detectable LTα1β2 expression (30). This observation is consistent with our finding that IL-7Rα signaling contributes to the induction of LTα1β2 on CD4+ CD3− cells found in developing mesenteric LNs and therefore we propose that both IL-7Rα signaling and TRANCE receptor signaling contribute to LTα1β2 induction during LN development.
Our findings are consistent with the possibility that CXCL13 is needed for efficient recruitment and/or clustering of IL-7Rαhi lymphoid tissue–inducing cells in the earliest LN anlagen and that IL-7Rα is needed for maximal expression of LTα1β2 on these cells. In the absence of CXCL13, other chemokines, including CCL21 and CCL19, may recruit sufficient IL-7Rαhi cells to the mesenteric LN anlagen such that the necessary LTα1β2 signaling is provided for events to proceed. When mice lack IL-7Rα, the lymphoid tissue–inducing cells have reduced amounts of LTα1β2 expression and the combined action of CXCL13 and additional chemokines might be able to recruit/cluster enough of the cells to ensure a sufficient LTα1β2 signal is provided for development to proceed. We propose that when both CXCL13 and IL-7Rα are lacking, the combined effect of reduced cell recruitment/clustering and reduced LTα1β2 expression on each cell results in insufficient LTα1β2 signaling for LN development. Thus, we establish for the first time the broad contribution of CXCL13 and IL-7Rα signaling to the development of all LNs and we suggest that although these molecules make distinct contributions, their ultimate contribution to the provision of LTα1β2 is partially redundant. Finally, our experiments establish that CCR7 ligands contribute to the development of peripheral and cervical LNs.
Acknowledgments
We thank Martin Lipp, Hideki Nakano, and Ninan Abraham for mice, Jeff Browning for staining reagents, Paul Dazin for cell sorting, and Ying Xu for excellent technical assistance. We also thank Theresa Lu and Takaharu Okada for critical reading of the manuscript.
S.A. Luther was supported by a Human Frontiers Science Program postdoctoral fellowship and is currently a research associate of the HHMI. K.M. Ansel was a predoctoral fellowship of the HHMI and is currently supported by the Damon Runyon Cancer Research Foundation (DRG-1682). J.G. Cyster is an Assistant Investigator of HHMI and a Packard Fellow. This work was supported in part by National Institutes of Health grant AI45073.
K.M. Ansel's present address is Center for Blood Research, Harvard Medical School, 200 Longwood Avenue, Boston, MA 02115.
References
- 1.Fu, Y.-X., and D.D. Chaplin. 1999. Development and maturation of secondary lymphoid tissues. Annu. Rev. Immunol. 17:399–433. [DOI] [PubMed] [Google Scholar]
- 2.Mebius, R.E., P. Rennert, and I.L. Weissman. 1997. Developing lymph nodes collect CD4+CD3− LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 7:493–504. [DOI] [PubMed] [Google Scholar]
- 3.Nishikawa, S., K. Honda, H. Hashi, and H. Yoshida. 1998. Peyer's patch organogenesis as a programmed inflammation: a hypothetical model. Cytokine Growth Factor Rev. 9:213–220. [DOI] [PubMed] [Google Scholar]
- 4.Fukuyama, S., T. Hiroi, Y. Yokota, P.D. Rennert, M. Yanagita, N. Kinoshita, S. Terawaki, T. Shikina, M. Yamamoto, Y. Kurono, et al. 2002. Initiation of NALT organogenesis is independent of the IL-7R, LTβR, and NIK signaling pathways but requires the Id2 gene and CD3− CD4+CD45+ cells. Immunity. 17:31–40. [DOI] [PubMed] [Google Scholar]
- 5.Finke, D., H. Acha-Orbea, A. Mattis, M. Lipp, and J. Kraehenbuhl. 2002. CD4(+)CD3(−) cells induce Peyer's patch development. Role of alpha4beta1 integrin activation by CXCR5. Immunity. 17:363–373. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida, H., K. Honda, R. Shinkura, S. Adachi, S. Nishikawa, K. Maki, K. Ikuta, and S.I. Nishikawa. 1999. IL-7 receptor alpha+ CD3(−) cells in the embryonic intestine induces the organizing center of Peyer's patches. Int. Immunol. 11:643–655. [DOI] [PubMed] [Google Scholar]
- 7.Honda, K., H. Nakano, H. Yoshida, S. Nishikawa, P. Rennert, K. Ikuta, M. Tamechika, K. Yamaguchi, T. Fukumoto, T. Chiba, et al. 2001. Molecular basis for hematopoietic/mesenchymal interaction during initiation of Peyer's patch organogenesis. J. Exp. Med. 193:621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peschon, J.J., P.J. Morrissey, K.H. Grabstein, F.J. Ramsdell, E. Maraskovsky, B.C. Gliniak, L.S. Park, S.F. Ziegler, D.E. Williams, C.B. Ware, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 180:1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmsen, A., K. Kusser, L. Hartson, M. Tighe, M.J. Sunshine, J.D. Sedgwick, Y. Choi, D.R. Littman, and T.D. Randall. 2002. Cutting edge: organogenesis of nasal-associated lymphoid tissue (NALT) occurs independently of lymphotoxin-alpha (LT alpha) and retinoic acid receptor-related orphan receptor-gamma, but the organization of NALT is LT alpha dependent. J. Immunol. 168:986–990. [DOI] [PubMed] [Google Scholar]
- 10.Förster, R., A.E. Mattis, E. Kremmer, E. Wolf, G. Brem, and M. Lipp. 1996. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 87:1037–1047. [DOI] [PubMed] [Google Scholar]
- 11.Ansel, K.M., V.N. Ngo, P.L. Hyman, S.A. Luther, R. Förster, J.D. Sedgwick, J.L. Browning, M. Lipp, and J.G. Cyster. 2000. A chemokine driven positive feedback loop organizes lymphoid follicles. Nature. 406:309–314. [DOI] [PubMed] [Google Scholar]
- 12.Luther, S.A., T. Lopez, W. Bai, D. Hanahan, and J.G. Cyster. 2000. BLC expression in pancreatic islets causes B cell recruitment and lymphotoxin-dependent lymphoid neogenesis. Immunity. 12:471–481. [DOI] [PubMed] [Google Scholar]
- 13.Gunn, M.D., S. Kyuwa, C. Tam, T. Kakiuchi, A. Matsuzawa, L.T. Williams, and H. Nakano. 1999. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 189:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tilney, N.L. 1971. Patterns of lymphatic drainage in the adult laboratory rat. J. Anat. 109:369–383. [PMC free article] [PubMed] [Google Scholar]
- 15.Rennert, P.D., J.L. Browning, and P.S. Hochman. 1997. Selective disruption of lymphotoxin ligands reveals a novel set of mucosal lymph nodes and unique effects on lymph node cellular organization. Int. Immunol. 9:1627–1639. [DOI] [PubMed] [Google Scholar]
- 16.Rennert, P.D., D. James, F. Mackay, J.L. Browning, and P.S. Hochman. 1998. Lymph node genesis is induced by signaling through the lymphotoxin beta receptor. Immunity. 9:71–79. [DOI] [PubMed] [Google Scholar]
- 17.Reif, K., E.H. Ekland, L. Ohl, H. Nakano, M. Lipp, R. Forster, and J.G. Cyster. 2002. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 416:94–99. [DOI] [PubMed] [Google Scholar]
- 18.Luther, S.A., A. Bidgol, D.C. Hargreaves, A. Schmidt, Y. Xu, J. Paniyadi, M. Matloubian, and J.G. Cyster. 2002. Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis. J. Immunol. 169:424–433. [DOI] [PubMed] [Google Scholar]
- 19.Forster, R., A.E. Mattis, E. Kremmer, E. Wolf, G. Brem, and M. Lipp. 1996. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 87:1037–1047. [DOI] [PubMed] [Google Scholar]
- 20.Förster, R., A. Schubel, D. Breitfeld, E. Kremmer, I. Renner-Muller, E. Wolf, and M. Lipp. 1999. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 99:23–33. [DOI] [PubMed] [Google Scholar]
- 21.Vassileva, G., H. Soto, A. Zlotnik, H. Nakano, T. Kakiuchi, J.A. Hedrick, and S.A. Lira. 1999. The reduced expression of 6Ckine in the plt mouse results from the deletion of one of two 6Ckine genes. J. Exp. Med. 190:1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther, S.A., H.L. Tang, P.L. Hyman, A.G. Farr, and J.G. Cyster. 2000. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc. Natl. Acad. Sci. USA. 97:12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adachi, S., H. Yoshida, K. Honda, K. Maki, K. Saijo, K. Ikuta, T. Saito, and S.I. Nishikawa. 1998. Essential role of IL-7 receptor alpha in the formation of Peyer's patch anlage. Int. Immunol. 10:1–6. [DOI] [PubMed] [Google Scholar]
- 24.Leonard, W.J. 2002. TSLP: finally in the limelight. Nat. Immunol. 3:605–607. [DOI] [PubMed] [Google Scholar]
- 25.Cao, X., E.W. Shores, J. Hu-Li, M.R. Anver, B.L. Kelsall, S.M. Russell, J. Drago, M. Noguchi, A. Grinberg, E.T. Bloom, et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity. 2:223–238. [DOI] [PubMed] [Google Scholar]
- 26.Park, S.Y., K. Saijo, T. Takahashi, M. Osawa, H. Arase, N. Hirayama, K. Miyake, H. Nakauchi, T. Shirasawa, and T. Saito. 1995. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 3:771–782. [DOI] [PubMed] [Google Scholar]
- 27.von Freeden-Jeffry, U., P. Vieira, L.A. Lucian, T. McNeil, S.E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, D., R.E. Mebius, J.D. MacMicking, S. Jung, T. Cupedo, Y. Castellanos, J. Rho, B.R. Wong, R. Josien, N. Kim, et al. 2000. Regulation of peripheral lymph node genesis by the tumor necrosis factor family member TRANCE. J. Exp. Med. 192:1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laky, K., L. Lefrancois, E.G. Lingenheld, H. Ishikawa, J.M. Lewis, S. Olson, K. Suzuki, R.E. Tigelaar, and L. Puddington. 2000. Enterocyte expression of interleukin 7 induces development of gammadelta T cells and Peyer's patches. J. Exp. Med. 191:1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida, H., A. Naito, J. Inoue, M. Satoh, S.M. Santee-Cooper, C.F. Ware, A. Togawa, and S. Nishikawa. 2002. Different cytokines induce surface lymphotoxin-alphabeta on IL-7 receptor-alpha cells that differentially engender lymph nodes and Peyer's patches. Immunity. 17:823–833. [DOI] [PubMed] [Google Scholar]