Mycobacterium marinum Escapes from Phagosomes and Is Propelled by Actin-based Motility (original) (raw)
Abstract
Mycobacteria are responsible for a number of human and animal diseases and are classical intracellular pathogens, living inside macrophages rather than as free-living organisms during infection. Numerous intracellular pathogens, including Listeria monocytogenes, Shigella flexneri, and Rickettsia rickettsii, exploit the host cytoskeleton by using actin-based motility for cell to cell spread during infection. Here we show that Mycobacterium marinum, a natural pathogen of fish and frogs and an occasional pathogen of humans, is capable of actively inducing actin polymerization within macrophages. M. marinum that polymerized actin were free in the cytoplasm and propelled by actin-based motility into adjacent cells. Immunofluorescence demonstrated the presence of host cytoskeletal proteins, including the Arp2/3 complex and vasodilator-stimulated phosphoprotein, throughout the actin tails. In contrast, Wiskott-Aldrich syndrome protein localized exclusively at the actin-polymerizing pole of M. marinum. These findings show that M. marinum can escape into the cytoplasm of infected macrophages, where it can recruit host cell cytoskeletal factors to induce actin polymerization leading to direct cell to cell spread.
Keywords: mycobacteria, macrophage, Arp2/3, WASP, VASP
Introduction
Organisms of the genus Mycobacterium cause the human disease tuberculosis, as well as tuberculosis-like diseases in cattle, deer, voles, and fish. Although they are recognized as classical intracellular pathogens of macrophages, the mechanism by which mycobacteria invade and persist in host cells is not well understood. Mycobacterium marinum causes a systemic tuberculosis-like disease in its natural hosts, fish and frogs, and a localized disease in immunocompetent humans, both marked by the presence of a granulomatous host response, a hallmark of the human systemic diseases caused by mycobacteria, tuberculosis, and leprosy. Like Mycobacterium tuberculosis, M. marinum exists in vivo in host macrophages, leading to aggregation of infected cells and ultimately granuloma formation (1). M. marinum is closely related to M. tuberculosis not only in its pathology but also genetically (2), and has been used increasingly as a model for understanding the pathogenesis of tuberculosis (3–5).
The list of bacterial pathogens known to initiate actin-based motility is diverse and thus far includes the Gram-positive bacterium Listeria monocytogenes and the Gram-negative bacteria Shigella flexneri and Rickettsia rickettsii (6). These pathogens share the ability during intracellular infection to enter the host cell cytoplasm, induce actin polymerization, and use actin-based motility for spread between host cells. Direct cell to cell spread allows these pathogens to circumvent some host immune responses, e.g., antibody and complement.
In contrast to Listeria, Shigella, and Rickettsia, pathogenic mycobacteria are widely believed not to enter the cytoplasm, but to exist exclusively within phagosomes. The infecting mycobacteria alter phagosome maturation so that these membrane-bound compartments become suitable environments for survival and proliferation of the pathogen (7). Here we demonstrate that intracellular M. marinum not only enters the cytoplasm of infected macrophages, but also has the ability to be propelled by actin-based motility through induction of actin polymerization using host cytoskeletal factors. In addition to extending the ability to induce actin polymerization to a distinct type of bacterium, these studies raise the possibility that escape from the phagosome and direct cell to cell spread might be significant for the pathogenesis of M. marinum infection.
Materials and Methods
Macrophages.
Macrophages were derived from the bone marrow of either 129/Sv or C57BL/6 mice as previously described (8). Cells were harvested 10–18 d after plating and allowed to adhere to fibronectin-coated coverslips (Becton Dickinson) for infection with M. marinum the next day. The fish macrophage cell line CLC was maintained as previously described (9) and seeded onto fibronectin-coated coverslips and infected similarly to bone marrow–derived macrophages (BMDMs).
Bacteria and Infection.
M. marinum expressing green fluorescent protein (GFP) were generated by transforming M. marinum with a GFP expression plasmid as previously described (5). Wild-type (strain M) or GFP-expressing M. marinum were cultured in Middlebrook 7H9 (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% ADC enrichment (Fisher Scientific).
For infection, bacteria were washed twice in serum-free cell culture media and disrupted into single bacilli by passage through a 26-gauge needle. Immediately before infection, BMDMs and CLC were washed with serum-free medium. M. marinum were added to the cells at a multiplicity of infection (MOI) of 1, centrifuged at 500 g for 10 min, and incubated at 32°C (BMDM) or 28°C (CLC) in 5% CO2. After 2 h, the infected cells were washed with serum-free medium to remove extracellular bacteria. BMDMs or CLC were incubated further in DMEM with 2% FBS, 20 mM Hepes, 2% CMG14–12 SN (10) at 32°C in 5% CO2 (BMDM) or MEM with 2% FBS supplemented with 10% essential amino acids at 28°C in 5% CO2 (CLC) for 48 h before microscopy.
Intercellular spreading assays were performed in confluent monolayers of A549 cells infected with M. marinum at an MOI of 0.1 essentially as previously described (11). In some wells, culture medium contained 40 μg/ml amikacin, a concentration that we have found does not affect the growth of intracellular M. marinum, but effectively kills extracellular bacteria. Media was changed every 2 d and monolayers were examined for pattern of infection 8 d later.
In Vitro Actin Polymerization.
Mouse brain extracts were prepared as previously described (12). For cell-free extract studies, M. marinum were isolated from BMDMs infected for at least 48 h. Bacteria were centrifuged, washed, and added to the extract with an ATP-containing energy mix, rhodamine-actin, 1% Triton X-100, 2% methyl cellulose, and a mix of glucose oxidase, catalase, and glucose, and examined microscopically after 30 min or 1 h at room temperature.
Microscopy.
Time lapse video sequences were taken at 32°C using a 60× objective on a Nikon Eclipse TE 300 inverted microscope. Images were acquired at 2- or 5-s intervals with a MicroMax cooled CCD camera (Princeton Instruments) with IPLabs acquisition software (Scanalytics). Average rates of bacterial movement were determined by analysis of multiple sequential images and a stage micrometer using Adobe Photoshop.
2 μM CM-DiI (Molecular Probes) was used to label intracellular membranes by adding to infected BMDMs for 1 h before fixation. Labeled, infected BMDMs were fixed with 3.7% paraformaldehyde and permeabilized with 0.1% Triton X-100. Alexa Fluor phalloidin (Molecular Probes) was added to coverslips for 20 min at room temperature to stain for F-actin. To localize host cytoskeletal proteins, infected BMDMs were fixed as described above and permeabilized with cold methanol. Subsequent indirect immunofluorescence was performed with anti-arp3 (13), anti–p34-Arc (14), anti–vasodilator-stimulated phosphoprotein (VASP; reference 15), anti–Wiskott-Aldrich syndrome protein (WASP; Santa Cruz Biotechnology, Inc.), anti-actin (Sigma-Aldrich), and species-appropriate Alexa Fluor secondary antibodies (Molecular Probes). To determine the kinetics of actin tail formation, BMDMs were fixed and stained with phalloidin at various times after infection, and total bacteria per cell and bacteria with actin tails were enumerated. For electron microscopy, BMDMs were examined 48 h after infection by transmission electron microscopy (TEM; reference 16) or freeze fracture electron microscopy (FFEM; reference 17).
Online Supplemental Material.
Time lapse images demonstrate the movement of M. marinum in macrophages in Video 1. Video 2 shows the movement of GFP-expressing M. marinum in a macrophage subsequently fixed and stained for F-actin. Direct cell to cell spread of M. marinum propelled by actin-based motility is illustrated in Video 3. Videos 1–3 are available at http://www.jem.org/cgi/content/full/jem.20031072/DC1.
Results and Discussion
M. marinum Induce Actin Polymerization in Macrophages and in Cell-free Extracts.
During studies of phagosome maturation, we found that some M. marinum were motile within BMDMs (Fig. 1 A and Video 1, available at http://www.jem.org/cgi/content/full/jem.20031072/DC1). The average rate of motile M. marinum in BMDMs was 10.69 μm/min (SD = 1.86; n = 8), comparable to that of intracellular Listeria and Shigella found previously (6). Associated with motile bacteria were phase-dense “tails” that had the appearance of polymerized actin. To determine whether bacterial motility indeed was actin based, BMDMs were infected with M. marinum expressing GFP and stained for F-actin, which demonstrated the presence of actin tails (Fig. 1 B and Video 2, available at http://www.jem.org/cgi/content/full/jem.20031072/DC1). We also found actin tails after infection of the murine macrophage cell lines J774 A.1 and RAW 264.7 (not depicted), and the fish macrophage cell line CLC (Fig. 2 A). CLC cells have been used previously for M. marinum infection (9). This finding demonstrates that M. marinum polymerizes actin in macrophages of a natural host. Phagocytosed heat-killed M. marinum were not motile in any cell type, indicating that actin polymerization was an active process induced by viable intracellular bacteria.
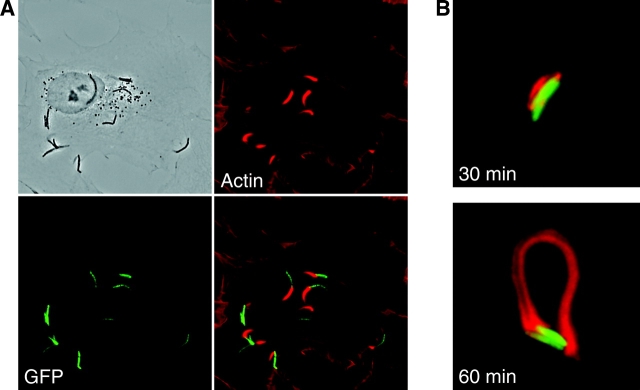
Figure 1.
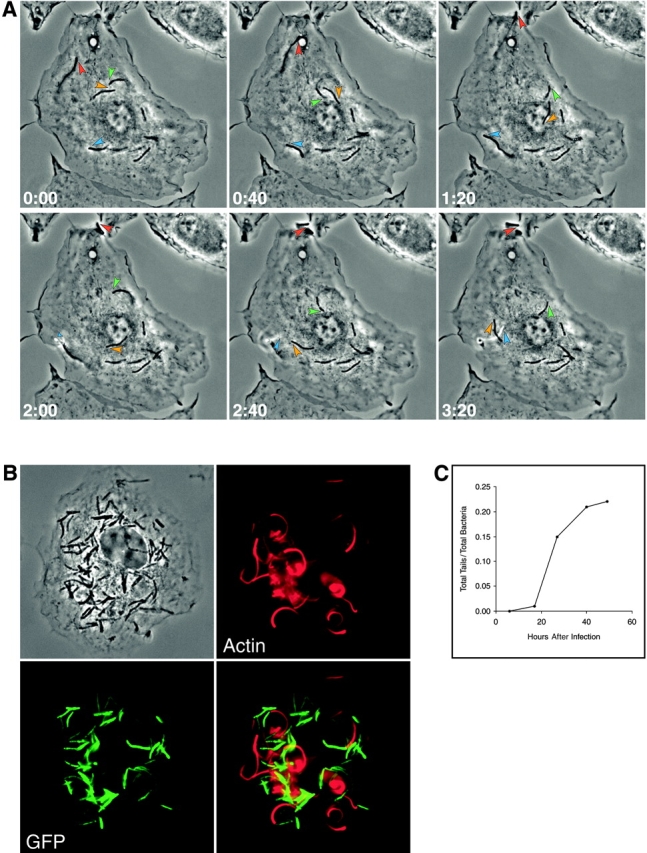
M. marinum is propelled by actin-based motility in primary macrophages. (A) Time lapse images show movement of M. marinum within macrophages. Select motile bacteria are followed with colored arrowheads (also see corresponding Video 1, available at http://www.jem.org/cgi/content/full/jem. 20031072/DC1). (B) A macrophage (shown in phase contrast in the top left) infected with _M. marinum_–expressing GFP (bottom left) was stained for F-actin (top right). A merged image (bottom right) demonstrates the association of actin tails with the bacterial pole. (C) The appearance of actin tails behind M. marinum as a function of time after infection of primary macrophages is shown. The y axis is the fraction of the total intracellular bacteria that have actin tails. Data shown are from one of two detailed experiments with similar results.
Figure 2.
M. marinum induces actin polymerization in a fish macrophage cell line and is capable of polymerizing actin in vitro. (A) CLC cells (shown in phase contrast in the top left) were infected with _M. marinum_–expressing GFP (bottom left). 48 h after infection, the cells with fixed and stained for F-actin (top right). A merged image is shown in the bottom right. (B) _M. marinum_–expressing GFP (green) grown in macrophages for 48 h were isolated and added to mouse brain extracts. Within 30 min, M. marinum polymerized actin (red) in diffuse clouds surrounding the bacteria (top) and by 1 h, M. marinum polymerized actin into tails at its pole (bottom).
Actin tails first appeared ∼15 h after BMDM infection (Fig. 1 C). The number of bacteria with actin tails increased until ∼20% of all intracellular bacteria demonstrated actin tails 48 h after initiation of infection, after which there was marked toxicity to the BMDM monolayer. At 48 h, 90% of BMDMs contained at least one actin-associated M. marinum. The average was about six mycobacteria with actin tails per macrophage (not depicted). Thus, there is an initial lag before actin tail formation by intracellular M. marinum, but over time nearly all infected cells contain M. marinum with actin tails.
In addition to polymerizing actin during intracellular infection, Listeria, Shigella, and Rickettsia are able to polymerize actin in cell-free extracts (18). M. marinum grown in standard broth conditions did not induce actin polymerization in cell-free extracts. However, M. marinum isolated after 2 d of growth in BMDMs were able to polymerize actin in Xenopus egg (not depicted) and mouse brain extracts, forming actin clouds surrounding bacteria after 30 min of incubation and tails after 1 h (Fig. 2 B). This suggests that expression of the bacterial surface molecule(s) required for M. marinum actin polymerization is enhanced in the intracellular milieu, consistent with the lag in appearance of actin tails in BMDMs (Fig. 1 C). Expression of the actin-nucleating proteins ActA of Listeria and IcsA of Shigella are also up-regulated during intracellular infection (19, 20), so enhancement of bacterial actin polymerization by the intracellular milieu might be a general phenomenon for intracytoplasmic pathogens.
M. marinum That Polymerize Actin Are in the Host Cell Cytoplasm.
Because intracellular mycobacteria are believed to exist exclusively within a phagosome whereas all bacteria known to initiate actin-based motility escape from phagosomes and are free in the cytoplasm, we investigated the subcellular location of the motile M. marinum using both light and electron microscopy. Using the lipid marker DiI under conditions that label intracellular membranes, we found that most M. marinum were in a membrane-bound compartment labeled with DiI (Fig. 3 A). However, none of the M. marinum with actin tails was associated with DiI staining, indicating either that these bacteria are in a membrane-bound compartment distinct from that occupied by nonmotile bacteria and not labeled by DiI, or that the motile bacteria are in the cytoplasm.
Figure 3.
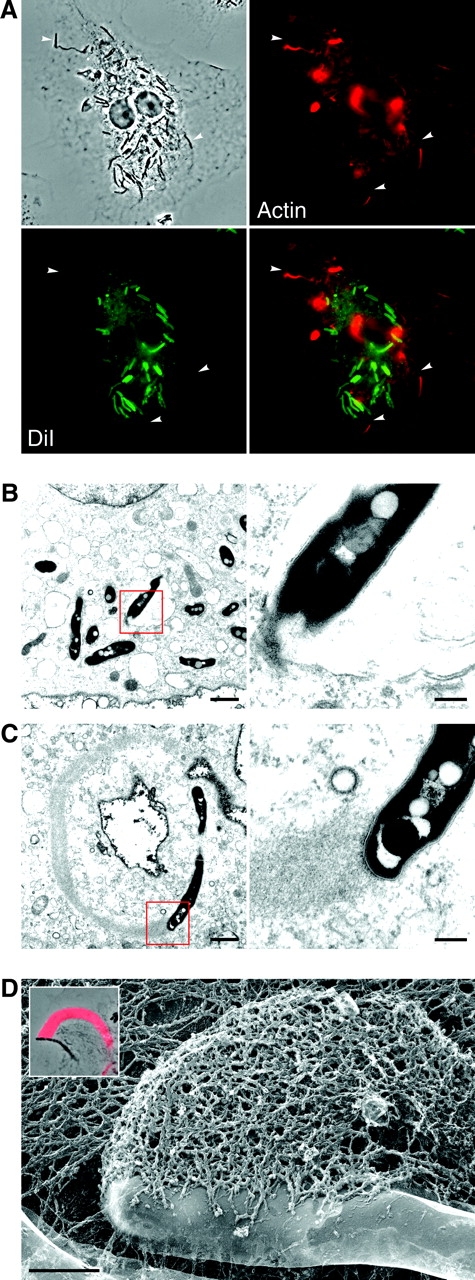
M. marinum with actin tails are found free in the host cell cytoplasm. (A) Macrophages were infected with M. marinum (shown in phase contrast in the top left) and stained with DiI (bottom left), a membrane marker, and for F-actin (top right). A merged image is shown in the bottom right. Arrowheads indicate three bacteria with actin tails that are in the plane of focus and not surrounded by DiI. (B and C) Macrophages were infected with M. marinum and observed by TEM. (B) Many M. marinum are found in membrane-bound compartments and show no evidence of actin polymerization. Detail of boxed area is shown at right to highlight the bacterial cell wall and the host membrane lipid bi-layer. (C) An example of M. marinum that polymerizes actin. The detail at right demonstrates the close apposition of actin filaments to the bacterial cell wall. Bars: left, 1.0 μm; right, 0.2 μm. (D) An FFEM image shows the intimate association of M. marinum with its actin tail. Bar, 0.5 μm. Although more often found at a pole, actin polymerization can occur at the side of a bacterium. Inset, a phase contrast image of another M. marinum with actin polymerized at the side and fluorescently labeled F-actin superimposed in red.
TEM demonstrated that although most bacteria were separated from the cytoplasm by an electron-transparent region limited by a host cell membrane (Fig. 3 B), bacteria associated with actin tails were found in electron-dense regions indistinguishable from the cytoplasm, not surrounded by host cell membrane (Fig. 3 C). High resolution FFEM, used to better visualize the ultrastructural details of this relationship, clearly demonstrated the intimate association of actin with the M. marinum surface (Fig. 3 D). Thus, motile M. marinum are found free in the cytoplasm of host cells.
M. marinum Recruit WASP to the Site of Actin Polymerization.
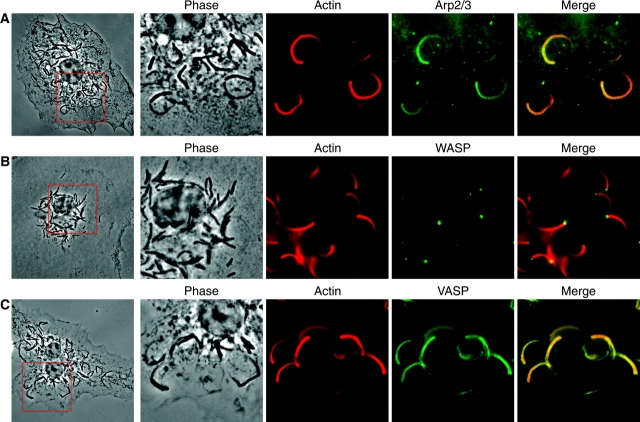
In TEM and FFEM images (Figs. 3, C and D), the actin in tails behind M. marinum appeared to be polymerized in a branched pattern, similar to the actin polymerized by Listeria and Shigella, but unlike the parallel bundles of actin in Rickettsia tails (18, 21). This difference has been correlated with presence or absence of the Arp2/3 complex, an essential component of one major mechanism for actin nucleation and for branching of actin filaments. Immunofluorescence using antibodies recognizing Arp3 (not depicted) or p34-Arc (Fig. 4 A), subunits of the Arp2/3 complex, demonstrated the presence of the Arp2/3 complex in M. marinum actin tails.
Figure 4.
Arp2/3, WASP, and VASP localize in the actin tails of M. marinum. Macrophages were infected with M. marinum and stained with antibodies for actin (red) and for the host cell proteins. (A) p34-Arc, subunit of the Arp2/3 complex, (B) WASP, and (C) VASP (all shown in green). For orientation, the entire macrophage is shown at the left and details of the boxed area are shown to the right. Images reveal that the Arp2/3 complex and VASP are located throughout the actin tail of M. marinum, whereas WASP is located exclusively at the pole at which the actin tail is formed.
During normal actin remodeling in host cells, the Arp2/3 complex is activated by members of the WASP family, which includes WASP, expressed only in hematopoietic cells including macrophages, and N-WASP, expressed ubiquitously. Listeria and Shigella have evolved independent mechanisms to induce actin polymerization that converge on this step in the activation of the Arp2/3 complex. The Listeria protein ActA directly activates the Arp2/3 complex by mimicking WASP, whereas the Shigella protein IcsA indirectly activates the Arp2/3 complex by recruiting N-WASP to the bacterial surface (6). WASP staining of _M. marinum_–infected macrophages revealed the localization of WASP exclusively at the pole of M. marinum where the actin tail formed (Fig. 4 B). Thus, intracellular growth induces M. marinum recruitment of WASP to its surface, an event that would be sufficient to induce branching actin polymerization and initiate intracellular motility using the Arp2/3 complex. Although this is similar to induction of actin polymerization by Shigella, IcsA binds only to N-WASP and Shigella is incapable of forming actin tails in macrophages that predominantly express WASP (22).
To explore further the similarity between M. marinum and other pathogens that induce actin polymerization, we examined the localization of VASP in the actin tails of cytoplasmic M. marinum. VASP localizes to regions of dynamic actin rearrangements in host cells, and Listeria ActA recruits VASP by a direct interaction that induces localization of VASP to the interface of Listeria with its actin tail (23). In contrast, IcsA does not bind VASP, leading to VASP staining throughout the actin tail behind Shigella (18) due to VASP's association with F-actin, rather than preferential recruitment to the bacteria–actin interface. Similar to Shigella, VASP was present throughout the length of the actin tail of M. marinum (Fig. 4 C), suggesting that it has no direct interaction with the accelerator of actin polymerization on the bacterial surface. Based on the staining of WASP and VASP, the mechanism of M. marinum induction of actin polymerization is more similar to that of Shigella than Listeria, even though mycobacteria are often grouped phylogenetically more closely with Gram-positive organisms.
M. marinum Spread from Cell to Cell Directly.
In phase contrast time lapse microscopy, motile M. marinum primarily moved in arcs within the cell boundaries (see bacteria marked with orange and green arrowheads in Fig.1 A and Video 1, available at http://www.jem.org/cgi/content/full/jem.20031072/DC1). Occasionally at earlier and often at later times, M. marinum was observed to move beyond the cell boundary on membranous stalks toward adjacent cells (see bacteria marked with red and blue arrowheads in Fig. 1 A and Videos 1 and 3, available at http://www.jem.org/cgi/content/full/jem.20031072/DC1). This phenomenon is reminiscent of Listeria and Shigella that use actin-based motility for intercellular spread between host cells without exposure to the extracellular milieu. Consistent with this role for actin-based motility, Video 3 illustrates an event of actin-based motility-dependent direct cell to cell spread of a single bacterium.
Additional evidence that M. marinum is capable of direct cell to cell spread was provided by the pattern of bacterial growth in monolayers of host cells in the presence of amikacin to kill extracellular bacteria (Fig. 5) . 8 d after infection with a low MOI, small foci of cells were visibly infected by the GFP-labeled M. marinum, consistent with spread from an initially infected cell to adjacent cells. These results suggest a role for actin-based motility in direct cell to cell spread of M. marinum. A similar mechanism is a known virulence factor for Listeria (24). It may have a similar role in the pathogenesis of M. marinum infection.
Figure 5.
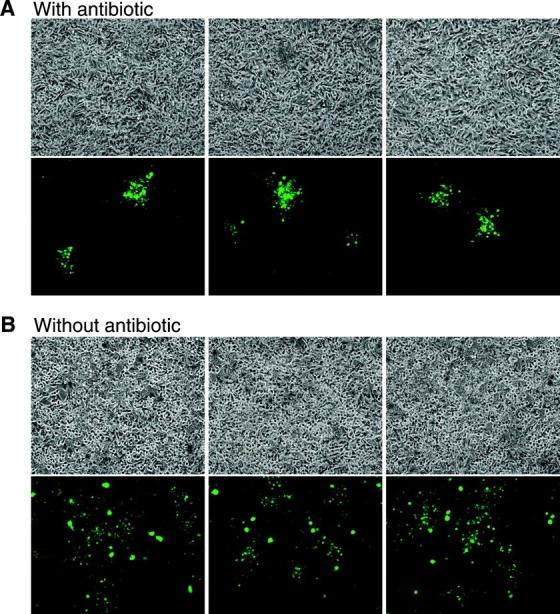
Focal growth of intracellular M_. marinum_ in the presence of antibiotics is evidence of direct cell to cell spread. A confluent cell monolayer was infected with GFP-expressing M. marinum and growth was assessed in the (A) presence or (B) absence of amikacin to kill extracellular bacteria. (A) The top row depicts the cell monolayer in phase contrast, and directly below the corresponding fluorescence image demonstrates the focal pattern of GFP-expressing M. marinum. Three representative fields are shown. (B) In parallel experiments where the media did not contain antibiotics, the pattern of M. marinum growth is diffuse.
These data show that M. marinum can escape from phagosomes and recruit host cell cytoskeletal factors in the cytoplasm to induce actin polymerization resulting in intracellular motility and direct cell to cell spread. This observation adds M. marinum to the phylogenetically diverse list of pathogens that have found it beneficial to exploit the host cytoskeleton to spread to adjacent cells without leaving the cytoplasm. Only a minority of M. marinum exhibits this behavior. The characteristics distinguishing cytoplasmic, motile bacteria from those that remain in phagosomes are unknown.
Does the ability to escape from phagosomes and initiate actin-dependent cell to cell spread extend to other mycobacteria? There is a controversial report of TEM visualization of M. tuberculosis free in the cytoplasm (25), as well as one report of direct M. tuberculosis cell to cell spread in tissue culture (26). However, unlike M. marinum, M. tuberculosis has been extensively studied for decades without evidence for actin-based motility. If this has any role in M. tuberculosis infection, it likely is at a site in vivo or at a time after initial infection that has thus far escaped close scrutiny.
Acknowledgments
We thank J.D. Cirillo for the CLC cell line, D.A. Portnoy and J.S. Cox for valuable discussions, and J. Skoble and I.C. Koo for critical reading of this manuscript and encouragement.
This work was supported by grants AI33348 and AI55614 to E.J. Brown from the National Institutes of Health. L.M. Stamm is supported by the University of California San Francisco Medical Scientist Training Program. L.-Y. Gao is supported by the institutional training grant 5T32AI007334-15.
The online version of this article contains supplemental material.
Abbreviations used in this paper: BMDM, bone marrow–derived macrophage; FFEM, freeze fracture electron microscopy; GFP, green fluorescent protein; MOI, multiplicity of infection; TEM, transmission electron microscopy; VASP, vasodilator-stimulated phosphoprotein; WASP, Wiskott-Aldrich syndrome protein.
References
- 1.Davis, J.M., H. Clay, J.L. Lewis, N. Ghori, P. Herbomel, and L. Ramakrishnan. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 17:693–702. [DOI] [PubMed] [Google Scholar]
- 2.Springer, B., L. Stockman, K. Teschner, G.D. Roberts, and E.C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramakrishnan, L., N.A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 288:1436–1439. [DOI] [PubMed] [Google Scholar]
- 5.Gao, L.Y., R. Groger, J.S. Cox, S.M. Beverley, E.H. Lawson, and E.J. Brown. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71:922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg, M.B. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell, D.G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569–577. [DOI] [PubMed] [Google Scholar]
- 8.Roach, T.I., S.E. Slater, L.S. White, X. Zhang, P.W. Majerus, E.J. Brown, and M.L. Thomas. 1998. The protein tyrosine phosphatase SHP-1 regulates integrin-mediated adhesion of macrophages. Curr. Biol. 8:1035–1038. [DOI] [PubMed] [Google Scholar]
- 9.El-Etr, S.H., L. Yan, and J.D. Cirillo. 2001. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 69:7310–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeshita, S., K. Kaji, and A. Kudo. 2000. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 15:1477–1488. [DOI] [PubMed] [Google Scholar]
- 11.Castro-Garza, J., C.H. King, W.E. Swords, and F.D. Quinn. 2002. Demonstration of spread by Mycobacterium tuberculosis bacilli in A549 epithelial cell monolayers. FEMS Microbiol. Lett. 212:145–149. [DOI] [PubMed] [Google Scholar]
- 12.May, R.C., M.E. Hall, H.N. Higgs, T.D. Pollard, T. Chakraborty, J. Wehland, L.M. Machesky, and A.S. Sechi. 1999. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr. Biol. 9:759–762. [DOI] [PubMed] [Google Scholar]
- 13.Welch, M.D., A. Iwamatsu, and T.J. Mitchison. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 385:265–269. [DOI] [PubMed] [Google Scholar]
- 14.Welch, M.D., A.H. DePace, S. Verma, A. Iwamatsu, and T.J. Mitchison. 1997. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, G.A., J.A. Theriot, and D.A. Portnoy. 1996. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J. Cell Biol. 135:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taunton, J., B.A. Rowning, M.L. Coughlin, M. Wu, R.T. Moon, T.J. Mitchison, and C.A. Larabell. 2000. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuser, J. 1981. Preparing biological samples for stereomicroscopy by the quick-freeze, deep-etch, rotary-replication technique. Methods Cell Biol. 22:97–122. [DOI] [PubMed] [Google Scholar]
- 18.Gouin, E., H. Gantelet, C. Egile, I. Lasa, H. Ohayon, V. Villiers, P. Gounon, P.J. Sansonetti, and P. Cossart. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112:1697–1708. [DOI] [PubMed] [Google Scholar]
- 19.Moors, M.A., B. Levitt, P. Youngman, and D.A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg, M.B., J.A. Theriot, and P.J. Sansonetti. 1994. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infect. Immun. 62:5664–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Kirk, L.S., S.F. Hayes, and R.A. Heinzen. 2000. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect. Immun. 68:4706–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki, T., H. Mimuro, S. Suetsugu, H. Miki, T. Takenawa, and C. Sasakawa. 2002. Neural Wiskott-Aldrich syndrome protein (N-WASP) is the specific ligand for Shigella VirG among the WASP family and determines the host cell type allowing actin-based spreading. Cell. Microbiol. 4:223–233. [DOI] [PubMed] [Google Scholar]
- 23.Skoble, J., D.A. Portnoy, and M.D. Welch. 2000. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J. Cell Biol. 150:527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brundage, R.A., G.A. Smith, A. Camilli, J.A. Theriot, and D.A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA. 90:11890–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonough, K.A., Y. Kress, and B.R. Bloom. 1993. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect. Immun. 61:2763–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrd, T.F., G.M. Green, S.E. Fowlston, and C.R. Lyons. 1998. Differential growth characteristics and streptomycin susceptibility of virulent and avirulent Mycobacterium tuberculosis strains in a novel fibroblast-mycobacterium microcolony assay. Infect. Immun. 66:5132–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]