Epstein-Barr Virus–Infected Resting Memory B Cells, Not Proliferating Lymphoblasts, Accumulate in the Peripheral Blood of Immunosuppressed Patients (original) (raw)
Abstract
When Epstein-Barr virus (EBV) infects B cells in vitro, the result is a proliferating lymphoblast that expresses at least nine latent proteins. It is generally believed that these cells are rigorously controlled in vivo by cytotoxic T cells. Consistent with this, the latently infected cells in the peripheral blood of healthy carriers are not lymphoblasts. Rather, they are resting memory B cells that are probably not subject to direct immunosurveillance by cytotoxic T lymphocytes (CTLs). When patients become immunosuppressed, the viral load increases in the peripheral blood. The expansion of proliferating lymphoblasts due to the suppressed CTL response is believed to account for this increase and is considered to be a major risk factor for posttransplant lymphoproliferative disease (PTLD) and AIDS-associated B cell lymphoma. Here we show that there is an increase in the numbers of latently infected cells in the peripheral blood of immunosuppressed patients. However, the cells are not proliferating lymphoblasts. They are all latently infected, resting, memory B cells—the same population of infected cells found in the blood of healthy carriers. These results are discussed in the context of a model for EBV persistence that explains why PTLD is usually limited to the lymph nodes.
Keywords: Epstein-Barr virus, B cell, latency, immunosuppression
Epstein-Barr virus (EBV) is a B lymphotropic herpesvirus (for a general review, see reference 1). It will latently infect any resting primary B cell in vitro, irrespective of its phenotype. As a consequence, the resting B cell becomes activated into a latently infected lymphoblast that will proliferate indefinitely. This is referred to as the lymphoblastoid form of latency and is associated with the expression of nine known latent proteins (for a general review, see reference 2), several of which have potent growth-promoting activity and can act as oncogenes.
The pathogenic potential of EBV, suggested by the in vitro studies, contrasts with the behavior of the virus in vivo. Greater than 90% of the human population is benignly infected for life. The persistent infection is only apparent as stable levels of viral shedding in the saliva 3 and infected cells in the blood 4 and lymphoid tissue. Furthermore, persistently infected cells in vivo bear no resemblance to the proliferating blasts seen with in vitro infection. In the peripheral blood of healthy carriers, the virus is restricted to latently infected 5, memory B cells 6 that do not express the growth-promoting latent genes 7 and are not phenotypically lymphoblastoid 8. The cells are not proliferating, but resting in G0 9 and do not constitute a pathogenic threat. We have proposed 6 10 that EBV infects and activates B cells to become proliferating blasts in vivo in order to differentiate into memory cells. This provides a mechanism for the virus to gain access to the memory compartment, which we have suggested as a site of long-term persistence.
It is generally assumed that immunosuppression leads to an increased number of latently infected cells in the blood. However, this belief is based on two assays 11 12 13 14 15 16 17 that both have significant limitations (for a discussion, see reference 8) such that it is only possible to conclude with certainty that there is a higher viral genome burden. Whether this increased burden is due to an expansion of latently infected cells, viral reactivation, or a combination of the two is unknown. The proposed increase in latently infected cells is believed to be in the form of B lymphoblasts. These blasts are targets for cytotoxic T cells in healthy carriers 18, but are thought to proliferate unchecked in the blood when the cellular immune response is suppressed. The expansion of these cells may lead to posttransplant lymphoproliferative disease (PTLD),1 which ultimately can result in tumors that resemble in vitro–infected B lymphoblasts (for a review, see reference 14). These proliferating blasts survive only because the immune response is compromised, since PTLD usually resolves upon removing immunosuppression 19. Surprisingly, most studies of EBV in immunosuppressed patients have been restricted to a very small subset of patients, namely those with PTLD. The presence of EBV-infected lymphoblasts in the blood of immunosuppressed patients who lack overt PTLD has been described only once and for a single patient 20. Given the finding that EBV is restricted to memory cells in the peripheral blood of healthy carriers, we decided it was time to reassess the status of virally infected cells in the peripheral blood of immunosuppressed patients. We have used recently developed methods to precisely quantitate and characterize the infected cells in the blood of patients who were immunosuppressed as a consequence of organ transplantation. The patient population was clinically healthy at the time of study and showed no signs of PTLD. In this study, we demonstrate unambiguously for the first time that there is an expansion of latently infected B cells in the blood of immunosuppressed patients who are otherwise clinically healthy. The cells are not infected proliferating lymphoblasts, but rather the same population of resting, memory B cells found in healthy carriers. The only exception is that viral reactivation is frequently detected in the blood of immunosuppressed patients but not in healthy carriers. In these cases, viral replication, not latently infected cells, provides the largest contribution to the increased genome burden.
Materials and Methods
Cell Lines and Primary Cells.
Namalwa (American Type Culture Collection) is an EBV+ BL line containing one or two copies of the viral genome. This was used as a positive control for the EBV-specific DNA PCR. BJAB is an EBV− B cell lymphoma used as a negative control for all PCR-related experiments. For reverse transcriptase (RT)-PCR of latent membrane protein (LMP)-2a, LMP-1, and EBV nuclear antigen (EBNA)-2, the LCL line IB4 was used as a positive control. Rael, an EBV+ BL line, was used as a positive control for EBNA-1(Qp) RT-PCR. All cell lines were maintained in 5% CO2 with 10% FCS, RPMI 1640 with penicillin and streptomycin.
Patients attending the Transplantation Clinic at the New England Medical Center were recruited for blood donation. The patients, including both kidney and liver recipients, were usually 1–2 mo posttransplantation. They all received cyclosporin A or FK506, combined with azathioprine, mycophenolate mofetil, or sirolimus (Table ). All of the patients received prednisone and were under active immunosuppression, but were otherwise clinically healthy, at the time they were studied and showed no evidence of PTLD. Healthy volunteers were recruited from Tufts University School of Medicine. The patient and control populations were matched for age distribution, sex, and minority representation.
Table 1.
Summary of the Patients Described in This Study
| Patient | Organ | Rejection | Immuno-suppression | Antiviral | |
|---|---|---|---|---|---|
| Primary | Secondary | ||||
| 1 | Liver | + | FK | AZA | Ga |
| 2 | Liver | + | CSA | MMF | Ga |
| 3 | Liver | + | FK | MMF | Ga |
| 4 | Kidney | + | CSA | AZA | − |
| 5 | Liver | − | CSA | AZA | Ac |
| 6 | Liver | − | CSA | MMF | Ga |
| 7 | Liver | + | CSA | AZA | Ga |
| 8 | Kidney | + | CSA | MMF | − |
| 9 | Liver | − | CSA | AZA | Ac |
| 10 | Kidney | + | FK | AZA | Ac |
| 11 | Liver | + | CSA | AZA | Ac |
| 12 | Kidney | − | FK | SIR | Ac |
| 13 | Liver | − | CSA | AZA | Ga |
| 14 | Kidney | − | CSA | AZA | Ga |
| 15 | Kidney | + | FK | AZA | Ac |
| 16 | Liver | − | FK | AZA | Ga |
| 17 | Liver | + | CSA | AZA | − |
| 18 | Liver | − | FK | AZA | Ga |
| 19 | Kidney | + | CSA | AZA | Ga |
| 20 | Kidney | − | CSA | AZA | Ac |
| 21 | Liver | + | CSA | AZA | Ga |
| 22 | Kidney | − | CSA | SIR | Ga |
| 23 | Liver | + | FK | AZA | Ga |
| 24 | Liver | + | FK | AZA | Ga |
| 25 | Liver | − | FK | AZA | Ga |
| 26 | Liver | + | FK | AZA | Ga |
| 27 | Liver | − | CSA | AZA | Ga |
| 28 | Liver | − | FK | AZA | Ga |
Cell Fractionations.
PBMCs were isolated and fractionated using the MACS® (Miltenyi Biotec) cell separation system as described previously 6. The antibodies and concentrations used were 0.018 μg/ml anti-CD19–biotin, 0.015 μg/ml anti-IgD–biotin (Southern Biotechnology Associates), 0.03 μg/ml anti-CD23–biotin (The Binding Site), and 1 μg/ml each goat anti-IgG, -IgA, and -IgM (Southern Biotechnology Associates). Positively selected fractions were typically >90% pure for the desired marker. Contamination of a specific B cell population with an undesired B cell population was always <5%.
FACS® Analysis and Antibodies.
Fractionated populations were analyzed on a Becton Dickinson FACScan™ using Lysis II Software. Anti-CD20–FITC (Dako), a pan-B cell marker, and anti-IgD–PE (The Binding Site) were used to assay the purity and depletion of the desired populations. MOPC21 (IgG1 isotype control; Sigma Chemical Co.) and MOPC121 (IgG2b isotype control; Sigma Chemical Co.) were used as negative controls.
EBV-specific DNA PCR.
Isolated populations were distributed to the wells of a V-bottomed microtiter plate (Nunc) at the desired cell number. The cells were pelleted, the cell pellets lysed, and the extract subjected to DNA PCR specific to the W repeat region of the EBV genome exactly as described previously 6. PCR products were resolved on a 2% Nuseive agarose (FMC Corp.), 1% Seakem LE agarose (FMC Corp.) gel, and Southern blotted to Nytran Plus as described by the manufacturer (Schleicher and Schuell). Specific products were detected using random primed labeled, purified PCR product from Namalwa cells as described previously 6.
Cell Cycle Analysis.
Analysis of the cell cycle status of a given population was performed by labeling cells with propidium iodide in the presence of the nonionic detergent Triton X-100 exactly as described previously 9. The cells were sorted based on DNA content using a Becton Dickinson FACStarPlus™, into a V-bottomed microtiter plate. DNA PCR was performed as described above.
The calculation of expected versus observed numbers of EBV-infected cells in the S-G2-M B cell populations has been described previously 9 and was performed as follows. In Table , we used the measured frequency of EBV-infected B cells in the G0-G1 population to estimate the expected frequency of EBV-infected cells in the S-G2-M population if all of the infected cells were proliferating and therefore all of the positive cells in the G0-G1 fraction were in G1. This number was then compared with the number of signals obtained experimentally. To make the estimate, the following calculation was performed taking patient 2 from Table as an example:
Table 3.
Analysis of the Cell Cycle Status of EBV-infected Cells in the Peripheral Blood of Immunosuppressed Patients
| Patient | Frequency | Frequency in G0-G1 | Percent S-G2-M | No. of S-G2-M cells tested | No. observed in S-G2-M | No. expected in S-G2-M |
|---|---|---|---|---|---|---|
| 2 | 8,100 | 8,000 | 1.07 | 600 | 2 | 36 |
| 9 | 1,500 | 1,200 | 0.6 | 2,040 | 2 | 33 |
| 18 | 200 | 260 | 1.22 | 8,800 | 1 | 34 |
| Total | ||||||
| 5 | 83 |
1.07% of the B cell population was in S-G2-M. Therefore, for every 106 G0-G1 B cells, there are 1.07/99 × 106 = 1.08 × 104 cells in S-G2-M. The frequency of EBV-infected cells was 800/106 in the G0-G1 population. In a typical cycling EBV+ lymphoblastoid cell line, the percentage of cells in S-G2-M is 45 ± 4%. Therefore, if we assume that the EBV-infected cells have a similar cell cycle distribution, then for every 800 EBV+ G0-G1 cells there should be 45/55 × 800 = 655 EBV+ cells in S-G2-M. Therefore, we should find 655 EBV+ cells in 1.08 × 104 S-G2-M cells. In this experiment, we analyzed a total of 6 × 102 S-G2-M cells. Therefore, we expect 655 × (1.08 × 104)/(6 × 102) = 36 infected cells in the S-G2-M population we tested. We actually obtained two signals.
Gardella Gel Analysis.
To resolve linear viral genomes from circular viral genomes, the hallmarks of lytic and latent infection, respectively, PCR-modified Gardella gel analysis was used as described previously 5. In brief, whole cells are lysed in situ in the wells of a 0.75% low melt agarose gel, and the linear and episomal forms of the genome are separated based on their differential mobility in agarose gels: linear genomes migrate faster because they “snake” through the gel. The lanes were sliced into sections, and the agarose from each slice was digested using β-agarase (FMC Corp.). DNA was precipitated, and EBV-specific DNA PCR was performed for each slice as described above. The relative migration points for linear and circular DNA were identified using virions and PBL B cells, respectively, as controls.
RNA Purification and RT-PCR.
RNA was isolated from 5 × 106 primary cells or 2 × 105 culture cells, and cDNA synthesis was performed as described previously 6. 5 μl of cDNA was then used in PCR for EBNA-1(Qp), EBNA-2, LMP-1, and LMP-2a. This allowed us to test for all four transcripts from one pot of cDNA. EBNA-1(Qp) PCR was carried out using the amplimers described by Schaefer et al. 21. Reaction concentrations were 50 mM KCl, 20 mM Tris, pH 8.3, 2.5 mM MgCl2, 0.2 mM dNTPs, and 20 μM of each primer. Reaction conditions were 15 s at 95°C, 30 s at 62°C, and 30 s at 72°C for 40 cycles, followed by a 5-min 72°C extension step. For EBNA-2 PCR, amplimers described by Chen et al. were used 22. Reaction concentrations and temperature conditions were the same as for EBNA-1(Qp). LMP-1 PCR amplimers were as described by Chen et al. 22, and reaction concentrations were 50 mM KCl, 20 mM Tris, pH 8.3, 3.0 mM MgCl2, 0.2 mM dNTPs, 20 μM each primer. Reaction conditions were 95°C for 15 s, 65°C for 30 s, and 72°C for 30 s, followed by a 5-min 72°C extension step. LMP-2a PCR amplimers were as described by Tierney et al. 23, and reaction concentrations were 50 mM KCl, 10 mM Tris, pH 8.3, 2.0 mM MgCl2, 0.2 mM dNTPs, and 20 μM of each primer. Temperature conditions were 95°C for 15 s, 55°C for 30 s, and 72°C for 45 s for 40 cycles, followed by a 5-min 72°C extension step. PCR products were electrophoresed on 2% Nuseive agarose (FMC Corp.), 1% Seakem LE agarose (FMC Corp.) gel, and Southern blotted to Nytran Plus as described by the manufacturer (Schleicher and Schuell). Products were detected by probing with the appropriate purified PCR product (from Rael for EBNA-1, and from IB4 for EBNA-2, LMP-1, and LMP-2a) that was random primed labeled (Boehringer Mannheim) as described by the manufacturer. In every case, it was possible to detect the mRNA from a single infected cell (IB4 or Rael) in the presence of 5 × 106 uninfected PBL B cells.
Results
The Infected Cells in the Blood of Immunosuppressed Patients Are Memory B Cells.
To identify the type of latently infected cells in the peripheral blood of immunosuppressed patients, we first characterized their cell surface phenotype. PBLs were fractionated using biotinylated antibodies and the MACS® magnetic bead technique. The antibodies used recognized CD19, a pan-B cell marker; CD23, a marker expressed on naive B cells and strongly expressed on B cell blasts activated by EBV infection; IgD, which is expressed only on naive B cells; and the A, G, and M isotypes of Ig, which are expressed on the surface of memory cells in the absence of IgD.
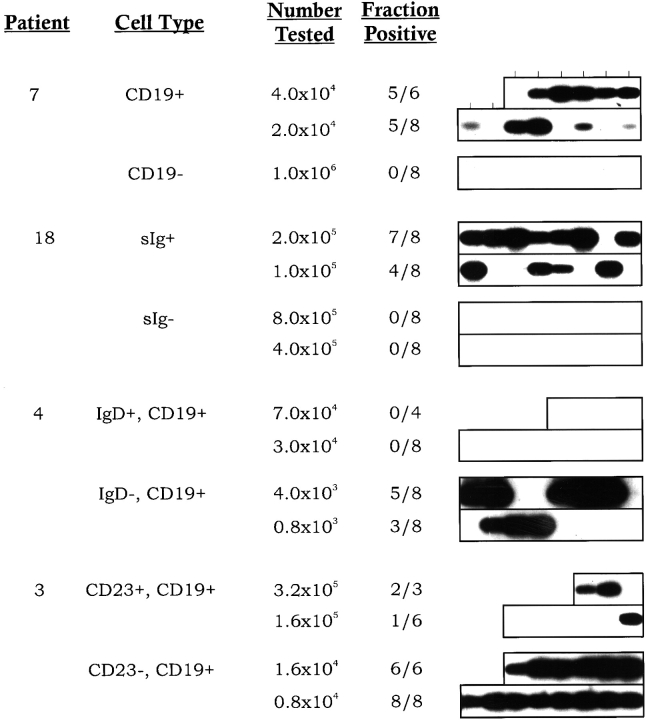
In the first round of experiments, CD19 was used to separate the B and non-B cell populations, and an anti–IgG+IgA+IgM antibody was used to separate cells expressing surface Ig (sIg) from those lacking sIg. The frequency of virus-infected cells in each population was then assayed by limiting dilution DNA PCR. An example of one experiment for each marker is shown in Fig. 1, and a summary of the quantitation for two patients for each marker is shown in Table . As expected, essentially all of the virus-infected cells were B cells (CD19+) expressing sIg. Less than 1% of the infected cells were in the non-B (CD19−) sIg− compartment. This number was so small, it could be accounted for by low levels of contamination of the CD19− cells with B cells. The B cells (CD19+) were then further fractionated on the basis of IgD expression to see if there were infected cells in the naive compartment, and CD23 expression to test if the infected cells expressed the lymphoblastoid phenotype. The results of one experiment for each marker are shown in Fig. 1, and the quantitation for two patients with each marker is summarized in Table . Essentially all of the infected cells reside in the CD23− and IgD− populations. There was no significant number of infected cells detected in either the CD23+ or IgD+ populations. This is not the expected phenotype of a B lymphoblast. Rather, it is the same as the latently infected cells found in the blood of healthy carriers—memory B cells. Therefore, we conclude that the surface phenotype of infected cells in the peripheral blood of immunosuppressed individuals is the same as that found in healthy carriers.
Figure 1.
The cell surface phenotype of EBV-infected cells in the blood of immunosuppressed individuals. Peripheral lymphocytes were fractionated using mAbs and the MACS® separation technique. For CD19 and sIg, whole PBMCs were fractionated for the specific marker. For CD23 and IgD, the cells were first selected for expression of the desired marker and the remaining marker-negative B cells were then selected using the pan-B cell marker CD19. The cells were serially diluted, and replicates of each dilution were subjected to DNA PCR for the W repeat of the viral DNA using a technique that will detect a single viral genome in 106 uninfected cells (reference 4). The PCR products were detected by Southern blotting with a specific probe for the W repeat. The position of each lane containing a sample is indicated at the top. Two negative and one positive control were performed for each dilution of cells (not shown). For full experimental details, see Materials and Methods. The frequency of virus-infected cells was calculated using Poisson statistics, and the results are summarized in Table . The cell surface phenotype of the cells tested, the number of cells tested per sample, and the fraction of samples positive at each cell number are given.
Table 2.
The Cell Surface Phenotype of EBV-infected Cells in the Peripheral Blood of Immunosuppressed Patients
| Cell type | Patient | Frequency | |
|---|---|---|---|
| Enriched | Depleted | ||
| CD19 | 3 | >6,100 | 10 |
| 7 | 1,550 | <10 | |
| sIg | 16 | 750 | <8 |
| 18 | 400 | <1 | |
| CD23 | 3 | 360 | >10,000 |
| 23 | <12 | 925 | |
| IgD | 4 | <10 | 3,100 |
| 14 | 60 | 1,800 |
The Infected Memory Cells Are Resting.
The cell surface phenotype of the infected cells in the blood of immunosuppressed individuals, CD19+sIg+IgD−CD23−, is the same as that found for healthy carriers, namely resting, memory B cells 6. To confirm that the infected cells in immunosuppressed patients were indeed resting, purified B cells were permeabilized and stained for DNA content with the dye propidium iodide. The cells were fractionated on the basis of DNA content into G0/G1 and S/G2/M populations, and the frequency of virus-infected cells in each population was assessed. The results of three such experiments, on patients with widely ranging frequencies, are summarized in Table . Only a small fraction, ∼1%, of the peripheral blood B cells were in S/G2/M. This is probably a result of the immunosuppressive drugs. If the virus-infected cells were in cycle, there should be a large enrichment of virus-positive cells in the S/G2/M population. However, EBV was not found in the S/G2/M population of B cells from immunosuppressed patients. In fact, so few positive signals were obtained from the S/G2/M population that it was impossible to measure a frequency. This was not an artifact of the techniques used, because EBV-infected lymphoblasts could be quantitatively detected in the S/G2/M population when spiked into the peripheral B cells before analysis (not shown, and reference 9). The frequencies of infected cells in the G0/G1 populations could be measured and were indistinguishable from the values obtained for the unfractionated cells. These results imply that most or all of the virus-infected cells are in the G0/G1 population. To further quantitate the accuracy of this conclusion, we used the frequency of virus-infected cells, measured in the G0/G1 population, to predict the number of positives we would expect to find in the S/G2/M population if the cells had been in cycle. This is the same analysis we have used for healthy donors 9. As can be seen from Table , the signals obtained represented ∼5% of those expected or ∼10% of the cells in cell cycle (taking into account the cycling cells in G1). Therefore, we estimate that ≥90% of the infected cells are resting. We conclude that the population of EBV-infected cells found in the peripheral blood of immunosuppressed patients has the phenotype of resting, memory B cells.
Viral Replication Is Frequently Detected in the Blood of Immunosuppressed Patients.
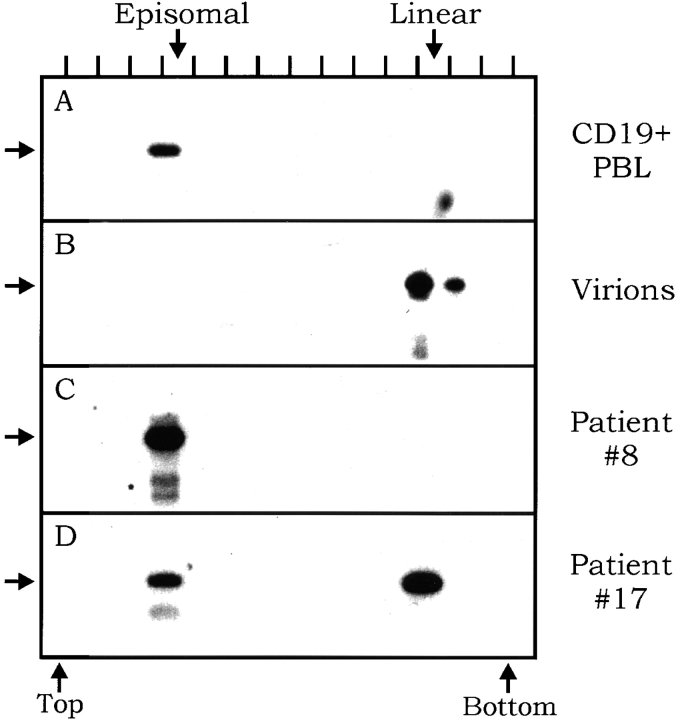
If the phenotype of infected cells in the blood of immunosuppressed individuals is unchanged from that found in healthy carriers, can we be sure that the number of infected cells is changed? Previous studies could not distinguish whether there was truly an increase in the number of infected cells or simply an increase in the fraction of cells replicating the virus. Therefore, we decided to test the relative contributions of latent and lytic genomes to the viral burden. This was done by using a technique, DNA PCR–modified Gardella gels, that can distinguish the linear form of the viral genome, characteristic of cells replicating the virus, from the circular or episomal form characteristic of latent infection. Using this approach, we have shown that infection in the peripheral blood of healthy carriers is tightly latent (7 of 7 tested), with no viral replication detected 5. An example of such an analysis on a healthy carrier is shown in Fig. 2 A, and the results for virion DNA, which is linear, are shown in Fig. 2 B for comparison. The results for two immunosuppressed patients are shown in Fig. 2C and Fig. D, and a summary of all the patients tested is shown in Table . It is apparent from these studies that in half the patients (nos. 1, 8, 11, and 15), all of the cells are latently infected (Fig. 2 C, for example). However, the other patients (nos. 10, 12, and 17) demonstrated a substantial number of linear genomes (Fig. 2 D, for example) in addition to the latent episomal genomes. Comparison of the relative signals in the episomal and linear location indicates that linear genomes were the major contributor to the viral genome burden in these individuals. Our analysis cannot distinguish the fraction of cells that might be replicating the virus. However, comparison with results obtained with cell lines 5 24 suggests that the relative ratio of linear and episomal genomes could be accounted for by a very small (<10%) fraction of the cells replicating the virus at any one time. We do not believe that failure to detect linear genomes in some patients represents a sampling variation or that the appearance of linear genomes is an artifact due to shearing, since when certain patients were retested, the same results were obtained. For example, patient 8 was tested three times over the course of 8 mo under continuous immunosuppression, and never showed detectable levels of linear genomes. The detection of linear genomes was not a function of the number of infected cells present, since the same number of B cells was loaded into the gel for each patient tested, yet there was no correlation between the frequency of infected cells and the presence of linear genomes (Table ).
Figure 2.
Gardella gel analysis of peripheral B cells from immunosuppressed individuals. The gel resolves the faster running linear genomes from slower running episomal genomes. Each sample lane was excised and cut into a series of slices. DNA was extracted from each slice and subjected to EBV-specific DNA PCR. The PCR products from each gel slice were detected by Southern blotting. The position of each lane is indicated at the top. The location of the top and bottom slices of the Gardella gels and the expected migration point of linear and episomal forms of the genome are indicated. The expected location of the PCR product is shown by an arrow. For full experimental details, see Materials and Methods. (A) Analysis of peripheral blood B cells from a healthy carrier as a positive control for the migration of episomes. It has been shown previously that only the episomal form of the virus is detected in these samples. (B) Analysis of whole EBV virions as a positive control for the migration of the linear form of the genome. (C) Analysis of peripheral blood B cells from an immunosuppressed individual who only carries latently infected cells. (D) Analysis of peripheral blood B cells from an immunosuppressed individual who has both the latent (episomal) and lytic (linear) forms of the viral genome.
Table 4.
Detection of the Episomal and Linear Forms of the EBV Genome Characteristic of Latent and Lytic Infection, Respectively
| Patient | Frequency | Episomal | Linear |
|---|---|---|---|
| 1 | 16,700 | + | − |
| 8 | 1,500 | + | − |
| 10 | 1,400 | + | + |
| 11 | 1,300 | + | − |
| 12 | 1,000 | + | + |
| 15 | 800 | + | − |
| 17 | 670 | + | + |
We conclude from these studies that in approximately half of the patients studied, the viral genome burden can be entirely accounted for by latently infected cells. However, the remaining patients have both latently and lytically infected cells, with linear genomes contributing the largest component of the viral genome burden.
The Absolute Number of EBV-infected Cells Increases after Immunosuppression.
Previous studies have claimed that immunosuppression leads to an increase in the number of latently infected cells in the blood. However, these studies have relied on assays that could not distinguish an increase in the infected cell number from an increase in the fraction of cells replicating the virus. We have now shown that the infected cells are not proliferating, and that viral replication is a frequent occurrence in the blood of immunosuppressed patients. Therefore, it was possible that there was no change in the number of infected cells and that the previously described increase in viral burden was entirely due to increased rates of reactivation. We decided to definitively resolve this issue by precisely quantitating the number of virus-infected cells using the limiting dilution DNA PCR assay we have described previously 4. Because this assay involves the titration and detection of intact infected cells, it provides a measure of the number of virus-infected cells that is independent of the number of viral genomes per cell and therefore will not be artifactually biased by lytically infected cells that have high genome copy numbers. The results from 28 immunosuppressed allograft patients are summarized in Table . As shown previously, the frequency distribution in healthy carriers is skewed; therefore, it is not possible to calculate a meaningful mean value or standard deviation for comparison between populations. However, the data show a normal distribution when the ln frequencies are derived (Fig. 3 A); this allows the calculation of mean and standard deviation for the ln frequencies. The mean frequency of the ln values for healthy donors is 3.96. Calculating the anti-ln, this is 50 infected cells per 107 B cells, in agreement with our previously published estimates on a smaller subset of healthy carriers 4. When this analysis was applied to the data set from immunosuppressed patients, it was apparent that there are two discrete groups (Fig. 3 A). The first, represented by patients 1–19, also demonstrated a normal distribution of the ln frequencies, with a standard deviation indistinguishable from that of the healthy carriers (0.95 vs. 1.02). The mean frequency of the ln values was 7.36 or, from the anti-ln, 1,600 infected cells per 107 B cells, representing a 30-fold increase over the healthy carriers. The fraction of B cells in these patients varied from normal to ∼10% of normal, but in no case could the increase in the frequency of infected cells be accounted for by selective loss of uninfected cells. Therefore, we may conclude that there was also an increase in the absolute numbers of virus-infected cells. The relative ratio of naive to memory B cells, as judged by sIgD expression, was not different from the control population (Fig. 3 B). To confirm that there was an increase in infected cells due to immunosuppressive drugs, the frequency of virus-infected cells was tested in several patients before and after immunosuppression. In every case, the frequencies before immunosuppression were within the range observed for the control population, and after immunosuppression the frequencies were higher than the highest value seen with the controls (not shown).
Table 5.
Comparison of the Frequencies of EBV-infected B Cells in the Peripheral Blood of Healthy Controls and Immunosuppressed Patients
| Control | Frequency | Patient | Frequency |
|---|---|---|---|
| 1 | 425 | 1 | 16,700 |
| 2 | 320 | 2 | 8,100 |
| 3 | 110 | 3 | 6,500§ |
| 4 | 95 | 4 | 5,200 |
| 5 | 90 | 5 | 2,900 |
| 6 | 85 | 6 | 1,700 |
| 7 | 85 | 7 | 1,550 |
| 8 | 75 | 8 | 1,500 |
| 9 | 55 | 9 | 1,500 |
| 10 | 50 | 10 | 1,400 |
| 11 | 40 | 11 | 1,300 |
| 12 | 30 | 12 | 1,000 |
| 13 | 30 | 13 | 1,000 |
| 14 | 30 | 14 | 825 |
| 15 | 25 | 15 | 800 |
| 16 | 25 | 16 | 750 |
| 17 | 25 | 17 | 670 |
| 18 | 15 | 18 | 400 |
| 19 | 10 | 19 | 300 |
| 20 | 0∥ | 20 | 200 |
| 21 | 0∥ | 21 | 100 |
| 22 | 70 | ||
| 23 | 62 | ||
| 24 | 50 | ||
| 25 | 40 | ||
| 26 | 25 | ||
| 27 | 8 | ||
| 28 | 3 |
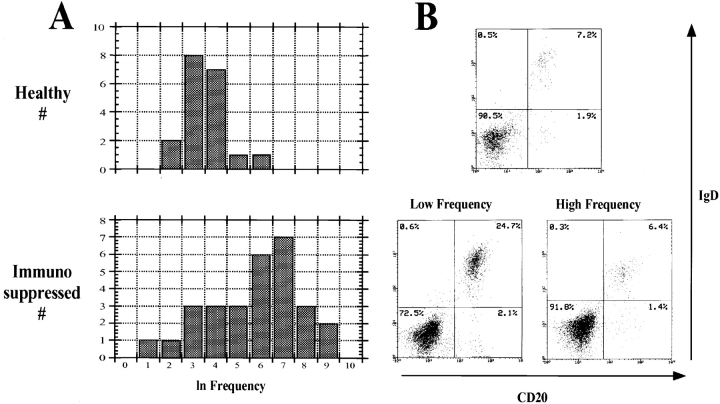
Figure 3.
Frequencies of virus-infected cells in healthy control and immunosuppressed populations. (A) The graphs show the distribution in the frequencies for each population plotted as their ln values. The resulting curve, for healthy donors, approximates a normal distribution with a mean frequency of 50 infected cells per 107 B cells. The immunosuppressed patients fall into two groups with means of 36 and 1,600, respectively. (B) FACS® analysis of representative individuals from the healthy controls and from each group of immunosuppressed patients. Note that the level of IgD+ B cells in the representative high frequency patient (6.4%) is within the normal range compared with the healthy control (7.2%), whereas it is highly elevated in the low frequency patient (24.7%). In comparison, the memory cells (IgD−CD20+) are within normal range for both patients (1.4 and 2.1 vs. 1.9% in the control).
The second group is represented by patients 20–28. Although too small to draw precise conclusions, this group had a mean frequency of virus-infected cells (36 vs. 50) similar to the healthy controls. The simplest interpretation is that this group of patients did not respond to the immunosuppressive drugs; however, they did not show elevated rates of rejection. Confirmation that this was a discrete population came from the observation that they exhibited a large increase in the numbers of uninfected naive B cells (Fig. 3 B; three to six times that in the controls), which was never seen in the high frequency group. This increase counterbalanced a modest increase in infected memory cells, resulting in no overall change in the frequency of virus-infected B cells. We have attempted to correlate the status of the two groups of patients with medications, type and origin of the transplanted organ, diagnosis at the time of transplant, sex, ethnicity, and occurrence of rejection episodes. The only factor that correlated was the use of mycophenolate mofetil with the occurrence of high frequencies of virus-infected cells (see Table ). No factor could be associated specifically with the low frequency patients.
We conclude that there is an increase in the frequency of infected B cells in the blood of only two thirds of patients receiving immunosuppressive drugs at the New England Medical Center.
Viral Gene Expression.
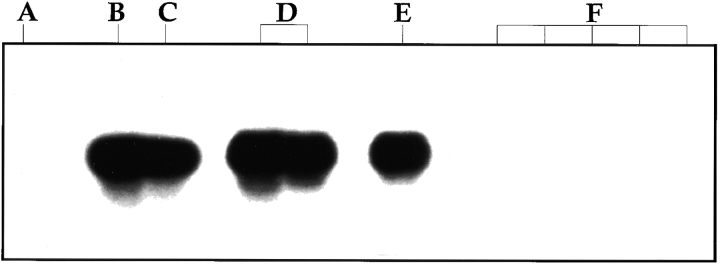
RT-PCR analysis of virally infected cells from the peripheral blood of healthy carriers suggests that the only viral latent genes detected are LMP-2a and possibly EBNA-1 7 9 22 23, although the latter result is controversial. LMP-2a contains the same signaling immunoreceptor tyrosine-based activation motif (ITAM) found in the B cell receptor 25. Because of this, we have suggested that LMP-2a could play a role in the long-term maintenance of the infected memory cells by supplying a surrogate B cell receptor survival signal 6. Immunosuppressed individuals also carry the virus in the resting memory population, but at a higher frequency. Therefore, we expected to detect LMP-2a with ease in these individuals. However, as shown in Table , LMP-2a was only found in one of the four patients tested, and this was the patient with the highest frequency of virus-infected cells. The results for two patients are shown in Lanes A and C of Fig. 4, and the results for all four patients are summarized in Table . Lane A shows the result from a patient whose cDNA was tested from a pool of peripheral blood B cells estimated to contain ∼20 infected cells. No LMP-2a message was detected. Lane C is from a patient whose B cell sample was estimated to contain ∼1,700 infected cells. In this case, the signal obtained had an intensity lying between that obtained with one and five cells from the EBV-immortalized cell line IB4 (lanes B or D vs. E). The cell line IB4 was used as a positive control because, in studies not shown, the RT-PCR signal obtained for LMP-2a was similar for IB4 and a number of other in vitro–immortalized lymphoblastoid lines. Note that in control studies, performed for every RT-PCR, the LMP-2a message can be readily detected in a single cell from the IB4 cell line in the presence of 5 × 106 uninfected PBL B cells (lane E). The negative PCR signals obtained were not due to an artifact or inhibitory contaminants in the samples, because we could readily detect five IB4 cells when they were mixed with the patient cells before analysis (see lane B, for example). cDNA from the LMP-2a− patients was also tested for expression of the EBNA-2 gene, which is required for expression of the full panoply of latent genes found in the lymphoblastoid type of latent infection, and EBNA-1(Qp), which is derived from a unique promoter that allows the expression of EBNA-1 only in Burkitt lymphoma cells. Again, no signal was obtained, although the techniques readily detected a single spiked EBV-infected cell, IB4 for EBNA-2 and Rael for EBNA-1(Qp), in the presence of 5 ×106 uninfected PBL B cells. These results suggest that the vast majority of infected cells in the blood of immunosuppressed individuals may be transcriptionally silent for the known latent genes.
Table 6.
Expression of the EBV-encoded LMP-2a Gene in Peripheral Blood Cells from Immunosuppressed Patients
| Patient | No. of EBV+ cells tested | LMP-2a | EBNA-2 | EBNA-1 (Qp) |
|---|---|---|---|---|
| 6 | 1,700 | + | ND | ND |
| 13 | 1,000 | − | − | − |
| 20 | 5 | − | − | − |
| 25 | 20 | − | − | − |
Figure 4.
RT-PCR analysis for the expression of LMP-2a mRNA in infected B cells from immunosuppressed individuals. CD19+ B cells were purified by the MACS® system. RNA was extracted using Trizol, and RT-PCR was performed for LMP-2a. (A) cDNA from a sample containing an estimated 20 infected cells from patient 25. (B) As in A, but spiked with five cells from the EBV-immortalized lymphoblastoid cell line IB4. (C) cDNA from a sample containing an estimated 1,700 infected cells from patient 6. (D) cDNA from 5 IB4 cells in 2 × 105 EBV− BJAB cells. (E) cDNA from 1 IB4 cell in 2 × 105 EBV− BJAB cells. (F) cDNA from 2 × 105 EBV− BJAB cells.
Discussion
The main conclusion of this paper is that latently infected resting, memory B cells expand in the peripheral blood of immunosuppressed individuals. This surprising result means that there is no expansion of EBV-infected proliferating lymphoblasts in the blood, as had been thought previously.
Most previous studies on the increased viral burden in the immunosuppressed have focused on the small subpopulation of individuals that have PTLD, whose systems are perturbed not only by immunosuppression but also by the presence of malignancy. This is the first study to characterize the phenotype of cells that expand in the periphery of patients that are immunosuppressed but otherwise clinically healthy. In fact, this is the first study to demonstrate quantitatively that the number of latently infected cells increases in the peripheral blood of most, but not all, patients after immunosuppression and that the cells are all B lymphocytes. The limitations of the previous studies are best exemplified by our demonstration that in approximately half of the patients studied, viral replication contributes a larger portion of the viral genome burden than does the increase in latently infected cells (compare linear and episomal signal intensity in Fig. 2 D).
Our results cannot exclude the possibility that small (<10%) numbers of proliferating infected lymphoblasts are present in the peripheral circulation. We have attempted to confirm the presence of these cells by testing enriched CD23+ B cells for expression of EBNA-2 and LMP-1, latent genes characteristically expressed in the lymphoblastoid form of latency. However, we have only detected expression of these genes in one out of eight patients tested. Therefore, it seems likely that in most patients the small numbers of infected cells detected by DNA PCR in the CD23+ and cycling populations are due to low levels of cross-contamination.
We have shown previously that EBV-infected cells in the peripheral blood of healthy carriers are restricted to the memory compartment. This cannot be because infected naive cells are preferentially lysed by CTLs, because we have now shown that a similar restriction applies even after immunosuppression. Therefore, some other mechanism must account for the specific association of EBV with memory cells in the peripheral blood. In addition, the number and frequency of latently infected cells increase in the periphery of immunosuppressed individuals, yet the cells themselves are resting. Therefore, the production of latently infected memory cells must occur elsewhere. The explanation we favor is based on the idea that EBV uses the normal pathways of B cell activation and differentiation to establish and maintain a latent infection. In this model, EBV-infected lymphoblasts are generated from newly infected B cells in secondary lymphoid tissue such as the tonsil. We propose that these latently infected blasts behave like normal B blasts and remain in the lymph nodes, where they follow the normal pathways of B cell activation and differentiation to gain access to the long-lived memory compartment, which we have proposed as the site of long-term persistence. Upon immunosuppression, there is more infectious virus produced in the lymphoid tissue, and therefore more cells are infected and become proliferating lymphoblasts. These blasts expand due to the lack of a cellular immune response, and eventually leave the lymph nodes as increased numbers of differentiated memory B cells. If our hypothesis is correct, then EBV-associated hyperproliferation due to an impaired CTL response should also be limited to the lymph nodes. This is what is observed with PTLDs, the majority of which are limited to the lymph nodes and disappear when immunosuppression is lifted 26. Escape from the lymph nodes into the peripheral circulation would be atypical behavior for a B blast, and would be expected to be associated with the acquisition of genetic defects. Consistent with this, extranodal PTLDs are less common, associated with genetic anomalies, more malignant, and less likely to respond to reductions in immunosuppression 26.
A second, but not mutually exclusive, explanation for the results in this paper is that the latently infected resting memory B cells are themselves under immunosurveillance, for example due to expression of LMP-2a 27. An impeded CTL response would allow higher numbers of these cells to be tolerated. The RT-PCR analysis presented here argues against this possibility. Although we can detect LMP-2a message in mixtures where one IB4 cell is diluted with 5 × 106 uninfected PBL B cells, we were unable to detect it in as many as 1,000 infected cells from immunosuppressed patients. This suggests either that the transcript copy number in the resting memory cells is extremely low or that the major population that expands in the immunosuppressed is transcriptionally silent for LMP-2a. We have also failed to detect two other markers of latent gene expression, EBNA-2 and EBNA-1(Qp). EBNA-2 is essential for the expression of all the latent genes expressed in the lymphoblastoid form of latency. Its absence was expected, since EBNA-2 activity would be inconsistent with the resting state of the cells. These results suggest that the major infected population in the blood of immunosuppressed patients may not express the known latent proteins and cannot therefore be detected by immunosurveillance.
The one major difference in the behavior of the virus between healthy carriers and immunosuppressed patients is the presence of viral replication in the peripheral blood of some patients. We have not tested our patients for lytic gene expression; however, Prang et al. 28 reported finding immediate early and early but not late transcripts of the lytic cycle in the peripheral blood of healthy carriers. This is consistent with our finding that viral genome replication is not detectable. They also demonstrated strong CTL responses to the immediate early proteins 29 and proposed that cells in the periphery occasionally enter the lytic cycle spontaneously, but are eliminated by CTLs before they produce infectious virus. A direct prediction of this idea is that upon immunosuppression a small fraction of the infected cells in the peripheral blood will spontaneously replicate the virus, and indeed this is what we have found.
In conclusion, we have shown that immunosuppression does not lead to the appearance of latently infected blasts in the blood. Furthermore, our experiments lend further support to the idea that EBV has evolved to efficiently exploit the normal biology of B lymphocytes to establish and maintain latency in the memory compartment.
Acknowledgments
We thank Allan Parmalee and Glenn Paradise for performing the flow cytometry, and members of the Transplant Unit at New England Medical Center for help in obtaining blood samples and for stimulating discussions. We are particularly indebted to all of the patients who participated in the study. This work could not have been completed without their cooperation.
The authors' work was supported by U.S. Public Health Service grants AI18757 and CA65883.
Footnotes
1used in this paper: EBNA, EBV nuclear antigen; LMP, latent membrane protein; PTLD, posttransplant lymphoproliferative disease; RT, reverse transcriptase; sIg, surface Ig
G.J. Babcock and L.L. Decker contributed equally to the experiments described in this paper.
References
- Rickinson A.B., Kieff E. Epstein-Barr virus. In: Fields B.N., Knipe D.M., Howley P.M., editors. Virology, Vol. 2, 3rd. ed. 2 vols. Raven Press; New York: 1996. pp. 2397–2446. [Google Scholar]
- Kieff E. Epstein-Barr virus and its replication. In: Fields B.N., Knipe D.M., Howley P.M., editors. Virology, Vol. 2, 2nd. ed. 2 vols. Raven Press; New York: 1996. pp. 2343–2396. [Google Scholar]
- Yao Q.Y., Rickinson A.B., Epstein M.A. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int. J. Cancer. 1985;35:35–42. doi: 10.1002/ijc.2910350107. [DOI] [PubMed] [Google Scholar]
- Khan G., Miyashita E.M., Yang B., Babcock G.J., Thorley-Lawson D.A. Is EBV persistence in vivo a model for B cell homeostasis? Immunity. 1996;5:173–179. doi: 10.1016/s1074-7613(00)80493-8. [DOI] [PubMed] [Google Scholar]
- Decker L.L., Klaman L.D., Thorley-Lawson D.A. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J. Virol. 1996;70:3286–3289. doi: 10.1128/jvi.70.5.3286-3289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Decker L.L., Volk M., Thorley-Lawson D.A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- Qu L., Rowe D.T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J. Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita E.M., Yang B., Lam K.M., Crawford D.H., Thorley-Lawson D.A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- Miyashita E.M., Yang B., Babcock G.J., Thorley-Lawson D.A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson D.A. EBV persistence in vivo. Invading and avoiding the immune response. In: Medveczky P.G., Friedman H., Bendinelli M., editors. Herpesviruses and Immunity. Plenum Press; New York: 1998. pp. 207–229. [Google Scholar]
- Kenagy D.N., Schlesinger Y., Weck K., Ritter J.H., Gaudreault-Keener M.M., Storch G.A. Epstein-Barr virus DNA in peripheral blood leukocytes of patients with posttransplant lymphoproliferative disease. Transplantation. 1995;60:547–554. doi: 10.1097/00007890-199509270-00005. [DOI] [PubMed] [Google Scholar]
- Lam K.M., Syed N., Whittle H., Crawford D.H. Circulating Epstein-Barr virus-carrying B cells in acute malaria. Lancet. 1991;337:876–878. doi: 10.1016/0140-6736(91)90203-2. [DOI] [PubMed] [Google Scholar]
- Martinez O.M., Villanueva J.C., Lawrence-Miyasaki L., Quinn M.B., Gish R., Cox K., So S., Esquivel C.O., Krams S.M. Molecular markers of Epstein-Barr virus infection in the circulation of transplant recipients. Transplant Proc. 1995;27:1211–1212. [PubMed] [Google Scholar]
- McKnight J.L., Cen H., Riddler S.A., Breinig M.C., Williams P.A., Ho M., Joseph P.S. EBV gene expression, EBNA antibody responses and EBV+ peripheral blood lymphocytes in post-transplant lymphoproliferative disease. Leuk. Lymphoma. 1994;15:9–16. doi: 10.3109/10428199409051672. [DOI] [PubMed] [Google Scholar]
- Telenti A., Marshall W.F., Smith T.F. Detection of Epstein-Barr virus by polymerase chain reaction. J. Clin. Microbiol. 1990;28:2187–2190. doi: 10.1128/jcm.28.10.2187-2190.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q.Y., Rickinson A.B., Gaston J.S., Epstein M.A. In vitro analysis of the Epstein-Barr virushost balance in long-term renal allograft recipients. Int. J. Cancer. 1985;35:43–49. doi: 10.1002/ijc.2910350108. [DOI] [PubMed] [Google Scholar]
- Lewin N., Aman P., Masucci M.G., Klein E., Klein G., Oberg B., Strander H., Henle W., Henle G. Characterization of EBV-carrying B-cell populations in healthy seropositive individuals with regard to density, release of transforming virus and spontaneous outgrowth. Int. J. Cancer. 1987;39:472–476. doi: 10.1002/ijc.2910390411. [DOI] [PubMed] [Google Scholar]
- Khanna R., Burrows S.R., Moss D.J. Immune regulation in Epstein-Barr virus-associated diseases. Microbiol. Rev. 1995;59:387–405. doi: 10.1128/mr.59.3.387-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzl T.E., Nalesnik M.A., Porter K.A., Ho M., Iwatsuki S., Griffith B.P., Rosenthal J.T., Hakala T.R., Shaw B.W., Jr., Hardesty R.L. Reversibility of lymphomas and lymphoproliferative lesions developing under cyclosporin-steroid therapy. Lancet. 1984;1:583–587. doi: 10.1016/s0140-6736(84)90994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornef M.W., Wagner H.J., Fricke L., Bein G., Kirchner H. Immunocytochemical detection of Epstein-Barr virus antigens in peripheral B lymphocytes after renal transplantation. Transplantation. 1995;59:138–140. doi: 10.1097/00007890-199501150-00025. [DOI] [PubMed] [Google Scholar]
- Schaefer B.C., Strominger J.L., Speck S.H. A simple reverse transcriptase PCR assay to distinguish EBNA1 gene transcripts associated with type I and II latency from those arising during induction of the viral lytic cycle. J. Virol. 1996;70:8204–8208. doi: 10.1128/jvi.70.11.8204-8208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zou J.Z., di Renzo L., Winberg G., Hu L.F., Klein E., Klein G., Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J. Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney R.J., Steven N., Young L.S., Rickinson A.B. Epstein-Barr virus latency in blood mononuclear cellsanalysis of viral gene transcription during primary infection and in the carrier state. J. Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers W.C., Klein G. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J. Virol. 1976;18:151–155. doi: 10.1128/jvi.18.1.151-155.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaufils P., Choquet D., Mamoun R.Z., Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:5105–5112. doi: 10.1002/j.1460-2075.1993.tb06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles D.M., Cesarman E., Chadburn A., Frizzera G., Chen J., Rose E.A., Michler R.E. Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders. Blood. 1995;85:552–565. [PubMed] [Google Scholar]
- Khanna R., Slade R.W., Poulsen L., Moss D.J., Burrows S.R., Nicholls J., Burrows J.M. Evolutionary dynamics of genetic variation in Epstein-Barr virus isolates of diverse geographical originsevidence for immune pressure-independent genetic drift. J. Virol. 1997;71:8340–8346. doi: 10.1128/jvi.71.11.8340-8346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prang N.S., Hornef M.W., Jager M., Wagner H.J., Wolf H., Schwarzmann F.M. Lytic replication of Epstein-Barr virus in the peripheral bloodanalysis of viral gene expression in B lymphocytes during infectious mononucleosis and in the normal carrier state. Blood. 1997;89:1665–1677. [PubMed] [Google Scholar]
- Bogedain C., Wolf H., Modrow S., Stuber G., Jilg W. Specific cytotoxic T lymphocytes recognize the immediate-early transactivator Zta of Epstein-Barr virus. J. Virol. 1995;69:4872–4879. doi: 10.1128/jvi.69.8.4872-4879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]