The Intestinal T Cell Response to α-Gliadin in Adult Celiac Disease Is Focused on a Single Deamidated Glutamine Targeted by Tissue Transglutaminase (original) (raw)
Abstract
The great majority of patients that are intolerant of wheat gluten protein due to celiac disease (CD) are human histocompatibility leukocyte antigen (HLA)-DQ2+, and the remaining few normally express HLA-DQ8. These two class II molecules are chiefly responsible for the presentation of gluten peptides to the gluten-specific T cells that are found only in the gut of CD patients but not of controls. Interestingly, tissue transglutaminase (tTG)-mediated deamidation of gliadin plays an important role in recognition of this food antigen by intestinal T cells. Here we have used recombinant antigens to demonstrate that the intestinal T cell response to α-gliadin in adult CD is focused on two immunodominant, DQ2-restricted peptides that overlap by a seven-residue fragment of gliadin. We show that tTG converts a glutamine residue within this fragment into glutamic acid and that this process is critical for T cell recognition. Gluten-specific T cell lines from 16 different adult patients all responded to one or both of these deamidated peptides, indicating that these epitopes are highly relevant to disease pathology. Binding studies showed that the deamidated peptides displayed an increased affinity for DQ2, a molecule known to preferentially bind peptides containing negatively charged residues. Interestingly, the modified glutamine is accommodated in different pockets of DQ2 for the different epitopes. These results suggest modifications of anchor residues that lead to an improved affinity for major histocompatibility complex (MHC), and altered conformation of the peptide–MHC complex may be a critical factor leading to T cell responses to gliadin and the oral intolerance of gluten found in CD.
Keywords: HLA-DQ2, modification, gluten, oral tolerance, mucosal immunity
Introduction
Celiac disease (CD) is the most common food-sensitive enteropathy in humans, with an incidence as high as 1/300 in some European countries. It is a chronic inflammatory disease that has become a valuable model for the study of HLA-associated diseases because there is a well defined HLA class II association 1 2, it is relatively easy to obtain biopsies from the disease-affected organ, and the antigen that precipitates disease is known.
The observation that CD is strongly associated with DQ2 encoded by DQA1*0501/DQB1*02 or DQ8 encoded by DQA1*03/DQB1*0302 implies that CD4+ T cells play a central role in disease pathogenesis. Indeed, gluten-specific, CD4+ intestinal T cells can be isolated from intestinal biopsies of CD patients but not of controls 3 4. These CD4+ cells are TCR-α/β1 and typically of the Th1 phenotype secreting large amounts of IFN-γ 5, a cytokine that has been linked to mucosal damage 6. A striking and significant aspect of the intestinal T cell recognition of gluten is that it is predominantly restricted by DQ2 or DQ8 3 7, the very same molecules that immunogenetic studies have identified as conferring susceptibility to CD. These two molecules share an unusual preference for binding peptides containing negatively charged residues at relative positions P4, P6, or P7 for DQ2 8 9 10 and P1, P4, or P9 for DQ8 11 12. Initially, it was difficult to reconcile the DQ2 peptide binding motif with the DQ2-restricted presentation of multiple gluten peptides, as gluten has remarkably few negatively charged residues. This puzzle was explained by the recent demonstration that gluten, which has an exceptionally high glutamine content, becomes a far better T cell antigen once it been deamidated 13. Furthermore, this conversion of glutamine to glutamic acid has been shown to be mediated in an ordered and specific fashion by tissue transglutaminase (tTG; references 14 and 15), an enzyme that is also the major target of autoantibodies in CD 16.
To understand the disease pathology in CD and to explain its association with DQ2, it will be critical to dissect the relationship between the function and specificity of tTG, the processing of gliadin, and the binding of gliadin peptides to DQ2. A first step in this process will be to identify the gliadin epitopes recognized by CD patients and to assess their relative immunological importance. A comparison of multiple gliadin epitopes may identify common motifs that are targeted by tTG or that influence MHC class II binding and will allow for the significance of deamidation on T cell recognition to be more fully examined at a molecular level. However, the identification of gliadin T cell epitopes has been hampered by the heterogeneous nature of this antigen. Wheat gluten can be roughly separated into the glutenins and the gliadins according to alcohol solubility. The majority of the gluten-specific T cells appear to recognize the alcohol-soluble gliadin fraction 3 13 17. The gliadins can be further separated according to primary sequence into the α-, γ-, and ω-gliadins, with numerous variants present within each group 18. No DQ2-restricted intestinal T cell α-gliadin epitope has yet been described; a single γ-gliadin–derived peptide has been identified, but this is recognized by intestinal T cells from a minority of patients and does not appear to be a dominant epitope 13. To circumvent the inherent problems of using natural gliadins as antigens 19, we have cloned, sequenced, and expressed a panel of recombinant α-gliadins from a commercial Nordic wheat cultivar 20. This study exploits these antigens to characterize the response to the α-gliadins using a panel of T cell clones (TCCs) and T cell lines (TCLs) established from intestinal biopsies taken from DQ2+ CD patients.
Materials and Methods
Production of _α_-Gliadin Recombinants.
The production of α-gliadins has been described in detail elsewhere 20. In brief, mature α-gliadin genes were cloned from both cDNA and genomic DNA from the commercial Nordic wheat strain, Mjolner. 11 full-length clones were identified and expressed in Escherichia coli using the pET 17xb expression system (Novagen, Inc.). Gliadin was extracted from the E. coli by lysing in 70% ethanol at 60°C. Bacterial cell debris was then removed by centrifugation, and gliadin was precipitated by addition of NaCl to 1 M. Coomassie staining of these preparations run on SDS-PAGE revealed single dominant bands of appropriate weight with only minor contamination. For purification of T cell–active fragments, 10 mg of α-9 dissolved in 8 M urea/0.4 M NH4HCO3, pH 8.0, was reduced and alkylated 21, dialysed against 0.1 M NH4HCO3/0.1 mM CaCl2, and digested with chymotrypsin (1:100 wt/wt). This was passed over a Superdex Peptide HR 10/30 column (Amersham Pharmacia Biotech) in a 0.1 M NH4HCO3 buffer, vacuum dried, and resuspended in 100 μl of 5 mM Tris/0.8 mM CaCl2 with 100 μg/ml of guinea pig tTG (Sigma Chemical Co.). T cell–reactive fragments were further separated by anion exchange chromatography (Mono-Q PC 1.6/5) equilibrated with 5 mM Tris/HCL buffer, pH 5.5, and developed with a gradient ending at 50 mM NaCl, followed by reverse-phase HPLC (μRPC C2/C18; Pharmacia) using a gradient running from 100% buffer A (0.1% TFA in H2O) to 100% buffer B (80% acetonitrile, 19.9% H2O, 0.1% TFA). Mono-Q and reverse-phase HPLC were run on a SMART system (Pharmacia).
Preparation of Antigens.
Digestion of crude gliadin with either pepsin and trypsin 3 or chymotrypsin 14 was done as described. The overlapping A-gliadin peptides were identical to those previously described 22. All remaining peptides were supplied by Research Genetics and were >80% pure. Acid/heat treatment was achieved by dissolving peptides in 100 μl of 0.01 M CH3COOH, pH 1.8, and heating at 96°C until dry. Human tTG was expressed as a GST fusion protein in E. coli using the vector construct and protocols described by Lai et al. 23. Treatment with human and guinea pig tTG (Sigma Chemical Co.) was performed at 37°C in PBS plus 0.8 mM CaCl2 using 50–200 μg/ml of tTG for 3 h.
Gliadin-specific T Cells.
T cells were grown and assayed in RPMI 1640 media (GIBCO BRL) supplemented with 15% inactivated pooled human serum, penicillin/streptomycin, and 0.01 M 2-ME. Biopsies from the Norwegian patients were challenged overnight in an organ culture chamber by immersion in gliadin antigen 3 14. Biopsies from patients CD377, CD414, CD416, CD419, CD420, CD421, and CD424 were challenged with chymotrypsin-digested gliadin. The remaining patients were challenged with a pepsin and trypsin digest of gliadin. Four different batches of gliadin were used: gliadin extracted from commercial gluten from Fluka AG or Sigma Chemical Co., extracted from the Norwegian commercial flour Regal (Regal Mølle a.s.), or extracted from flour prepared from the wheat strain Kadett. Biopsies from patient CD427 were taken from an untreated patient at the time of diagnosis. The remaining Norwegian patients were on gluten-free diets. Biopsies were treated with collagenase A to produce a single-cell suspension that was added to 96 U-bottomed plates containing equal numbers of irradiated autologous PBMCs together with 10 U/ml IL-2 and cultured in 5% CO2 at 37°C. These TCLs were restimulated weekly with PHA and IL-2 3 and typically tested between weeks 2 and 4.
The Dutch TCLs established from pediatric biopsies were established using a different restimulation strategy. In brief, biopsies were incubated twice in 1 mM dithiothreitol at room temperature for 10 min, washed, and then incubated at 37°C for at least 1 h in 0.75 mM EDTA while being gently agitated. The biopsy cells were then washed twice before being placed in one well of a 96-well culture plate containing RPMI media supplemented with 10% human serum, gentamicin, and 20 μg/ml of pepsin and trypsin–digested gliadin (Fluka AG). These cultures were restimulated with irradiated autologous PBMCs that had been prepulsed with 20 μg/ml of pepsin and trypsin–digested gliadin every 7–10 d and typically tested after 3–4 wk. For one of the TCLs, the antigen was treated with tTG before restimulation. The TCL derived from a Dutch adult patient was generated using the protocol described in reference 17.
Proliferation Assays.
Duplicate or triplicate wells containing irradiated APCs (5 × 104 cells per well; DR3+DQ2+ B lymphoblastoid cell lines) were prepulsed overnight with peptide or digested gliadin in a volume of 100 μl. 5 × 104 T cells per well in 50 μl were added the following day, and [3H]thymidine was added 2 d later. Plates were harvested after a further 12–16-h incubation, and [3H]thymidine incorporation was counted on a Betaplate Counter (Wallac Turku). All assays were performed two or more times with similar results each time.
HLA-DQ2 Peptide Binding Assay.
An 125I-labeled indicator peptide (KPLLIIAEDVEGEY) was used in a competitive binding assay using affinity-purified HLA-DQ2(α1*0501,β*0201) molecules as described 8. Binding affinity is presented in the figures as the concentration of unlabeled gliadin peptide required to inhibit indicator peptide binding by 50% (IC50).
Mass Spectrometry.
Matrix-assisted laser desorption/ionization (MALDI) spectra were acquired on a Reflex II time-of-flight mass spectrometer (Bruker Daltonik). For tandem mass spectrometry (MS/MS) experiments 24, a nanoelectrospray ion trap (Esquire, Bruker Daltonik) or a nanoelectrospray Q-TOF (Micromass) was used.
Results
Gluten-specific Intestinal TCCs Recognize a Limited Number of Recombinant α-Gliadins.
To investigate the nature and number of DQ2-restricted T cell epitopes present within the α-gliadin family, we tested a panel of 11 different α-gliadin recombinants for their ability to stimulate 7 intestinal TCCs derived from 4 CD patients (Table ). The recombinant antigens were digested with chymotrypsin to solubilize them and then pretreated with tTG to deamidate them before addition to the proliferation assays. The clones from patients CD412, CD370, and CD387 were chosen because they all recognized a purified natural α-gliadin but exhibited different response patterns toward two purified natural γ-gliadins. Clones CD380 E3 and CD380 E37 were randomly chosen. Testing of the α-recombinants without prior treatment with tTG failed to stimulate any of the clones (data not shown). However, after tTG treatment, five of the recombinant α-gliadins were recognized by two or more of the TCCs. The recombinant α-2 stimulated all of the TCCs efficiently, whereas strong proliferative responses to α-8, α-9, α-10, and α-11 were detected for only two clones from patient CD387. The remaining six gliadins failed to stimulate any of the recombinant gliadins tested, suggesting that this panel of TCCs recognizes a limited number of epitopes.
Table 1.
Recognition of a Panel of tTG-treated Recombinant α-Gliadin Antigens by Seven TCCs
| TCC | + | α-1 | α-2 | α-3 | α-4 | α-5 | α-6 | α-7 | α-8 | α-9 | α-10 | α-11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 412 R3 | 13.8 | 0.9 | 32.3 | 1.0 | 1.0 | 1.2 | 1.1 | 1.5 | 1.4 | 1.3 | 2.3 | 2.4 |
| 412 R5.32 | 15.7 | 1.2 | 19.0 | 0.9 | 1.1 | 1.2 | 1.2 | 1.1 | 1.2 | 0.9 | 1.6 | 0.9 |
| 370 R2.3 | 17.7 | 1.1 | 5.0 | 0.9 | 1.1 | 0.3 | 1.2 | 1.0 | 1.4 | 1.3 | 4.3 | 4.6 |
| 387 E9 | 52.6 | 1.0 | 25.0 | ND | 0.8 | ND | 1.2 | 1.2 | 6.1 | 25.3 | 23.7 | 19.4 |
| 387 E34 | 25.1 | 0.9 | 25.1 | ND | 1.0 | ND | 1.3 | 1.1 | 5.0 | 15.2 | 15.3 | 16.2 |
| 380 E3 | 2.4 | 1.0 | 2.2 | ND | 0.9 | ND | 1.3 | 1.1 | 1.1 | 1.0 | 0.6 | 1.0 |
| 389 E37 | 6.5 | 1.1 | 3.7 | 1.1 | 1.2 | 1.6 | 1.4 | 1.0 | 2.8 | 2.1 | ND | ND |
Identification of a DQ2-restricted T Cell Epitope in Recombinant Gliadin α-9.
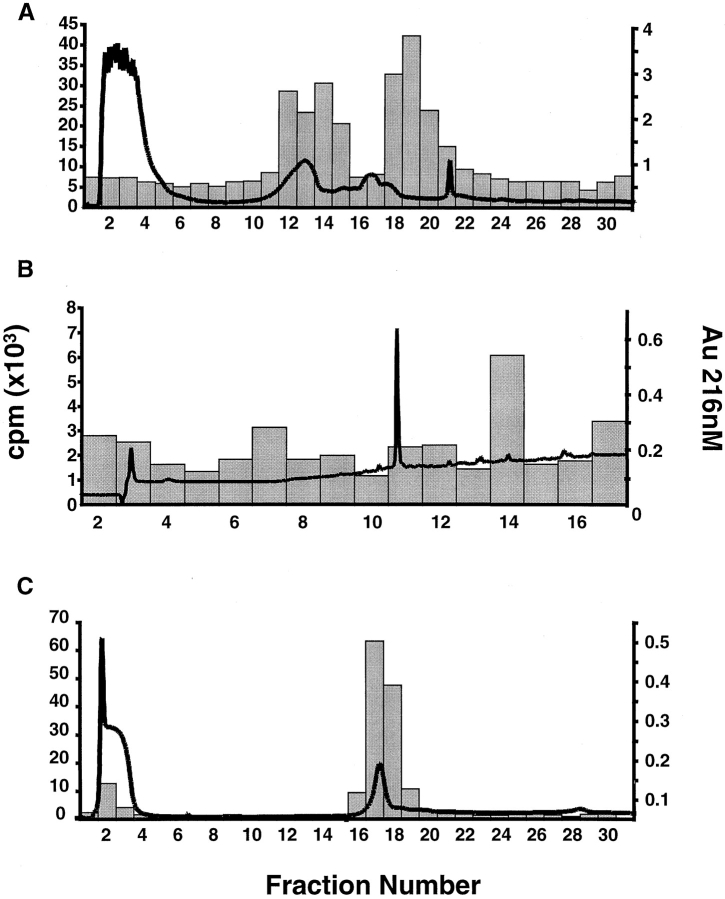
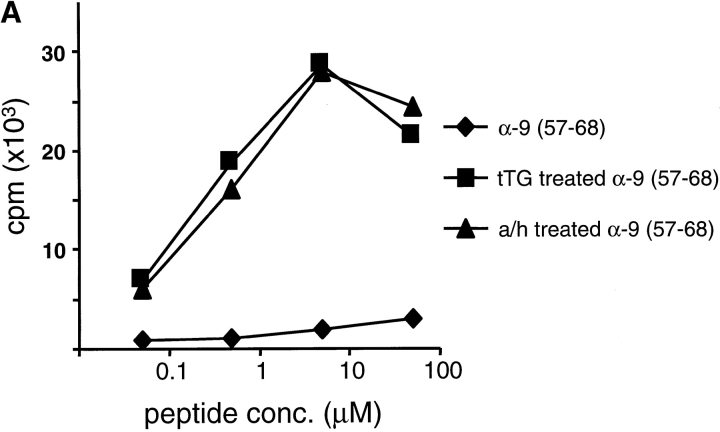
A peptide fragment containing the T cell epitope present in the α-9 recombinant was isolated by a series of biochemical purification steps using T cell reactivity with CD387 E34 to identify the positive fractions (Fig. 1). The recombinant was digested with chymotrypsin and then separated using size exclusion chromatography. The resulting fractions were treated with recombinant human tTG and tested for recognition by the TCC CD387 E34. The fraction containing the smallest peptides that still efficiently stimulated the TCC (fraction 29) was then separated using ion exchange chromatography (Mono-Q; Fig. 1 A). Two T cell–reactive Mono-Q fractions were then subjected to reverse-phase HPLC separation. Both produced a single fraction capable of stimulating TCC CD387 E34 (Fig. 1 B). Analysis of these fractions by MALDI-TOF mass spectrometry yielded a single signal of 1,438.6 daltons. Analysis of the sequence for the α-9 recombinant identified no obvious chymotrypsin fragments of this mass. However, nine peptides were identified that were of an appropriate mass when deamidation of glutamine to glutamic acid (or pyroglutamic acid if glutamines were present at the NH2 terminus) were taken into consideration. A comparison between the T cell reactivity of the recombinants and the presence or absence of each of these candidate peptides excluded eight of the nine from being the probable peptide recognized by CD387 E34 (Table ). However, one peptide spanning residues 57–68 of the α-9 recombinant contains a sequence that is present in the α-2, α-8, α-9, α-10, and α-11 recombinants, all of which are recognized by CD387 E34. The remaining α-recombinants all contain sequence variation within this peptide and are not recognized by this TCC. The identity of the epitope in α-9 was confirmed by testing a synthetic peptide corresponding to the α-9(57–68) sequence for its ability to stimulate TCC CD387 E34. This peptide was inactive in its native form but potently stimulated the clone after treatment with tTG (Fig. 2 A). This peptide also stimulated clones CD387 E9, CD380 E3, and CD380 E37 in a similar fashion (data not shown). Clones CD380 E3 and CD380 E37 were the least reactive of the seven clones, which may explain why no proliferation was detected when they were tested with recombinants containing this peptide. Experiments using allogeneic APCs and blocking antibodies have confirmed that these clones recognize this peptide in the context of DQ2 (data not shown).
Figure 1.
Biochemical purification of a peptide fragment from α-9 recombinant stimulatory for TCC CD387 E34. A T cell–reactive superdex fraction of the tTG-treated α-9 recombinant chymotrypsin digest was separated by ion exchange chromatography (A). The absorbency at 216 nm is overlaid on a histogram depicting the proliferative response of TCC CD387 E34 to the resulting fractions. Fractions 18 and 19 were further separated by reverse-phase HPLC, each producing a small T cell–reactive peak in fraction 14 (fraction 19 shown in B). tTG treatment of synthetic peptide α-9(57–68) and separation by Mono-Q generates a new peak at fractions 17 and 18 that is recognized by TCC CD387 E34 (C).
Table 2.
Comparison of T Cell Recognition and Sequence for α-Gliadin Recombinants
Figure 2.
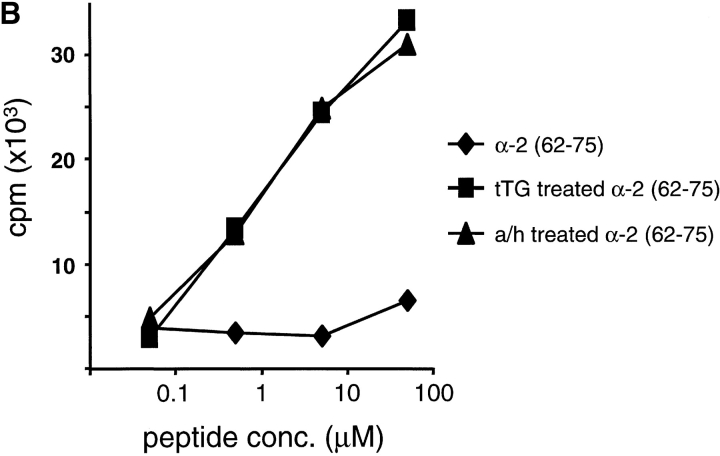
Recognition of peptides α-9(57–68) and α-2(62–75) by two TCCs. Peptides α-9(57–68) (A) and α-2(62–75) (B) were tested in their native state (♦) or after treatment with human tTG (▪) or acid/heat (a/h; ▴) for their ability to induce proliferation in (A) TCC CD387 E34 and (B) TCC CD412 R5.32. Responses are given in cpm.
Identification of a DQ2-restricted T Cell Epitope in Recombinant Gliadin α-2.
The TCC from patients CD370 and CD412 recognized the α-2 recombinant alone and so presumably recognizes a peptide that is absent in the other recombinants. Although several such regions are present in the α-2 recombinant, one region (amino acids 62–81) is noteworthy because it shares considerable homology with the α-9(57–68) epitope. This is due to the presence of a triple repeat of a 7-mer motif in the α-2 gliadin, a motif that is only present once in the α-9 recombinant. A 12-mer peptide, α-2(62–75), corresponding to two of these repeats was tested for its ability to stimulate the clone CD412 R5.32. This peptide efficiently stimulated this clone after treatment with tTG but not in its native form (Fig. 2 B). Clones CD412 R3 and CD370 R2.3 gave similar results when tested with this peptide. Proliferation assays using allogeneic APCs as well as blocking antibodies confirmed that this peptide is presented for T cell recognition by DQ2 (data not shown).
Deamidation of a Single Glutamic Acid Residue Is Critical for T Cell Recognition of the α-9(57–68) and α-2(62–75) Peptides.
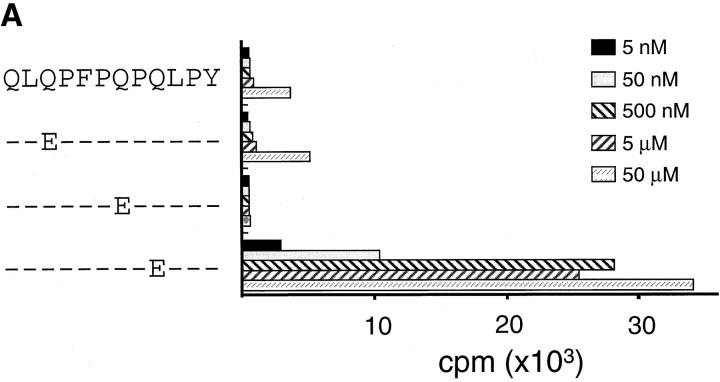
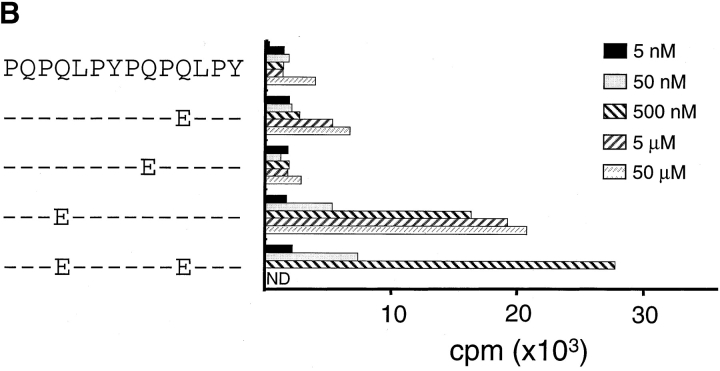
The α-2(62–75) and α-9(57–68) peptides were only recognized by the TCC after deamidation with tTG or in conditions known to promote nonenzymatic deamidation (i.e., heating in an acidic environment). To identify the number and position of glutamine residues targeted by tTG, we separated the α-2(62–75) and α-9(57–68) peptides by Mono-Q chromatography after first treating them with recombinant human tTG. Analysis of the α-9(57–68) peptide indicated that a single new peptide peak had been created by treatment with tTG. Mono-Q fractions containing this peak efficiently stimulated TCC CD387 E34 (Fig. 1 C). This fraction was methyl-esterified to label the deamidation site with a methyl-ester group, and this position was identified as residue 65 by MS/MS analysis (peptide α-9[57–68]E65; reference 25). Two peaks were created by the tTG treatment of the α-2(62–75) peptide, both of which were stimulatory for the TCC CD412 R5.32 (data not shown). Mass spectrometry analysis revealed that the least acidic of these two peaks contained a peptide in which the glutamine at position 65 had been deamidated (peptide α-2[62–75]E65). The more acidic peak contained a peptide with glutamines deamidated at residues 65 and 72. Confirmation that the deamidation of glutamine at position 65 was critical for T cell recognition of both peptides was given by testing synthetic peptides with glutamines substituted with glutamic acid. Peptides containing glutamic acid at position 65 stimulated TCCs CD412 R5.32 and CD387 E34 extremely efficiently; the remaining peptides could, at best, only stimulate the TCCs very weakly when high concentrations of peptide were used (Fig. 3). Testing for recognition of both the α-9(57–68)E65 and α-2(62–75)E65 peptides demonstrated that these epitopes were not cross-reactive for this panel of TCCs (data not shown).
Figure 3.
Testing of synthetic glutamine to glutamic acid–substituted peptides for their ability to stimulate TCCs. TCC CD387 E34 (A) and TCC CD412 R5.32 (B) were tested for recognition of a set of singly substituted peptides that had the glutamine residues substituted for glutamic acid residues.
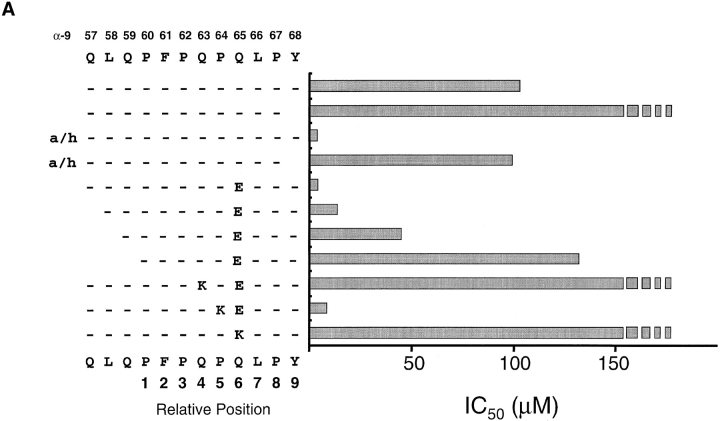
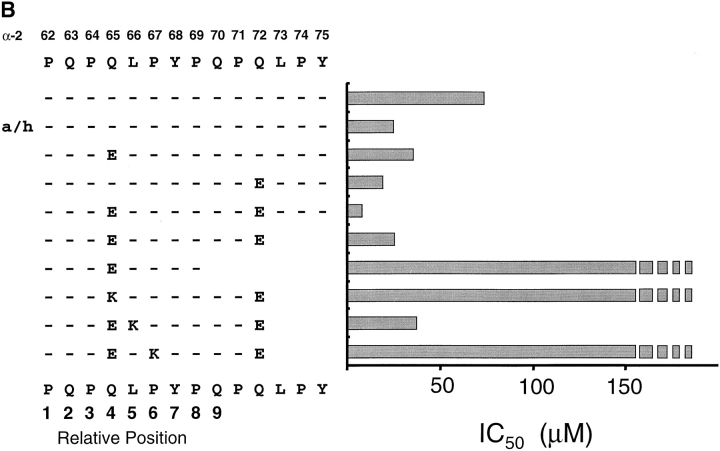
Binding of the α-9(57–68) and α-2(62–75) Peptides to DQ2.
The minimal core binding region and the binding frame for α-2(62–75) and α-9(57–68) peptides was characterized by measuring the affinity of a set of truncated and/or glutamic acid– and lysine-substituted peptides for DQ2 in a cell free binding assay (Fig. 4). The affinity for DQ2 of the α-2(62–75) and α-9(57–68) peptides increased dramatically when the peptides were first heated in an acidic environment or when a glutamic acid was substituted at the position identified as important for T cell recognition. Interestingly, substitution of glutamic acid residues at some other positions resulted in a comparable increase in affinity of the peptide for DQ2 but did not lead to T cell recognition (Fig. 4 and data not shown). This implies that recognition of these deamidated peptides is a combined result of an increase in peptide binding and a change in conformation of the peptide–MHC complex.
Figure 4.
Analysis of peptides α-9(57–68) and α-2(62–75) binding to DQ2. Acid/heat (a/h)-treated, glutamic acid–substituted, and truncated peptide variants of the (A) α-9(57–68) and (B) α-2(62–75) peptides were assessed for binding to DQ2 using a cell free competitive binding assay. The presumed binding frame for the two peptides is marked below the sequence. Broken bars indicate that peptide binds too poorly to DQ2 to be measured accurately in this assay.
Removal of the COOH-terminal tyrosine residue from the α-9(57–68) peptide effectively abolished DQ2 binding (Fig. 4 A) and T cell recognition (data not shown), strongly suggesting that this residue acts as an anchor in the P9 pocket. This would position residues 57, 58, and 59 at P3, P2, and P1, respectively. Successive truncation of the peptide at these positions leads to a progressive loss of affinity for DQ2 and T cell recognition (data not shown), most likely due to a loss in hydrogen bonding between the main chain residues in the peptide and DQ2. A similar progressive loss of affinity for DQ2 was seen for the α-2(62–75) peptide truncated at the COOH terminus. However, the removal of the glutamine at position 70 reduced the binding to undetectable levels, indicating that this residue act as the P9 anchor. T cell recognition of these truncated peptides gave results comparable to the binding data (data not shown).
Previous peptide binding studies have clearly demonstrated that positively charged residues inhibit peptide binding to DQ2 at relative positions P4, P6, or P7 but are well tolerated at P5, a position thought to point toward the T cell receptor 10. We therefore reasoned that the assumed binding frames for the α-2(62–75)E65 and α-9(57–68)E65 peptides could be confirmed by measuring DQ2 binding using lysine-substituted peptides at the tentative P4, P5, and P6 residues (Fig. 4 A). The binding to DQ2 of the α-9(57–68)E65 peptide dropped to below the accurate detection level for our assay when single lysine residues were substituted at positions 63 and 65. In contrast, the α-9(57–68)E65 peptide with a lysine substituted at position 64 showed only a marginal decrease in binding to DQ2. Similarly, the DQ2 binding of the α-2(62–75) peptide was inhibited by lysine substitutions at positions 65 and 67 but unaffected by lysine at position 66 (Fig. 4 B). The doubly deamidated peptide was chosen for these experiments because initial T cell experiments indicated that this tTG-created variant gave greater responses. Together, these data clearly show that despite overlapping by some seven residues, the α-2(62–75) and α-9(57–68) peptides bind in a different frame to DQ2, with the glutamic acid residue binding in the P4 and P6 pockets, respectively.
The α-9(57–68) and α-2(62–75) Peptides Are Commonly Recognized Gliadin Epitopes.
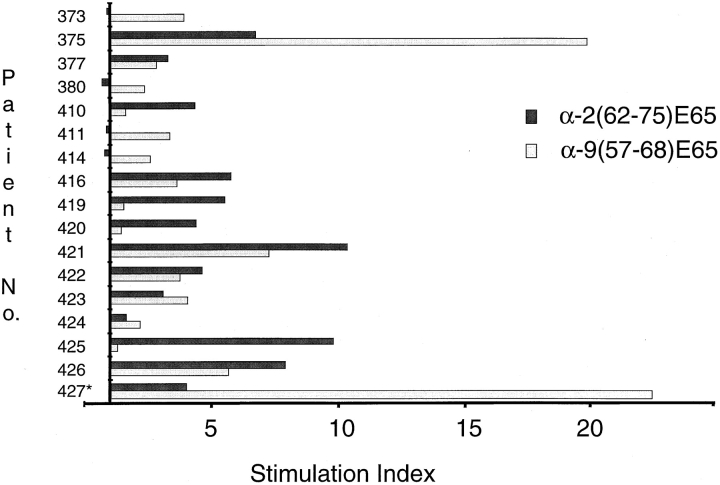
The peptide specificity of seven clones derived from four CD patients could be accounted for by the α-2(62–75)E65 and α-9(57–68)E65 peptides alone, suggesting that these may be dominant gliadin epitopes that are frequently recognized by CD patients. To further assess their importance, we screened a panel of gluten-specific, polyclonal intestinal TCLs established from 17 CD patients for recognition of the α-2(62–75)E65 and α-9(57–68)E65 peptides (Fig. 5). TCLs from 12 of these patients made clear responses to the α-9(57–68)E65 peptide (with a stimulation index ≥2), whereas 11 made responses to the α-2(62–75)E65 peptide. These responses were greater than or comparable with the responses to the pepsin and trypsin–digested gliadin at 1 mg/ml or tTG-treated chymotrypsin gliadin at 500 μg/ml (data not shown). A gliadin-reactive TCL from a DQ8+DQ2− Norwegian patient failed to respond to either peptide. When TCLs and TCCs were taken together, all 19 of the DQ2+ Norwegian patients were shown to have T cell responses to one or both of the α-2(62–75)E65 and α-9(57–68)E65 peptides (Table and Fig. 5). Recognition of tTG-treated α-2(62–75) and α-9(57–68) peptides was also analyzed using intestinal TCLs isolated from five Dutch children and a single Dutch adult (data not shown). Surprisingly, TCLs from only two patients responded to the peptides; the TCL from one child that was restimulated with tTG-treated, pepsin and trypsin–digested gliadin responded to both of the tTG-treated peptides, whereas the adult-derived TCL responded to the α-2(62–75) peptide after tTG treatment. There were no responses detected in the Dutch TCLs to the non–tTG-treated α-2(62–75) and α-9(57–68) peptides.
Figure 5.
Recognition of peptides α-9(57–68)E65 and α-2(62–75)E65 by polyclonal TCLs. 17 TCLs were tested for recognition of the synthetic α-9(57–68)E65 and α-2(62–75)E65 peptides at 5 μM. The y-axis crosses the x-axis at a stimulation index of 1, i.e., where no effect on T cell proliferation is observed.
The α-9(57–68) and α-2(62–75) Peptides Are the Immunodominant Intestinal T Cell Epitopes in α-Gliadin.
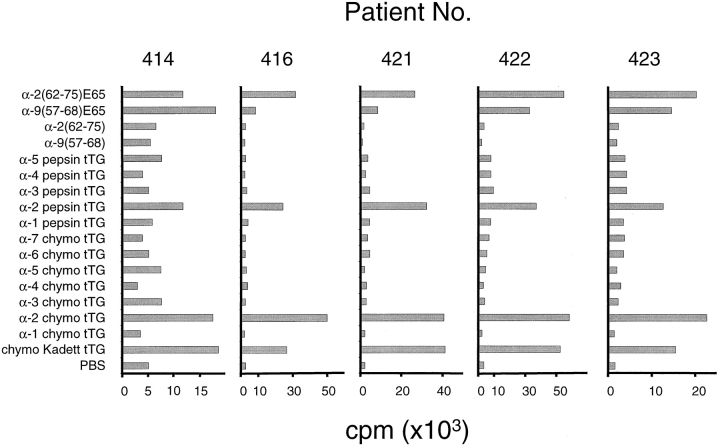
The α-9(57–68) and α-2(62–75) peptides are clearly major epitopes within α-gliadin. To investigate if other α-gliadin epitopes are present within the α-recombinants, we tested eight polyclonal gluten-specific TCLs isolated from DQ2+ adult CD patients for recognition of a panel of the α-gliadin recombinants. The recombinants were digested with either chymotrypsin or pepsin before treatment with tTG and addition to the proliferation assay. As expected, all of the TCLs recognized one or both of the α-9(57–68)E65 and α-2(62–75)E65 peptides and the α-2 recombinant that contains them (Fig. 6). In contrast, recombinants that displayed sequence variation within both of these epitopes failed to stimulate any of the TCLs tested. Furthermore, TCLs from six patients failed to recognize any of a series of native and tTG-treated overlapping peptides corresponding to residues 1–58 of A-gliadin (data not shown). Together, these data suggest that the α-9(57–68) and α-2(62–75) peptides are the only common α-gliadin epitopes recognized by T cells from the adult Norwegian CD population.
Figure 6.
Recognition of α-gliadin recombinants by a panel of polyclonal TCLs. Stimulation of polyclonal TCLs by α-9 (57–68) and α-2(62–75) peptides or tTG-treated α-recombinants that have been digested with pepsin or chymotrypsin. Peptides were tested at 5 μM. Recombinant α-gliadins and gliadin from the wheat variety Kadett were tested at 50 μg/ml. Data is shown for five representative TCLs out of eight that were tested.
Discussion
A molecular explanation for disease pathology and HLA association in CD has been difficult to address due to the lack of defined gliadin T cell epitopes. We therefore tested a panel of α-gliadin–recombinant antigens to characterize the T cell response to α-gliadin using gluten-specific TCCs and lines grown from intestinal biopsies of CD patients. Two distinct but overlapping epitopes have been identified that share a common seven–amino acid motif. Deamidation of the same glutamine residue situated within this 7-mer motif, a modification specifically performed by human tTG, is critical for the T cell recognition of these peptides. The two α-9(57–68)E65 and α-2(62–75)E65 peptides appear to be the dominant epitopes within the α-gliadin family, and between them they are recognized by most if not all adult CD patients. It is intriguing that the α-gliadin T cell response is focused on a single repetitive fragment of gliadin and that this is also targeted by tTG. It is possible that the specificity of tTG plays a more central role in the selection of gliadin T cell epitopes in the gut than has been appreciated.
In vivo and in vitro studies have identified α-gliadins as a major source of toxicity for CD patients 26 27 28 29. Our study confirms that these proteins are also a major focus of the intestinal T cell response to gluten. Notably, they do not correspond with the A-gliadin peptides previously identified as active in CD 22 30 31 32. The failure of the polyclonal intestinal TCLs to recognize either the native or tTG-treated α-gliadins lacking the α-9(57–68) and α-2 (62–75) sequence or native or tTG-treated A-gliadin peptides indicates that these patients do not make significant responses toward other α-gliadin peptides.
The disparity in recognition for the α-9(57–68) and α-2(62–75) peptides between the TCLs isolated in Norway and The Netherlands may well relate to differences in the restimulation strategy and antigen used for generating them. The Norwegian biopsies were challenged by immersion overnight in enzyme-digested gliadin in an organ culture system 3. We have recently found that under these conditions, the gliadin is deamidated in situ by endogenous tTG (Molberg, Ø., S.N. McAdam, K.E.A. Lundin, C. Kristiansen, H. Arentz-Hansen, K. Kett, and L.M. Sollid, manuscript in preparation). In contrast, the Dutch pediatric biopsies were first disrupted with dithiothreitol and EDTA and then cultured in a 96-well plate containing a relatively low concentration of pepsin and trypsin–digested gliadin. These cultures were then restimulated two to three times with irradiated autologous PBMCs that had been prepulsed with pepsin and trypsin–digested gliadin. Notably, this antigen failed to stimulate the Dutch peptide-specific TCLs unless it had been pre-treated with tTG, indicating that it was only minimally deamidated. Furthermore, the single pediatric line that responded to the peptides was the only Dutch TCL that had been restimulated with tTG-treated, pepsin and trypsin–digested gliadin. Thus, the protocol used to generate the TCLs from the pediatric biopsies should not efficiently expand T cells specific for the α-9 (57–68)E65 and α-2(62–75)E65 peptides. However, it is possible that there are also differences in gliadin specificity in the T cells with adult and pediatric patients. Ongoing work will investigate this possibility.
It is currently unclear how oral tolerance is established in humans. It is conceivable, however, that tTG-mediated deamidation plays a central role in the loss of oral tolerance to gliadin in CD. Deamidation is critical and necessary for the intestinal T cell recognition of the α-9(57–68) and α-2(62–75) peptides and of a DQ2-restricted γ-gliadin epitope 13. Enzymatic deamidation of these three peptides converts them from epitopes that bind weakly but significantly to DQ2 to epitopes with a reasonable but by no means exceptional affinity. It is notable that the γ-gliadin epitope shares no unusual sequence motif with the α-9(57–68) and α-2(62–75) peptides and that the modified glutamine residue for each of these epitopes occupies a different pocket within DQ2 33. This suggests that rather than recognition of a single “pathogenic motif,” it is the altered affinity of deamidated gliadin peptides for DQ2 that may be a critical factor involved in loss of tolerance. Concurrent with the increase in affinity for DQ2 induced by deamidation is a change in conformation of the gliadin–DQ2 complex. This is indicated by the failure of the TCCs to recognize the unmodified peptides or peptides containing glutamic acid residues at anchor residues not targeted by tTG but with affinity for DQ2 similar to that of the E65 peptides. Interestingly, the simple modification of glutamine residues that act as major TCR contact residues does not appear to be sufficient to break tolerance, as none of the modified glutamines are found at this position. This indicates that changing the DQ2/peptide conformation alone does not lead to a loss of tolerance and emphasizes the requirement for a change in affinity of the gliadin peptide with the DQ2 molecule.
A major role of tTG in vivo is the extracellular cross-linking of glutamine residues with primary amines via isopeptide bonds 34. Deamidation of glutamines is favored over their incorporation into isopeptide bonds when lysine/polyamine concentrations are low. The unusual ability of gliadins to act as excellent amine acceptor substrates for tTG 34 35 could lead to a local depletion in the concentration of lysine/polyamine and create microenvironments where deamidation of gliadin is likely. Gliadin fragments containing two glutamine residues targeted by tTG may well be deamidated and cross-linked to other lysine-containing proteins. Conditions may exist in the gut where T cell epitopes are both created and trapped locally by tTG, prohibiting their presentation by “tolerogenic” APCs in the gut. Alternatively, it may prevent these epitopes from spreading systemically as soluble antigen, a factor shown to be important in oral tolerance 36. An implication of this would be that administration of soluble deamidated gliadin peptides to CD patients should induce tolerance to gliadin. This therapeutic approach becomes practical with the identification of this immunodominant fragment and as it becomes apparent that fewer gliadin epitopes exist as was initially predicted 37.
If the gliadin fragments were cross-linked to tTG itself, a phenomenon known to occur in vitro, then gliadin-specific T cells may provide “help” for tTG-specific B cells. We have proposed that this mechanism may explain the intriguing observation that anti-tTG autoantibody production is dependent on gliadin exposure 38. It may therefore be significant that the motifs targeted by tTG and shared in the α-9(57–68) and α-2(62–75) epitopes is repeated in many α-gliadins.
An unusual feature of the α-9(57–68) and α-2(62–75) peptides is the presence of multiple proline residues at relative positions P1, P3, P6, and P8 and P1, P3, P5, and P8, respectively. These residues may serve to protect from proteolytic cleavage during luminal digestion and/or class II processing. It is unusual for natural class II ligands to have more than one proline residue within the core binding region 39. Moreover, class II peptide binding studies have rarely shown that proline residues have a positive influence on binding 40 41 42 43. It is tempting to speculate that DQ2 and DQ8 may be unique in their ability to accommodate peptides with repetitive proline residues and have a strong preference for binding peptides with negatively charged residues. Therefore, the basis of the HLA association in CD may be founded on the unique peptide binding properties of these two molecules.
Here we have demonstrated that intestinal T cell recognition of an immunodominant fragment of α-gliadin is dependent on the enzymatic modification of a single glutamine residue. These findings provide a new molecular perspective to analyze the mechanisms leading to wheat intolerance in CD that should yield new insights into the mechanisms of oral tolerance in humans.
Acknowledgments
We thank Christel Kristiansen, Tore Jensen, Eva Boretti, and Liv Mangchau for excellent technical assistance and Prof. Greenberg and Dr. Lai (Duke University), who generously provided reagents and advice for the production of human tTG. We also thank Profs. Sjöstrom and Norén for supplying purified natural gliadins, the patients and staff at gastroenterology units at Ullevål Hospital and the Rikshospitalet who kindly provided biopsy material, and Prof. Erik Thorsby for continued support.
This work is funded by research grants from the Research Council of Norway and the European Commission (BMH4-CT98-3087).
Footnotes
Abbreviations used in this paper: CD, celiac disease; TCC, T cell clone; TCL, T cell line; tTG, tissue transglutaminase.
References
- Sollid L.M., Markussen G., Ek J., Gjerde H., Vartdal F., Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ α/β heterodimer. J. Exp. Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid L.M., Thorsby E. HLA susceptibility genes in celiac diseasegenetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910–952. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- Lundin K.E.A., Scott H., Hansen T., Paulsen G., Halstensen T.S., Fausa O., Thorsby E., Sollid L.M. Gliadin-specific, HLA-DQ(α1*0501,β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 1993;178:187–196. doi: 10.1084/jem.178.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molberg Ø., Kett K., Scott H., Thorsby E., Sollid L.M., Lundin K.E.A. Gliadin-specific HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scand. J. Immunol. 1997;46:103–108. [PubMed] [Google Scholar]
- Nilsen E.M., Lundin K.E.A., Krajci P., Scott H., Sollid L.M., Brandtzaeg P. Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon-γ. Gut. 1995;37:766–776. doi: 10.1136/gut.37.6.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemioslo R.T., Lundin K.E.A., Sollid L.M., Nelufer J., Ciclitira P.J. Histological changes in small bowel mucosa induced by gliadin sensitive T lymphocytes can be blocked by anti-interferon gamma antibody. Gut. 1995;36:874–879. doi: 10.1136/gut.36.6.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin K.E.A., Scott H., Fausa O., Thorsby E., Sollid L.M. T cells from the small intestinal mucosa of a DR4, DQ7/DQ4, DQ8 celiac disease patient preferentially recognize gliadin when presented by DQ8. Hum. Immunology. 1994;41:285–291. doi: 10.1016/0198-8859(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Johansen B.H., Vartdal F., Eriksen J.A., Thorsby E., Sollid L.M. Identification of a putative motif for binding of peptides to HLA-DQ2. Int. Immunol. 1996;8:177–182. doi: 10.1093/intimm/8.2.177. [DOI] [PubMed] [Google Scholar]
- van de Wal Y., Kooy Y.C., Drijfhout J.W., Amons R., Koning F. Peptide binding characteristics of the coeliac disease-associated DQ(α1*0501, β1*0201) molecule. Immunogenetics. 1996;44:246–253. doi: 10.1007/BF02602553. [DOI] [PubMed] [Google Scholar]
- Vartdal F., Johansen B.H., Friede T., Thorpe C.J., Stevanovic S., Eriksen J.E., Sletten K., Thorsby E., Rammensee H.G., Sollid L.M. The peptide binding motif of the disease associated HLA-DQ (α1*0501, β1*0201) molecule. Eur. J. Immunol. 1996;26:2764–2772. doi: 10.1002/eji.1830261132. [DOI] [PubMed] [Google Scholar]
- Godkin A., Friede T., Davenport M., Stevanovic S., Willis A., Jewell D., Hill A., Rammensee H.G. Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin-dependent diabetes susceptibility allele HLA-DQ8 (DQ 3.2) Int. Immunol. 1997;9:905–911. doi: 10.1093/intimm/9.6.905. [DOI] [PubMed] [Google Scholar]
- Kwok W.W., Domeier M.L., Raymond F.C., Byers P., Nepom G.T. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J. Immunol. 1996;156:2171–2177. [PubMed] [Google Scholar]
- Sjöström H., Lundin K.E.A., Molberg Ø., Körner R., McAdam S.N., Anthonsen D., Quarsten H., Norén O., Roepstorff P., Thorsby E. Identification of a gliadin T-cell epitope in coeliac diseasegeneral importance of gliadin deamidation for intestinal T-cell recognition. Scand. J. Immunol. 1998;48:111–115. doi: 10.1046/j.1365-3083.1998.00397.x. [DOI] [PubMed] [Google Scholar]
- Molberg Ø., McAdam S.N., Körner R., Quarsten H., Kristiansen C., Madsen L., Fugger L., Scott H., Norén O., Roepstorff P. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998;4:713–717. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- van de Wal Y., Kooy Y., van Veelen P., Pena S., Mearin L., Papadopoulos G., Koning F. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 1998;161:1585–1588. [PubMed] [Google Scholar]
- Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E.O., Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- van de Wal Y., Kooy Y.M., van Veelen P.A., Pena S.A., Mearin L.M., Molberg Ø., Lundin K.E.A., Sollid L.M., Mutis T., Benckhuijsen W.E. Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc. Natl. Acad. Sci. USA. 1998;95:10050–10054. doi: 10.1073/pnas.95.17.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P.R., Tatham A.S., Kasarda D.D. Cereal proteins and coeliac disease. In: Marsh M.N., editor. Coeliac Disease. Blackwell Scientific Publications; Oxford, UK: 1992. pp. 305–348. [Google Scholar]
- Sjöström H., Friis S.U., Norén O., Anthonsen D. Purification and characterisation of antigenic gliadins in coeliac disease. Clin. Chim. Acta. 1992;207:227–237. doi: 10.1016/0009-8981(92)90121-6. [DOI] [PubMed] [Google Scholar]
- Arentz-Hansen E.H., McAdam S.N., Molberg Ø., Kristiansen C., Sollid L.M. Production of a panel of recombinant gliadins for the characterisation of T cell reactivity in coeliac disease. Gut. 2000;46:46–51. doi: 10.1136/gut.46.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempst P., Link A.J., Riviere L.R., Fleming M., Elicone C. Internal sequence analysis of proteins separated on polyacrylamide gels at the submicrogram levelimproved methods, applications and gene cloning strategies. Electrophoresis. 1990;11:537–553. doi: 10.1002/elps.1150110704. [DOI] [PubMed] [Google Scholar]
- Gjertsen H.A., Lundin K.E.A., Sollid L.M., Eriksen J.A., Thorsby E. T cells recognize a peptide derived from α-gliadin presented by the celiac disease-associated HLA-DQ (α1*0501,β1*0201) heterodimer. Hum. Immunol. 1994;39:243–252. doi: 10.1016/0198-8859(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Lai T.S., Slaughter T.F., Koropchak C.M., Haroon Z.A., Greenberg C.S. C-terminal deletion of human tissue transglutaminase enhances magnesium-dependent GTP/ATPase activity. J. Biol. Chem. 1996;271:31191–31195. doi: 10.1074/jbc.271.49.31191. [DOI] [PubMed] [Google Scholar]
- Fenn J.B., Mann M., Meng C.K., Wong S.F., Whitehouse C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Hunt D.F., Yates J.R., Shabanowitz J., Winston S., Hauer C.R. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchuk Z.M., Nelson D.L., Katz A.J., Bernardin J.E., Kasarda D.D., Hague N.E., Strober W. Gluten-sensitive enteropathy. Influence of histocompatibility type on gluten sensitivity in vitro. J. Clin. Invest. 1980;66:227–233. doi: 10.1172/JCI109848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekkens W., Haex A.J.C., Willighagen R.G.J. Some aspects of gliadin fractionation and testing by a histochemical method. In: Booth C.C., Dowling R.H., editors. Coeliac Disease. Churchill Livingstone; London: 1970. pp. 11–19. [Google Scholar]
- Hekkens W., Van der Aarsen C.J., Gilliams J.P., Lems-Van Kan P., Bouma-Frølich G. α-gliadin structure and degradation. In: Hekkens W., Pena A.S., editors. Coeliac Disease. H.E. Stenfert Kroese B.v; Leiden, The Netherlands: 1974. pp. 39–45. [Google Scholar]
- Kendall M.J., Schneider R., Cox P.S., Hawkins C.F. Gluten subfractions in coeliac disease. Lancet. 1972;2:1065–1067. doi: 10.1016/s0140-6736(72)92344-6. [DOI] [PubMed] [Google Scholar]
- de Ritis G., Auricchio S., Jones H.W., Lew E.J., Bernardin J.E., Kasarda D.D. In vitro (organ culture) studies of the toxicity of specific A-gliadin peptides in celiac disease. Gastroenterology. 1988;94:41–49. doi: 10.1016/0016-5085(88)90607-5. [DOI] [PubMed] [Google Scholar]
- Sturgess R., Day P., Ellis H.J., Lundin K.E., Gjertsen H.A., Kontakou M., Ciclitira P.J. Wheat peptide challenge in coeliac disease. Lancet. 1994;343:758–761. doi: 10.1016/s0140-6736(94)91837-6. [DOI] [PubMed] [Google Scholar]
- Wieser H., Belitz H.D., Idar D., Ashkenazi A. Coeliac activity of the gliadin peptides CT-1 and CT-2. Z. Lebensm. Unters. Forsch. 1986;182:115–117. doi: 10.1007/BF01454241. [DOI] [PubMed] [Google Scholar]
- Quarsten H., Molberg Ø., Fugger L., McAdam S.N., Sollid L.M. HLA binding and T cell recognition of a tissue transglutaminase modified gliadin epitope. Eur. J. Immunol. 1999;29:2506–2514. doi: 10.1002/(SICI)1521-4141(199908)29:08<2506::AID-IMMU2506>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Folk J.E. Mechanism and basis for specificity of transglutaminase-catalyzed ε-(γ-glutamyl) lysine bond formation. Adv. Enzymol. Relat. Areas Mol. Biol. 1983;54:1–56. doi: 10.1002/9780470122990.ch1. [DOI] [PubMed] [Google Scholar]
- Larre C., Chiarello M., Blanloeil Y., Chenu M., Gueguen J. Gliadin modifications catalysed by guinea pig liver transglutaminase. J. Food Biochem. 1993;17:267–282. [Google Scholar]
- Gütgemann I., Fahrer A.M., Altman J.D., Davis M.M., Chien Y.H. Induction of rapid T cell activation and tolerance by systemic presentation of an orally administered antigen. Immunity. 1998;8:667–673. doi: 10.1016/s1074-7613(00)80571-3. [DOI] [PubMed] [Google Scholar]
- Lundin K.E.A., Sollid L.M., Anthonsen D., Norén O., Molberg Ø., Thorsby E., Sjöström H. Heterogeneous reactivity patterns of HLA-DQ-restricted, small intestinal T-cell clones from patients with celiac disease. Gastroenterology. 1997;112:752–759. doi: 10.1053/gast.1997.v112.pm9041236. [DOI] [PubMed] [Google Scholar]
- Sollid L.M., Molberg Ø., McAdam S., Lundin K.E.A. Autoantibodies in coeliac disease. Tissue transglutaminase—guilt by association. Gut. 1997;41:851–852. doi: 10.1136/gut.41.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rotzschke O., Stevanovic S., Jung G., Rammensee H.G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- Jardetzky T.S., Gorga J.C., Busch R., Rothbard J., Strominger J.L., Wiley D.C. Peptide binding to HLA-DR1a peptide with most residues substituted to alanine retains MHC binding. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D., Arrhenius T., Sidney J., Del Guercio M.F., Albertson M., Wall M., Oseroff C., Southwood S., Colon S.M., Gaeta F.C. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J. Immunol. 1991;147:2663–2669. [PubMed] [Google Scholar]
- Hammer J., Valsasnini P., Tolba K., Bolin D., Higelin J., Takacs B., Sinigaglia F. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74:197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- Sette A., Sidney J., Oseroff C., Del Guercio M.F., Southwood S., Arrhenius T., Powell M.F., Colon S.M., Gaeta F.C., Grey H.M. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J. Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]