Identification of Tyrosinase-related Protein 2 as a Tumor Rejection Antigen for the B16 Melanoma (original) (raw)
Abstract
Recently, major advances have been made in the identification of antigens from human melanoma which are recognized by T cells. In spite of this, little is known about the optimal ways to use these antigens to treat patients with cancer. Progress in this area is likely to require accurate preclinical animal models, but the availability of such models has lagged behind developments in human tumor immunology. Whereas many of the identified human melanoma antigens are normal tissue differentiation proteins, analogous murine tumor antigens have not yet been identified. In this paper we identify a normal tissue differentiation antigen, tyrosinaserelated protein 2 (TRP-2), expressed by the murine B16 melanoma which was found by screening a cDNA library from B16 with tumor-reactive cytotoxic T lymphocytes (CTL). A peptide conforming to the predicted MHC class I H2-Kb binding motif, TRP-2181-188, was identified as the major reactive epitope within TRP-2 recognized by these anti-B16 CTLs. By site-directed mutagenesis, it was shown that alteration of this epitope eliminated recognition of TRP-2. It was further demonstrated that a CTL line raised from splenocytes by repeated stimulation in vitro with this peptide could recognize B16 tumor and was therapeutic against 3-d-old established pulmonary metastases. The use of TRP-2 in a preclinical model of tumor immunotherapy may be helpful in suggesting optimal vaccination strategies for cancer therapy in patients.
Anumber of studies have suggested that T cell responses to tumor-associated antigens can be of therapeutic benefit both in animals and in patients with cancer. Clinical trials demonstrated that 35% of patients with melanoma treated with specific, tumor-reactive, tumor-infiltrating lymphocytes and IL-2 achieved either partial or complete tumor regression (1). A class of nonmutated, melanocyte lineage normal differentiation proteins including tyrosinase (2, 3), tyrosinase-related protein (TRP)1-1 (4), melanoma antigen reactive with T cells (MART)-1 (5), and gp100 (6) proved to be widely recognized in an MHC class I–restricted fashion by tumor-reactive T cells from different patients. Current protocols are directed at using these widely expressed antigens in vaccination trials for patients with melanoma, although it has been difficult to rationally choose clinical therapeutic strategies because of the growing list of antigens and the multitude of immunizing vectors, adjuvants, and supportive cytokines which have been elucidated.
Accurate animal models may be valuable in better designing vaccination strategies. Existing models are limited by a relatively small number of known antigens, few of which are analogous to known antigens in human cancers. Murine tumor–associated antigens have been shown to include mutated proteins such as connexin 37 from the Lewis lung carcinoma, and proteins expressed only in tumor and testis or placenta such as P1A from the P815 mastocytoma. Although similar antigens have been found in human tumors, these antigens are either not shared (i.e., are unique to one patient), or are poorly immunogenic.
To better understand the immune response to cancer and to design immunotherapeutic strategies, efforts are underway to clone antigens from murine tumors which are recognized by T cells. In this paper, we report that T cells reactive with the B16 murine melanoma recognize a nonmutated product of the TRP-2 gene. This murine melanoma–associated antigen may be useful in designing vaccine and combination therapies to treat patients with melanoma.
Materials and Methods
Cell Lines.
The murine melanoma B16 is a spontaneously arising melanoma of C57BL/6 mice propagated by Dr. I.J. Fidler (M.D. Anderson Cancer Center, Houston, TX), which was cultured in complete media (CM) consisting of RPMI 1640 medium (Biofluids, Inc., Rockville, MD) supplemented with the following: 10% heat-inactivated fetal calf serum (Biofluids, Inc.), 0.1 mM nonessential amino acids and 1 mM sodium pyruvate (Biofluids, Inc.), 5 × 10−5 M 2-mercaptoethanol (GIBCO BRL, Gaithersburg, MD), 0.3% glutamine (National Institutes of Health Media Unit, Bethesda, MD), 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamicin, and 0.5 μg/ml fungizone. A B16 tumor line expressing the murine co-stimulatory molecule B7.1 was generated in our laboratory using a Moloney leukemia virusbased retroviral vector (LXSN) containing murine B7.1 (CD80). The B7.1 cDNA used was a gift from Dr. R. Germain (National Institute of Allergy and Infectious Diseases, Bethesda, MD). The transformed human embryonic kidney line 293 was purchased from the American Type Culture Collection (Rockville, MD), and grown in DMEM medium containing 5% heat-inactivated fetal calf serum, 0.3% glutamine, 0.01 M Hepes (National Institutes of Health Media Unit), 100 U/ml penicillin, and 100 μg/ml streptomycin. The murine thymoma EL-4 (and a variant transfected with the β-galactosidase [gal] gene, E22), and the murine colon carcinoma MC38, were grown in CM. The immortalized melanocyte line melan-A was a gift of Dr. V. Hearing (National Cancer Institute, Bethesda, MD), and was grown in Bennett's medium, pH 6.9, which consisted of MEM (GIBCO BRL), 5% heat-inactivated fetal calf serum, 1 mM sodium pyruvate, 25 mM sodium bicarbonate, 100 μM 2-mercaptoethanol, 200 nM PMA (Sigma Chemical Co., St. Louis, MO), 50 U/ml penicillin, and 50 μg/ml streptomycin.
Generation of Tumor-reactive T Cell Lines.
Female C57BL/6 mice were obtained from Charles River Laboratories (Raleigh, NC) and used when 6–12 wk old. Tumor-reactive T cell lines were generated as follows. Mice were immunized with 1 × 106 irradiated (20,000 cGy) B16 tumor cells and 50 μg Corynebacterium parvum. 2 wk later, spleens were harvested and crushed with the blunt end of a syringe in the presence of ACK lysis buffer. The suspension was passed through 100-gauge nylon mesh (Nitex), washed twice, and plated in 24-well plates at 4 × 106 per well with 2 × 105 irradiated B16 cells expressing murine B7.1 (B16·B7). Recombinant human IL-2 (Chiron Corp., Emeryville, CA) at 30 IU/ml was added after 2 d, and cultures were restimulated weekly with B16·B7 and refed every 3 d with medium containing IL-2. Several lines reactive with B16 were obtained in this manner, and one was selected for use based on its degree of reactivity and proliferation (designated line A).
Generation of Peptide-reactive T Cell Lines.
Splenocytes from normal mice or mice vaccinated with irradiated B16 and C. parvum were obtained as described above and cultured in CM with 5 μg/ml of purified peptide at a density of 4 × 106 per well in 24-well plates. On the second day of culture, IL-2 was added to a final concentration of 30 IU/ml. After 8–10 d of culture, nonviable cells were removed with a discontinuous lympholyte-M gradient (Accurate Chemical and Scientific Corp., Westbury, NY), and lymphocytes were washed and replaced in fresh CM with 30 IU/ ml of IL-2 at 8 × 105 cells/well. Stimulator cells were then generated by incubating 5 × 107 fresh splenocytes in CM with 5 μg/ml of peptide for 90 min, washing with HBSS (Biofluids Inc.), and resuspending in CM. After 2,000 cGy irradiation, 3 × 106 of these irradiated, peptide-pulsed splenocytes were added to each well of the culture to be stimulated. IL-2 and CM were partially replaced every 2–3 d, and repeated antigen stimulation was performed every 7–10 d. Cultures were typically tested and used after at least one in vitro restimulation.
Lymphocyte Adoptive Transfer Treatment Model.
Recipient C57BL/6 mice were first given 500 cGy whole body irradiation to suppress any endogenous responses to B16 or IL-2 and then were administered 3 × 105 B16 cells in HBSS by tail vein injection on day 0. On day three, 1–2 × 106 of the CTLs being evaluated for efficacy were administered intravenously and 6 × 104 IU IL-2 was administered intraperitoneally three times per day for 3 d. On days 13–17, mice were killed and the number of metastases were evaluated in a coded, blinded fashion. Differences in the numbers of metastases were analyzed by the Kruskal-Wallis test and all P values are two-tailed.
cDNA Expression Cloning and Screening.
Total RNA from B16 cells was extracted using the RNAzol B isolation system (Tel-Test, Friendswood, TX). mRNA was isolated using the Promega PolyATract mRNA isolation system (Promega Corp., Madison WI). Double-stranded cDNA was made using the NOVAGEN random primer directional cDNA library kit (Novagen, Inc., Madison, WI). After the addition of linkers, the cDNA was digested with EcoRI and HindIII to ensure proper unidirectional sense orientation during ligation. cDNA was size-selected at >800 bp on a 1.0% agarose gel before ligation. The cDNA was recovered using a Glass Select DNA Isolation Kit (5 Prime–3 Prime Inc., Boulder, CO), and cloned into the pcDNA3 vector (Invitrogen, San Diego, CA).
cDNA was electroporated into ElectroMAX DH10B Escherichia coli (GIBCO BRL) and grown in selection media for 48 h. Plasmid DNA was purified using the Wizard Series 9600 System (Promega Corp.) at a density of 25 clones/well in 96-well plates. 200 ng of each DNA pool and 2 μg lipofectamine (GIBCO BRL) were added to 4 × 104 293 cells previously transfected and selected to stably express the murine class I MHC molecules H2-Kb and H2-Db (designated 293KbDb) in a 96-well plate with 150 μl of transfection media consisting of DMEM with 10 mM Hepes and 0.3% glutamine. After 2 h, an equal volume of transfection media + 10% fetal calf serum was added to the lipid– DNA mixture. After overnight incubation, the media was removed and 4 × 104 line A cells were added to these 293KbDb cells in 250 μl of CM. After 24 h, 100 μl of supernatant was collected and the amount of mIFN-γ was measured in a standard ELISA (Endogen, Inc., Woburn, MA) in which 1 U mIFN-γ = 100 pg/ml. Individual cDNA clones were isolated from pools repeatedly showing reactivity above background. The cDNA inserts of the reactive clones were sequenced on an automated sequencer (ABI Prizm 310; Perkin-Elmer Corp., Foster City, CA). Sequences were compared to the Gen-Bank database using the BLAST search engine (7).
Northern Blot Analysis.
Total cellular RNA was isolated using RNAzol (Tel-Test). 20 μg of RNA were electrophoresed on a 0.6% formaldehyde-agarose gel and transferred to a nitrocellulose membrane. Radioactive probes were prepared using the Prime-It RmT Random Primer Kit (Stratagene, La Jolla, CA). After hybridization, membranes were washed at high stringency for 20 min at 65°C in 0.1× SSC + 0.1% SDS buffer.
Peptide Synthesis and Testing.
Peptides were synthesized on a multiple peptide synthesizer (Gilson AMS 422; Gilson Co., Inc., Worthington, OH) using standard F-moc chemistry. Larger quantities of selected peptides were obtained from Peptide Technologies Corporation (Gaithersburg, MD) and HPLC-purified to >98% purity. For testing, peptides were added at 10 μg/ml to stimulator cells and incubated for 90 min at room temperature. They were then added to effector cells and IFN-γ release measured.
Reverse Transcription–PCR for TRP-2.
Approximately 107 cultured cells were lysed with TRIzol Reagent (GIBCO BRL). Total RNA was collected, and first strand cDNA synthesis was performed with the SuperScript Preamplification System (GIBCO BRL) using oligo(dT)12-18 primers. PCR was performed using an optimally matched primer pair (5′-CAGAACTCAGGAGTGGAAGA-3′) and (5′-CAAGATGAGCGACGGTGTAA-3′) which generated a 1,691-bp fragment containing the entire protein coding region of murine TRP-2.
Site-directed Mutagenesis.
A Chameleon Double Stranded SiteDirected Mutagenesis Kit (Stratagene) was used to alter the nucleotide sequence for the amino acids at positions 5 and/or 8 of the TRP-2181-188 epitope VYDFFVWL of TRP-2. Two mutated TRP-2 clones were made, one used the primer (5′-GCAGCGTGTATGACTTTGATGTGTGGCTCC-3′) to change phenylalanine at position 5 to aspartic acid, and the other used the primer (5′-GCGTGTATGACTTTGATGTGTGGCGCCATTATTATTC-3′) to change position 5 as above and to also change the leucine at position 8 to tyrosine. Mutant clones were identified using restriction enzyme digestions with EcoRI and ScaI and confirmed by sequencing.
Results
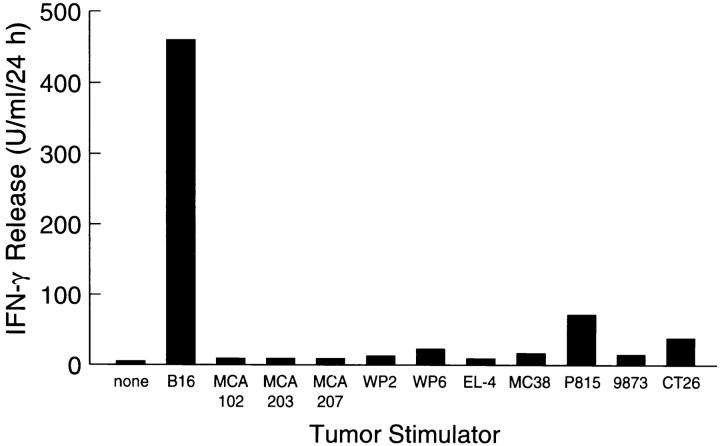
Several B16-reactive lines were obtained from the splenocytes of mice immunized with irradiated B16 tumor and cocultured with B16·B7 and IL-2. One line was selected based on its reactivity and in vitro expansion potential. This bulk lymphocyte culture (line A) showed preferential recognition of B16 tumor in vitro as measured by IFN-γ release (Fig. 1), and was specifically lytic for B16 (45% lysis in 4 h at an effector/target ratio of 5:1; data not shown). When adoptively transferred into mice bearing 3-d-old pulmonary metastases, line A significantly reduced the number of metastases compared to mice given IL-2 alone (P <0.001) or mice administered both IL-2 and CTL that recognized the β-gal protein (raised in a similar fashion to line A using the β-gal–expressing variant of EL-4, E22) (P <0.01) (Table 1).
Figure 1.
Stimulation of mINF-γ release from line A T cells by various murine tumor lines. 105 line A T cells and 105 tumor cells were coincubated in 2 ml of CM for 24 h. Aliquots of supernatant were assayed for mINF-γ. All targets were MHC type H2b except P815, 9873, and CT26 (H2d). Line A CTLs demonstrated relative specificity for the B16 tumor.
Table 1.
Treatment of 3-d-old Pulmonary Metastases from B16 by Adoptive Transfer of Anti-B16 Line A T Cells
| Cells Transferred | Systemic IL-2 | Metastases per mouse | Mean |
|---|---|---|---|
| None | − | >250, >250, >250, >250, >250 | >250 |
| None | + | 218, >250, >250, >250, >250, >250, >250, | >250 |
| >250, >250,>250, >250, >250, >250, >250 | |||
| Line A | + | 4, 23, 35, 60, 184 | 61* |
| anti–β-gal | + | >250, >250, >250, >250, >250 | >250 |
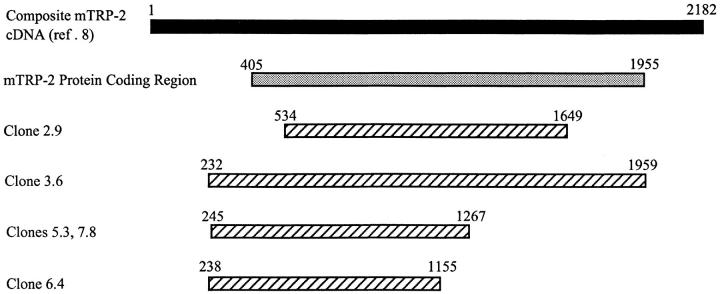
To identify the antigens of B16 which were being recognized by line A, a cDNA library from B16 tumor was screened by transiently transfecting pools of ∼25 cDNA clones into 293KbDb cells and coincubating these cells with line A CTLs. Upon screening ∼1,000 pools, five pools that consistently stimulated mIFN-γ release from line A T cells were identified. Individual clones that stimulated cytokine release from Line A T cells after transfection into target cells were then isolated and sequenced. All five clones were fragments of murine TRP-2. The locations and sizes of the five fragments are compared to the composite cDNA sequence of TRP-2 reported by I. Jackson et al. (8) in Fig. 2. Clone 3.6 contained the full-length protein coding sequence (bp 405–1955). Clone 2.9 represented a truncated fragment that did not contain the normal starting methionine of the TRP-2 protein. The clones 5.3 and 7.8 had identical sequences. A fifth sequence, 6.4, represented a truncated TRP-2 clone. A 622-bp sequence extending from mRNA bp 534 to 1155 was shared by all of the isolates.
Figure 2.
Alignment of TRP-2 inserts found by cDNA expression library screening with anti-B16 CTLs. The top line indicates the composite TRP-2 cDNA sequence described by Jackson et al. (8). The next line (shaded) represents the coding region for the TRP-2 protein. The location of the five TRP-2 cDNA clones identified are shown below (striped).
Northern blot analysis was performed to examine the patterns of tissue expression of TRP-2. A probe made from the clone 2.9 insert hybridized with a 2.2-kb product in the pigmented melanomas B16 (fresh and cultured) B16-F.10, M3, and JB/MS, as well as the pigmented immortalized murine melanocyte line, melan-A. The probe failed to hybridize to the nonpigmented melanoma K1735 as well as the following nonmelanoma tumors: Lewis lung carcinoma, the murine thymoma EL-4, the murine colon cancers MC38 and CT26, the hepatoma line Hepa-1, and the chemically induced sarcomas MCA 205 and 4JK. TRP-2 expression was not seen in normal tissues samples including brain, heart, liver, lung, spleen, and testis. Reverse transcriptase-PCR of mRNA from the normal melanocyte line melan-A followed by sequencing showed it to be identical with clone 3.6 from B16, but both of these sequences differed from the reported sequence of TRP-2 in Gen-Bank between nucleotides 1189 and 1192. The sequence of clone 3.6 (and melan-A) was GCTG, whereas the sequence reported for TRP-2 gene in Gen-Bank was CTGG. Although the reason for this discrepancy has not been resolved, it occurs outside of the 622-bp region shared by the five TRP-2 fragments that stimulated line A.
The MHC class I restriction element used by line A for the recognition of TRP-2 was investigated by transient transfection of TRP-2 into 293 cells expressing either H2-Kb and H2-Db, H2-Kb alone, or the human MHC molecule A2 (293 cells expressing H2-Db alone were not available). IFN-γ release by line A was similar when 293KbDb or 293Kb were transfected with TRP-2, but absent when the control 293-A2 was used (Table 2). These results supported the presence of at least one epitope recognized by line A in the context of H2-Kb.
Table 2.
Determination of MHC Restriction by Transfection of TRP-2 cDNA Clones into 293 Cells Expressing Different MHC Molecules
| IFN-γ Release | |||
|---|---|---|---|
| Transfected cDNA | Transfection targets | ||
| 293KbDb | 293Kb | 293-A2 | |
| U/ml* | |||
| Clone 5.3 | 7.0 | 10.7 | 0.5 |
| Clone 6.4 | 9.3 | 10.5 | 0.5 |
| Clone 7.8 | 8.1 | 7.3 | 0.5 |
| None | 0.6 | 0.6 | 0.5 |
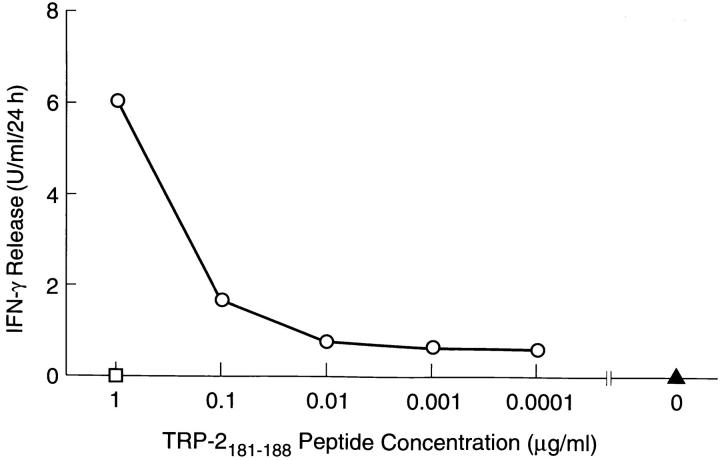
Peptides of eight amino acids from TRP-2 were then synthesized using the H2-Kb binding motif described by Rammensee (9, 10). 15 such peptides were identified in the normal reading frame of TRP-2. MC38 murine tumor cells expressing high levels of H2-Kb were incubated with each of these peptides and tested for their ability to stimulate line A T cells. Only one peptide, VYDFFVWL (TRP2181–188) mediated line A recognition of peptide-pulsed MC38. MC38 incubated with TRP-2181–188 caused a release of 6.0 U/ml/24 h of IFN-γ from line A versus ⩽1.0 U/ml/24 h for the other 15 peptides tested, and 0.6 U/ml/ 24 h for MC38 without peptide. The nucleotides coding for TRP-2181–188 are contained within the 622-bp sequence common to all five of the clones identified by cDNA library screening. Peptide concentrations of 100 ng/ml or higher were required for stimulation of cytokine release from line A T cells (Fig. 3).
Figure 3.
Release of mIFN-γ by line A when incubated with MC-38 (H2b) and varying amounts of TRP-2181–188 peptide (circles). No significant mIFN-γ release was seen when MC-38 was incubated with peptide without line A (square) or with line A without peptide (triangle).
To determine whether TRP-2 represented an immunodominant antigen of B16, 12 independent CTL cultures were generated from mice by using the same methodology used to produce line A, and were tested for their reactivity to TRP-2. Of these 12 cultures, 10 were reactive with TRP-2 by cytokine-release assay, which was best demonstrated by testing specific reactivity to target cells pulsed with the TRP-2181–188 peptide (data not shown). This suggested that not only was TRP-2 a dominant antigen, but that TRP-2181–188 was a dominant epitope of TRP-2 for H2b mice.
To pursue this latter finding, site-directed mutagenesis of the full-length TRP-2 clone 3.6 was used to determine whether other antigenic epitopes existed besides TRP-2181–188. Two separate modifications were made, the first altering both of the anchor residues at positions 5 and 8 of TRP-2181–188, while the second altered only the anchor residue at position 5 (position 8 is shared by a different overlapping peptide with an H2-Kb binding motif). The eight amino acid peptides beginning at residue 181 for those two mutated TRP-2 molecules were thus VYDFDVWL and VYDFDVWR, compared to the native sequence VYDFFVWL. The result of transfecting these two altered TRP-2 genes into 293KbDb is seen in Table 3. The alteration of one or both of the anchor residues abrogated the ability of T cells to recognize transfectants expressing these products. This suggests that recognition of the TRP-2181–188 T cell epitope accounts for all of the reactivity to TRP-2 by line A T cells.
Table 3.
Effect of TRP-2 Mutations within TRP-2181–188 on Recognition by Line A T Cells when Transfected into 293 Cells Expressing Murine MHC Class I Molecules
| IFN-γ Release | |||
|---|---|---|---|
| Transfected cDNA | Transfection targets | ||
| 293KbDb | 293Kb | 293 | |
| U/ml* | |||
| Clone 3.6 | 17.3 | 10.8 | 1.9 |
| Clone 3.6 mut 185 ‡ | 1.0 | 1.2 | 2.0 |
| Clone 3.6 mut 185 +188 § | 1.0 | 1.5 | 2.2 |
| pcDNA3 | 0.7 | 1.3 | 2.0 |
To determine if reactivity to TRP-2181–188 could account for the recognition of B16 by line A, TRP-2–specific reactivity was generated from the spleens of normal or B16vaccinated mice by weekly in vitro stimulation with TRP2181–188 peptide at 5 μg/ml as described in Materials and Methods. After two in vitro peptide stimulations, splenocytes were assayed by IFN-γ release for their ability to recognize peptide and B16 tumor (Table 4). Splenocytes from normal or immunized mice stimulated with the TRP-2181–188 peptide recognized 293KbDb pulsed with this peptide, but not parental 293 pulsed with TRP-2181–188. The same cultures also recognized B16 tumor. Lymphocytes from those same mice incubated in parallel with the H2-Kb–presented ovalbumin-derived peptide SIINFEKL (ova257–264) failed to recognize B16.
Table 4.
Reactivity of Anti–TRP-2181–188 and Anti-ovalbumin275–264 CTLs from Normal- and B16-immunized Mice with H2b-matched tumors and Peptide Pulsed Targets
| IFN-γ release | ||||||||
|---|---|---|---|---|---|---|---|---|
| Lymphocyte culture | Tumor targets | |||||||
| None | B16 | MCA 207 | MC-38 | 293 + ova257–264 | 293 + TPR-2181–188 | 293KbDb + ova257–264 | 293KbDb + TRP-2181–188 | |
| U/ml* | ||||||||
| Normal spleen‡ + ova257–264 | 36 | 92 | 8 | 4 | 35 | 29 | 74 | 42 |
| Immune spleen§ + ova257–264 | 1 | 43 | 50 | 9 | 9 | 14 | 91 | 18 |
| Normal spleen‡ + TRP-2181–188 | 0 | >600 | 20 | 9 | 6 | 3 | 3 | >600 |
| Immune spleen§ + TRP-2181–188 | 5 | >600 | 77 | 28 | 12 | 12 | 11 | >600 |
| Line A T cells | 0 | >600 | 24 | 7 | 0 | 0 | 0 | 134 |
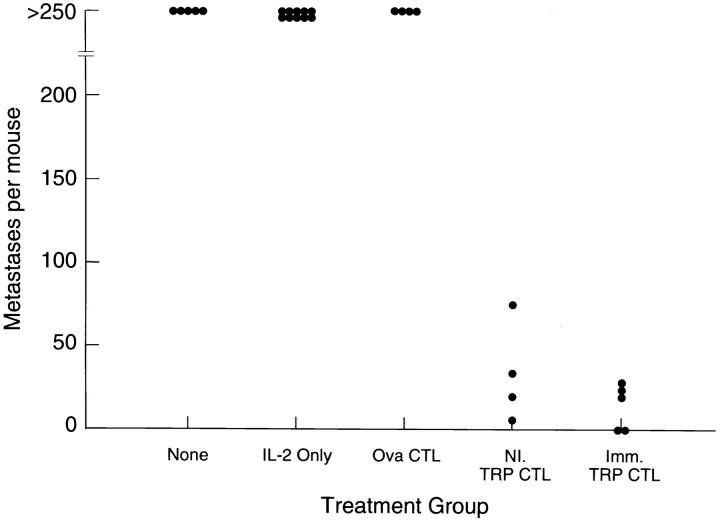
The therapeutic efficacy of CTL lines generated by in vitro stimulation with TRP-2181–188 was also examined. T cell lines generated from normal or B16 immune mice by stimulation with the TRP-2181–188 peptide were used to treat 3-d-old B16 lung metastases by intravenous administration of the CTL in combination with low dose IL-2. A highly significant therapeutic effect was seen (P <0.001) with cultures from normal or B16-immunized mice when stimulated with TRP-2181–188, but not with ova257–264 peptide (when compared to treatment with IL-2 alone) (Fig. 4).
Figure 4.
Treatment of 3-d-old pulmonary metastases from B16 by the intravenous administration of T cells reactive with TRP-2181–188 generated by in vitro peptide stimulation of splenocytes from normal or B16immunized mice. Low-dose systemic IL-2 was given with adoptive cell transfer. Therapeutic effects were seen from anti–TRP-2181–188 CTLs from normal mice (P = 0.0004 vs. IL-2 alone, and P = 0.014 vs. ovalbumin257–264 stimulated cultures), as well as from anti-B16 immunized mice (P = 0.0003 vs. IL-2 alone, and P = 0.010 vs. ovalbumin257–264 stimulated cultures).
Discussion
In the last five years, much progress has been made in the molecular identification of tumor cell proteins which are recognized by the cellular immune system in the context of the MHC (4–6, 11, 12). Work performed largely with patients with melanoma has identified nearly a dozen tumorassociated antigens which can be divided into several general categories. One group of antigens, characterized by the MAGE (13, 14), BAGE (15), and GAGE (16) families of proteins, are coded for by normal nonmutated genes, and seem to be expressed by tumor cell, testis, and sometimes placenta, but not other normal tissues. Another major family of antigens represents normal tissue differentiation proteins because they are expressed in melanoma, normal melanocytes, and pigmented retinal cells, and are typified by the proteins tyrosinase (2, 3), TRP-1 (4), gp100 (6), and MART-1 (5). Some tumors were also found to express a mutated version of a normal cellular protein which forms a “foreign” MHC-binding peptide specifically recognized by T cells. This category of mutated tumor-associated antigen is exemplified by both β-catenin (with a serine to phenylalanine mutation at position 37; reference 11), and the mutated form of cyclin-dependant kinase 4 (17).
Fewer tumor-associated antigens have been identified in murine tumors. Nevertheless, these antigens seem to conform to the categories identified for human tumor–associated antigens. The P1A protein of the P815 mastocytoma is analogous to the MAGE family of proteins in that it is encoded by a nonmutated gene which is expressed only in tumor, testis, and placenta (18–21). The Lewis lung carcinoma expresses a mutated form of the connexin 37 protein which is recognized by CD8+ T cells restricted by H2-Kb (22). In the present paper, the first murine tumor–associated antigen recognized by CD8+ T cells representing a normal tissue differentiation protein has been identified. As was the case with human gp100 and MART-1, a T cell line with known in vivo antitumor activity was used to screen a tumor cDNA library to identify this antigen (5, 6). In the case of mTRP-2, CTLs with specific activity against the TRP-2181–188 peptide were also effective in treating established B16 tumor, supporting the hypothesis that TRP-2 reactive T cells are at least in part responsible for the in vivo antitumor effects seen with the line A T cells.
Several future areas of investigation are generated by these data. After a single in vitro stimulation with the TRP-2181–188 peptide, splenocytes from normal mice, as well as from mice immunized with irradiated B16 tumor, showed reactivity to B16. It is not understood why naive mice have significant numbers of CTL precursors with reactivity to TRP-2. A search of the Gen-Bank database did not identify any other proteins which contained the amino acid sequence VYDFFVWL. Nevertheless, similar peptides from other immunogenic proteins capable of mimicking TRP-2181–188 may be involved in generating reactivity to this “self” protein (23). The peripheral tolerance mechanisms which prevent these precursors from becoming activated in vivo and mediating either B16 rejection or vitiligo are of interest. Overcoming these tolerizing influences may be critical to the future development of clinical immunotherapeutic strategies for patients with melanoma, and potentially for other tumors as well. The preclinical model delineated in this work appears to be well suited for studying a variety of proposed immunization strategies directed at achieving this goal. We are currently developing recombinant viral vectors containing mTRP-2, and are also studying combinations of anti–TRP-2 vaccination, co-stimulation, and systemic cytokines for the treatment of B16 melanoma.
Wang et al. have recently demonstrated that human TRP-2 is recognized as a melanoma-associated antigen by a T cell clone from a population of tumor infiltrating lymphocytes which mediated clinical tumor regression (24). This T cell clone recognized the hTRP-2197–205 peptide of TRP-2 in the context of HLA-A31. This finding supports the relevance of mTRP-2 as a model tumor antigen for preclinical studies, where it may prove useful in the planning of rational immunotherapy strategies for the treatment of patients with cancer.
Acknowledgments
We wish to thank Drs. Maria Parkhurst and Yifan Zhai for their advice and assistance.
Footnotes
M. Bloom is a Howard Hughes Medical Institute–National Institutes of Health Research Scholar.
1 Abbreviations used in this paper: CM, complete media; gal, galactosidase; MART, melanoma antigen reactive with T cells; TRP, tyrosinase-related protein.
References
- 1.Rosenberg SA, Yannelli JR, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, Seipp CA, Einhorn JH, White DE. Treatment of patients with metastatic melanoma with autologous tumorinfiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994;86:1159–1166. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 2.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins PF, El-Gamil M, Kawakami Y, Stevens E, Yannelli JR, Rosenberg SA. Recognition of tyrosinase by tumor-infiltrating lymphocytes from a patient responding to immunotherapy (erratum published 54:3952) Cancer Res. 1994;54:3124–3126. [PubMed] [Google Scholar]
- 4.Wang RF, Robbins PF, Kawakami Y, Kang XQ, Rosenberg SA. Identification of a gene encoding a melanoma tumor antigen recognized by HLA-A31-restricted tumor-infiltrating lymphocytes (erratum published 181:1261) J Exp Med. 1995;181:799–804. doi: 10.1084/jem.181.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Jackson IJ, Chambers DM, Tsukamoto K, Copeland NG, Gilbert DJ, Jenkins NA, Hearing V. A second tyrosinase-related protein, TRP-2, maps to and is mutated at the mouse slaty locus. EMBO (Eur Mol Biol Organ) J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 10.Stevanovic S, Rammensee HG. Identification of T cell epitopes using allele-specific ligand motifs. Behring Inst Mitt. 1994;95:7–13. [PubMed] [Google Scholar]
- 11.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated betacatenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulie PG, Somville M, Lehmann F, Hainaut P, Brasseur F, Devos R, Boon T. Precursor frequency analysis of human cytolytic T lymphocytes directed against autologous melanoma cells. Int J Cancer. 1992;50:289–297. doi: 10.1002/ijc.2910500220. [DOI] [PubMed] [Google Scholar]
- 13.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science (Wash DC) 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 14.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethe B, Brasseur F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boel P, Wildmann C, Sensi ML, Brasseur R, Renauld JC, Coulie P, Boon T, van der Bruggen P. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 16.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde KH, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science (Wash DC) 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 18.Amar-Costesec A, Godelaine D, Van den Eynde B, Beaufay H. Identification and characterization of the tumor-specific P1A gene product. Biol Cell. 1994;81:195–203. doi: 10.1016/0248-4900(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 19.Van den Eynde B, Lethe B, Van Pel A, De Plaen E, Boon T. The gene coding for a major tumor rejection antigen of tumor P815 is identical to the normal gene of syngeneic DBA/2 mice. J Exp Med. 1991;173:1373–1384. doi: 10.1084/jem.173.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lethe B, Van den Eynde B, Van Pel A, Corradin G, Boon T. Mouse tumor rejection antigens P815A and P815B: two epitopes carried by a single peptide. Eur J Immunol. 1992;22:2283–2288. doi: 10.1002/eji.1830220916. [DOI] [PubMed] [Google Scholar]
- 21.Van den Eynde B, Mazarguil H, Lethe B, Laval F, Gairin JE. Localization of two cytotoxic T lymphocyte epitopes and three anchoring residues on a single nonameric peptide that binds to H-2Ld and is recognized by cytotoxic T lymphocytes against mouse tumor P815. Eur J Immunol. 1994;24:2740–2745. doi: 10.1002/eji.1830241125. [DOI] [PubMed] [Google Scholar]
- 22.Mandelboim O, Berke G, Fridkin M, Feldman M, Eisenstein M, Eisenbach L. CTL induction by a tumour-associated antigen octapeptide derived from a murine lung carcinoma. Nature (Lond) 1994;369:67–71. doi: 10.1038/369067a0. [DOI] [PubMed] [Google Scholar]
- 23.Loftus D, Castelli C, Clay T, Squarcina P, Marincola F, Nishimura M, Parmiani G, Appella E, Rivoltini L. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART1(27–35) . J Exp Med. 1996;184:647–658. doi: 10.1084/jem.184.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R-F, Appella E, Kawakami Y, Kang X, Rosenberg SA. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184:2207–2216. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]