Ordering of Human Bone Marrow B Lymphocyte Precursors by Single-Cell Polymerase Chain Reaction Analyses of the Rearrangement Status of the Immunoglobulin H and L Chain Gene Loci (original) (raw)
Abstract
CD19+CD10+ human B lineage bone marrow cells were separated into cycling or resting cells, which differ in their expression of CD34, VpreB, recombination activating gene (RAG-1), and terminal deoxynucleotidyl transferase (TdT). Polymerase chain reaction analyses developed for DHJH and VκJκ, VκJκK(de) and VκK(de) rearrangements with DNA of single cells and a comparison with B lineage cell development in mouse bone marrow, allow to delineate the human B lymphocyte pathway of development as follows: CD34+VpreB +RAG-1+TdT+, DHJH-rearranged, κL germline cycling pre-B I cells → CD34−VpreB +μH chain+ (pre-B receptor+) RAG-1−TdT−, VHDHJH-rearranged, κL germline, cycling pre-B II cells → CD34−VpreB −, intracytoplasmic μH chain+ (pre-B receptor−) RAG-1+/− TdT−, VHDHJH-rearranged, mainly κL germline cycling pre-B II cells → CD34−VpreB − intracytoplasmic μH chain+, RAG-1+TdT−, VHDHJH-rearranged, VκJκ-rearranged, IgM−, resting pre-B II cells CD34+VpreB −, sIgM+, RAG-1+TdT−, VHDHJH- and VκJκ-rearranged IgM+ immature B cells → CD34−, CD10−, sIgM+/sIgD+ mature B cells. This order, for the first time established for human B lineage cells, shows striking similarities with that established for mouse B lineage cells in bone marrow.
The differential expression of genes as proteins on the surface and inside of B lymphocyte precursors has allowed a definition and separation of subpopulations of bone marrow cells on their way to mature, antigen-reactive B cells. In the mouse, these markers include the tyrosine kinase c-kit (CD117), the α chain of the IL-2 receptor (CD25), CD19, B220 (CD45), leukosialin (CD43), BP-1, components of the rearrangement machinery, i.e., terminal deoxynucleotidyl transferase (TdT)1, recombination activating gene (RAG)-1 and RAG-2, μ heavy (H) chains and light (L) chains, and the components of the surrogate L chain, VpreB and λ5 (1–6). In humans, markers include CD10, CD19, CD34, TdT, RAG-1 and RAG-2, the μH and L chains, as well as the surrogate L chains, (7–12).
In mice and humans, the surrogate L chain has been found on the surface of two distinct early precursor populations. On the earlier cell, the surrogate L chain is associated with a complex of glycoproteins; on the later cells, together with μH chains, it forms the pre-B receptors (11–16). In this paper, we show that a new mAb specific for human VpreB (16) detects these two early precursor B cell populations in human bone marrow.
Single-cell PCR analyses of the status of rearrangements in both alleles of the IgH and L chain gene loci have previously allowed to order the different B lineage subpopulations of mouse bone marrow in their developmental pathway (17). In this paper, we use the same technique to analyze the configurations of the IgH and L chain alleles in single cells of human B lineage subpopulations. This allows us for the first time to order these human precursors in their developmental pathway in bone marrow.
Materials and Methods
Cells
Human heparinized bone marrow was obtained by iliac crest aspiration from 53 individuals of different ages (between 9-mo and 76-yr old), healthy or free of hematological diseases, according to institutional guidelines established by the ethical committees at Kantonsspital and Kinderspital in Basel, Switzerland.
Bone marrow cells were isolated by Ficoll-Hypaque gradient (density = 1.077 g/ml) (Pharmacia, Uppsala, Sweden).
Dendritic cells were separated and cultured as described previously (18, 19).
Antibodies, Immunofluorescence, and Flow Cytometric Analysis
mAbs.
Surface staining of bone marrow cells was performed using the following antibodies: FITC-labeled anti-CD3 and antiCD4; biotin (BIO)-labeled anti-CD8; FITC-labeled anti-CD10 (CALLA), anti-CD13, anti-CD14, and anti-CD16; FITC-, PE- or BIO-labeled anti-CD19; FITC- or PE-labeled anti-CD34; FITC-labeled anti-CD56 and anti-CD57 (Becton Dickinson & Co., Mountain View, CA); BIO-labeled anti-CD40 (Caltag Laboratories, San Francisco, CA); FITC-labeled anti-CD33 (Dakopatts, Glostrup, Denmark); FITC-labeled anti-TdT (Immunotech, Marseille, France); and nonconjugated anti-VpreB (mouse IgM; SL688; 16).
Polyclonal Antibodies.
Polyclonal FITC-conjugated rabbit anti– human IgM F(ab)2 and polyclonal PE-conjugated rabbit anti–human κL and anti-λL F(ab)2 were purchased from Dakopatts (Glostrup, Denmark).
Polyclonal FITC-labeled goat anti–human κL and anti-λL were purchased from Tago Diagnostics, Inc. (Burlingame, CA).
Polyclonal PE- and BIO-conjugated goat anti–human IgM, polyclonal BIO-conjugated goat anti–human IgD, and polyclonal FITC- and PE-labeled goat anti–mouse IgM were obtained from Southern Biotechnology Associates, Inc. (Birmingham, AL).
Streptavidin.
Streptavidin-PE was purchased from Southern Biotechnology Associates. Streptavidin TRICOLOR was obtained from Caltag Laboratories.
Cell Surface Staining and Cell Sort
Three-color immunofluorescence analysis and sort were used for identification of the different B cell precursor populations. Cell-surface immunofluorescence, flow cytometric analysis, and cell sort were performed as described on a FACStar Plus® or FACSVantage® (Becton and Dickinson) at 4°C (20) within 4–8 h from aspiration of the material (21). Only cells exhibiting low forward angle and low right angle light scattering properties (lymphoid gate) were analyzed and, when needed, sorted. All the shown percentages refer to the total cells within this gate. To identify pre-B I precursors, bone marrow cells were stained with BIO-labeled anti-CD19 or anti-CD10 (and streptavidin TRICOLOR) and PE-labeled anti-CD34 antibodies. These cells could be sorted to be stained intracellularly (see below) with either FITC-labeled anti–human IgM or FITC-labeled anti-TdT antibodies after fixation and permeabilization. To evaluate VpreB expression on the surface of these cells, bone marrow samples were stained with BIO-labeled anti-CD19 or anti-CD10, FITClabeled anti-CD34, and purified anti-VpreB, followed by incubation with streptavidin TRICOLOR and PE-labeled anti–mouse IgM. Using this procedure for staining, we were also able to sort CD34−VpreB + cells and evaluate, in this population (pre-B II, VpreB +), the cytoplasmic expression of μH chain or nuclear expression of TdT, as well as the surface expression of κL or λL chains.
Furthermore, to identify large and small pre-B II VpreB − populations, bone marrow samples were stained with BIO-labeled anti-CD19 or anti-CD10, purified anti-VpreB, and FITC-labeled anti-κL and -λL chains (followed by streptavidin TRICOLOR and PE-labeled anti–mouse IgM). These cells were also discriminated by size on the side forward scatter profile. The two populations could be restained intracellularly for cytoplasmic μH chain and TdT expression (see Fig. 1).
Figure 1.
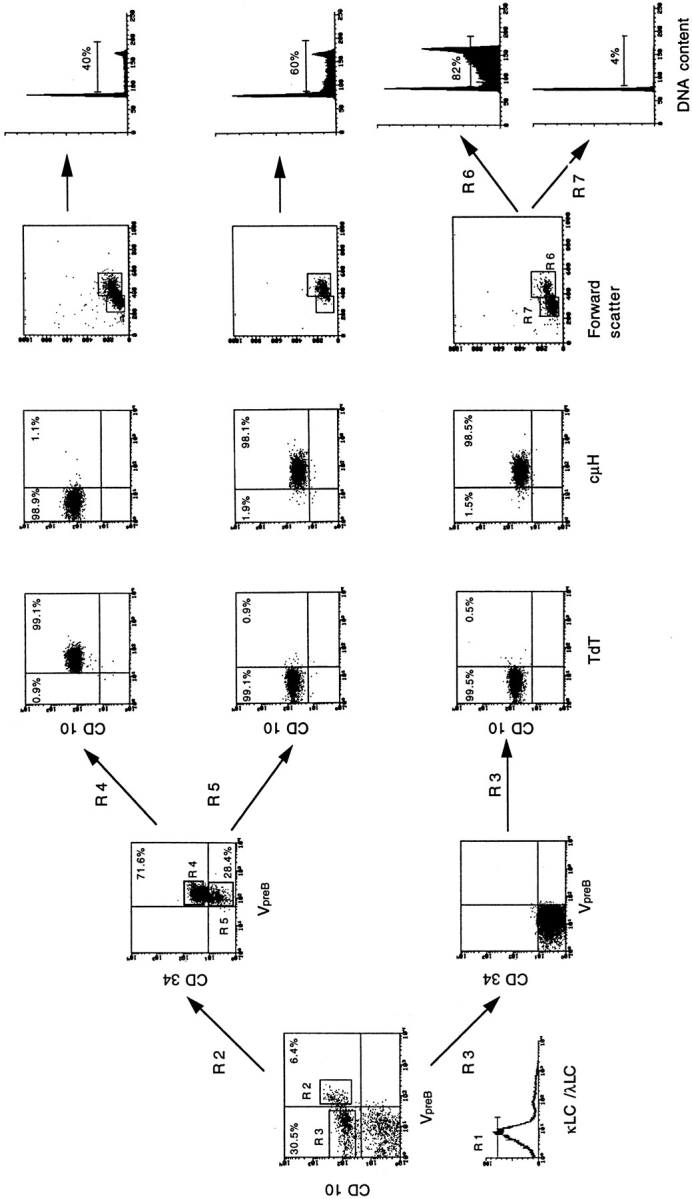
Surface phenotype and cell cycle status of pre-B cell subpopulations as defined by surface expression of VpreB, CD10, CD34, and κL and λL chains. Bone marrow lymphoid cells from a young donor were stained with purified anti-VpreB, BIO-labeled anti-CD10, and FITC-labeled anti–κL/λL chain antibodies, followed by incubation with streptavidin TRICOLOR and PE-labeled anti–mouse IgM, as described in Materials and Methods (left panel). Cells were gated for lymphoid cells. IgM− (κL−/λL−) (R1) CD10+ lymphoid precursors were FACS® sorted in two fractions (R2 and R3), and based on the surface expression of VpreB, were FACS® sorted and further analyzed for CD34 expression. A comparable separation could be achieved using CD19− instead of CD10− specific antibody, since we had seen that sIgM− CD10+ CD19+ cells coexpressed CD34, sIgM− CD34+ CD10+ cells coexpressed CD19, and sIgM− CD34+ CD19+ cells coexpressed CD10. CD34+ CD10+ CD19− cells were not detectable. Fraction R2 was further subdivided into CD34+VpreB + (R4) and CD34−VpreB + (R5). Cellular fractions R4, R5, and R3 were FACS® sorted, fixed, and stained intracellularly with FITC-labeled antibodies for intracytoplasmic TdT and μH chain expression, as described in Materials and Methods. To analyze TdT and μH chain expression in cells of fraction R4, a separate staining and sorting with BIO-labeled anti-CD10 and PElabeled anti-CD34 had to be performed. VpreB expression was tested by staining with purified anti-VpreB, followed by incubation with FITC-labeled anti–mouse IgM. The fraction (R3) was also subdivided based on CD34 expression and forward and side scatter profile (R6 and R7). The subpopulations R4, R5, R6, and R7 were sorted, fixed, and treated to prepare nuclei for analyses of DNA content, as described in Materials and Methods.
Immature B cells could be discriminated by staining with FITC-labeled anti-CD10, PE-labeled anti-κL and λL-chains, and BIO-labeled anti-IgD, followed by incubation with streptavidin TRICOLOR. Single cells were sorted using the FACStar Plus® equipped with an automatic cell deposition unit, directed into polycarbonat 96-well plates (Biozym Diagnostik GmbH, Oldendorf, Germany) containing 10 μl of PCR buffer (three times concentrated) or 5 μl of PBS. The plates were immediately frozen on dry ice and then stored at −70°C until use.
DNA and cDNA Preparation
To prepare DNA (17), 2 μl of proteinase K (5 mg/ml; Boehringer Mannheim GmbH, Mannheim, Germany) were added to the plates containing PCR buffer. Samples were then overlaid with PCR oil (Fluka, Buchs, Switzerland) and incubated for 1 h at 55°C. Proteinase K was subsequently inactivated for 10 min at 95°C. The plates were then stored at −20°C.
To prepare cDNA, single cells in PBS were heated up to 65°C for 2 min, and reverse transcription (RT) mix was added and incubated at 37°C for 1 h (Ten Boekel, E., F. Melchers, and A. Rolink, manuscript in preparation). The enzyme was inactivated for 2 min at 95°C.
Between 24 and 40 cells of each sorted population from three different bone marrows, from donors who were 9 mo to 13 yr old, were analyzed as follows.
PCR Analysis
PCR amplification of DH segments, Igκ gene and κ-deleting element (κ-de) rearrangements was carried out in two rounds, using degenerate oligonucleotide primers (as listed in Table 1a) to detect as many complementary sequences as possible. Nested primers (5′ and/or 3′ end) were used when needed, as shown in Table 1 a. The primers used to detect DH-JH rearrangements were designed to be complementary with the D element upstream of heptamer-nonamer sequences of the genes available on DNA sequence databases, since not all the DH genes are known. Hence, they were complementary to the members of 3 (DLR, DXP, and DQ52) of the 7 known families within the major DH gene locus, and they are expected to detect 10 of the 18 published sequences (22). They were also complementary to all the known DH genes within the minor locus (23). In detail, D primer 1 is complementary to DLR1, DLR2, DLR3, DLR4, and DLR5 genes. D primer 2 is complementary to DLR5, DLR6, DXP1, DXP′1, DXP4, D22/ 12, D21/7, D23/7, D21/9, D21/10, and D21/0.5 genes. D primer 3 is complementary to the DQ52 gene sequence. With these primers, two thirds of the published sequences can be expected to be detectable.
Table 1a.
Oligonucleotides Used for PCR Analysis of Germline IgH and Igκ Loci and Rearranged DH, Igκ, and κ−de Genes
| Sequence | Specificity | PCR round usage | |
|---|---|---|---|
| First | Second | ||
| 5′ primers | |||
| GTTTCTGTGCCCCTGGCTCAG | Germline IgH | + | + |
| GGATTTTGTGGGGGCT(C/T)GTGTCACTG | DH-primer 1* | + | − |
| ATTTTGTGGGGGCT(C/T)GTGTCACTGTG | DH-primer 1* | − | + |
| GGTTTGG(A/G)(A/G)TGAGGTCTGTGTCACTG | DH-primer 2‡ | + | − |
| TTTGGRRTGAGGTCTGTGTCACTGTG | DH-primer 2‡ | − | + |
| GTTTTTGGCTGAGCTGAGAACCACTG | DH-primer 3§ | + | − |
| TTTTGGCTGAGCTGAGAACCACTGTG | DH-primer 3§ | − | + |
| GCTCGAAAAGGGAGTTGAGCTTCAGC | Germline Igκ | + | + |
| AGTGGATCTGGGACAGA(C/T)TTCACTCTC | Vκ I/III | + | − |
| TGGATCTGGGACAGA(C/T)TTCACTCTCAC | Vκ I/III | − | + |
| TCTAACCGGGACTC(C/T)GGGGTCCC(A/T)GAC | Vκ II/IV | + | − |
| GGGTCCC(A/T)GAC(A/C)GATTCAG(C/T)GGCAG | Vκ II/IV | − | + |
| TGAGTGGCTTTGGTGGCCATGCCAC | J-Cκ intron | + | + |
| 3′ primers | |||
| TCCCAGTTCCCAAAGAAAGGCC | JH3 | + | − |
| TACCTGAAGAGACGGTGACCATTGT | JH3‖ | − | + |
| AACCGCAATGGCGCAGGAAACC | JH6 | + | − |
| ACCTGAGGAGACGGTGACCGTGGT | JH6‖ | − | + |
| ACCTGAGGAGACGGTGACCGTGGT | JH6‖ | − | + |
| CACCCAAAGCCACTGACTCTGGGAG | κ-de | + | − |
| AGGCCACAAACCCAGCAAAGCACC | κ-de | − | + |
| CTGGCCATCAGCCCCAAATTTCAGAAG | Jκ5 | + | − |
| TTAATCTCCAGTCGTGTCCCT | Jκ5 | − | + |
The four primers for the Vκ genes (Table 1a) were designed according to the consensus sequence derived from the alignments of the Vκ sequences of the 32 potentially functional genes and of the 16 genes with only minor defects (24). Within the two sets of primers, one was complementary to the sequences of the genes belonging to VκI and III families, in the sense that the primer had a complete concordance in sequence for 14 genes, only a 1-bp difference for 12 and a 2-bp difference for another 5 genes. The other set of primers was complementary to the genes in VκII and IV families, again in the sense that the sequence had a complete homology with six genes, only 2 bp difference with 6 and 4 bp difference (all in the 5′ region of the primer) with only three genes. No primers to detect families V, VI, and VII were designed since they are all nonfunctional. With such primers, at least 90% of the Vκ genes that are potentially functional or have only minor sequence defects can be expected to be detectable.
The PCR amplification conditions were as described by Ten Boekel et al. (17). PCR amplification of cDNA samples was carried out using the primers listed in Table 1b with the same strategy described above. The lengths of the different RTPCR products of the DNA and RNA analyses are listed in Table 2a and b, respectively_._ All the primer pairs are spanning intron sequences, allowing the distinction between expressed RNA from genomic DNA. Moreover, the primers used to detect B29 cDNA were also able to detect a splicing variant that lacks the entire exon III of the gene (25).
Table 1b.
Olgonucleotides Used for RT-PCR Assay
| Sequence | Specificity | PCR round usage | |
|---|---|---|---|
| First | Second | ||
| 5′ primers | |||
| GTGGCGTTGCTGCTGCTGCTCT | B29 | + | + |
| CCAAATTGCAGACATCTCAAC | RAG-1 | + | + |
| GCCGTCAGTGTGCTGGTTAAAGAGG | TdT | + | + |
| ATGTCCTGGGCTCCTGTCCT | VpreB | + | + |
| 3′ primer | |||
| GCTGTGCCAAGGTGCTGAATCC | B29 | + | + |
| CAACATCTGCCTTCACATCGATCC | RAG-1 | + | − |
| ACCATCCACAGGACCATGGACTGG | RAG-1 | − | + |
| TCTGCTTTGAGGAATATCCTCTTGG | TdT | + | − |
| AGAATCATCTTCCGCTCATGTGTGG | TdT | − | + |
| TGCAGTGGGTTCCATTTCTTCC | VpreB | + | − |
| GTAATACATAGCCTCGTCCTCAGG | VpreB | − | + |
Table 2a.
Approximate PCR Product Lengths of IgH and L Chain Analyses
| Rearrangements | Product lengths |
|---|---|
| Germline IgH | 903 |
| DH-primer 1- JH3 | 924 (D1, D2, D4, D5) 921 (D3) |
| DH-primer 1- JH3 | 714 (D1, D2, D4, D5) 711 (D3) |
| DH-primer 1- JH3 | 106 (D1, D2, D4, D5) 103 (D3) |
| DH-primer 1- JH6 | 1,152 (D1, D2, D4, D5) 1,149 (D3) |
| DH-primer 1- JH6 | 713 (D1, D2, D4, D5) 710 (D3) |
| DH-primer 1- JH6 | 120 (D1, D2, D4, D5) 117 (D3) |
| DH-primer 2- JH3 | 924 (DXP, D22, D23, D21/0.5) 923 (D21/9, 7)930 (D21/10) |
| DH-primer 2- JH3 | 714 (DXP, D22, D23, D21/0.5) 713 (D21/9, 7)720 (D21/10) |
| DH-primer 2- JH3 | 106 (DXP, D22, D23, D21/0.5) 105 (D21/9, 7)112(D21/10) |
| DH-primer 2- JH6 | 1,152 (DXP, D22, D23, D21/0.5) 1,151 (D21/9, 7)1,158 (D21/10) |
| DH-primer 2- JH6 | 713 (DXP, D22, D23, D21/0.5) 712 (D21/9, 7)719 (D21/10) |
| DH-primer 2- JH6 | 120 (DXP, D22, D23, D21/0.5) 119 (D21/9, 7)126 (D21/10) |
| DH-primer 3- JH3 | 904 |
| DH-primer 3- JH3 | 694 |
| DH-primer 3- JH3 | 86 |
| DH-primer 3- JH6 | 1,132 |
| DH-primer 3- JH6 | 693 |
| DH-primer 3- JH6 | 100 |
| Germline Igκ | 1,477 |
| Vκ I/III - Jκ1 | 1,436 |
| Vκ I/III - Jκ2 | 1,076 |
| Vκ I/III - Jκ3 | 774 |
| Vκ I/III - Jκ4 | 441 |
| Vκ I/III - Jκ5 | 125 |
| Vκ II/IV - Jκ1 | 1,461 |
| Vκ II/IV - Jκ2 | 1,101 |
| Vκ II/IV - Jκ3 | 799 |
| Vκ II/IV - Jκ4 | 466 |
| Vκ II/IV - Jκ5 | 150 |
| J-Cκ introl - κde | 500 |
| Vκ I/III - κde | 481 |
| Vκ II/IV - κde | 520 |
PCR Product Sequencing
The PCR products, obtained with VpreB-specific primers, were sequenced using the Ready Reaction Dye Determinator (PerkinElmer Corp.,Norwalk, CT) and an automated DNA sequencer (model 373; Applied Biosystems, Inc., Foster City, CA).
Intracellular Staining for FACS® Analysis
Cytoplasmic Staining.
For testing cytoplasmic μH protein expression, cells were fixed for 10 min in ice-cold 4% paraformaldehyde (dissolved in PBS; Fluka Chemie AG, Buchs, Switzerland) and then permeabilized with 0.2% Tween 20 (dissolved in PBS; Fluka Chemie) for 20 min at 37°C.
Subsequent staining and washing procedures, as well as FACS® analyses, were performed in the same way as described for surface staining of cells, using polyclonal FITC-conjugated rabbit anti– human IgM F(ab)2 (Dakopatts).
Nuclear Staining.
After performing the surface staining, as described before, the samples could be analyzed for the expression of the nuclear enzyme TdT (26).
The cells were fixed in 1 ml of an ice-cold solution containing 1% paraformaldehyde (dissolved in PBS), and were incubated for 2 min on ice. They were then permeabilized in a 1.5-vol of icecold absolute methanol and incubated for 20 min on ice. After washing, cells were resuspended in an appropriate volume of FCS (as a blocking serum) for 15 min, and a mixture of three FITCconjugated, anti-TdT mAbs (Immunotech) was added at a final concentration of 1:20 for 30 min on ice. FACS® analysis was performed as described before.
Cell Cycle Analysis
Sorted bone marrow cells were fixed in 70% ethanol at 4°C overnight. Fixed cells were resuspended in RNAse A (0.5 mg/ml; Boehringer Mannheim), and were incubated for 30 min, at 37°C. 1 mg/ml pepsin was then added and incubated for 15 min at 37°C to prepare nuclei. Finally, ethidium bromide (0.02 mg/ml in 0.2 M Tris, pH 8–8.5, and 0.5% BSA) was added, mixed, and the nuclei were incubated for another 15 min at room temperature. DNA content was analyzed using a FACScan® instrument equipped with the Doublet Discriminator Module.
Results
Detection of VpreB Protein on Bone Marrow Cells with the mAb 688
Cells from human bone marrow (from 24 donors, 10-mo to 76-yr old) were examined for the expression of VpreB protein, by surface staining with mAb 688 in three-color FACS® analysis initially for VpreB, CD10, and κL/λL chains. After removal of the surface κL/λL+ cells and a gating for lymphoid cells, cells were stained with CD34-specific antibody, and then sorted as shown in Fig. 1. CD19 expression could be used instead of CD10 expression to achieve this separation and enrichment of precursor B cells, since CD10+ are also CD19+ (not shown). Hence, from now on, we call all CD10+ precursors CD19+CD10+ cells.
The separation shown in Fig. 1 demonstrates that the majority (around two out of three) of VpreB + CD10+ B lineage cells did express CD34 on their surface (Fig. 1, R2). These triple-positive cells also expressed TdT, but not cytoplasmic μH chains (Fig. 1, R4). Note in the legend of the figure that a separate staining and sorting had to be performed for the R4 analysis.
An analysis by forward and side scatter gating revealed that the R4 population was composed of a mixture of small and large cells. Cell cycle analysis showed that ∼40% of them were in S and G2/M phases (Fig. 1, R4).
A small part of the CD10+VpreB +Ig κL chain− λL chain− cells did not express CD34 on their surface (Fig. 1, R2). These cells expressed μH chain, which is easily detectable in the cytoplasm. They did not express TdT (Fig. 1, R5). This cell population represents ∼5% of all CD10+ B lineage cells in the bone marrow. The vast majority of them, by forward scatter profile, were found to be large. More than 60% of these cells were in S and G2/M phases of the cell cycle (Fig. 1, R5).
With the remaining CD10+CD34− cells, in marrow that represented 45–60% of all of the CD10+ B lineage cells, VpreB could not be detected on the surface (Fig. 1, R3). This population consisted of 15–20% large and 80–85% small cells; while 70–90% of the large cells were in S and G2/M phases, only 3–5% of the small cells were in these phases of the cell cycle, i.e., resting.
VpreB could also not be detected on the surface of CD10+ CD34− sIgM+ κL chain+ or λL chain+ cells, nor on CD10− IgM+, IgD+ (κL chain+ or λL chain+) cells. The former population consisting of immature B cells represents 20–25% of the CD10+ B lineage cells in bone marrow. Less than 2% were in S and G2/M phases, i.e., practically all of them were resting cells. No CD3+, CD4+, CD8+, CD13+, CD14+, CD16+, CD33+, CD56+, or CD57+ cells showed detectable expression of VpreB on their surface (all data not shown).
We conclude that mAb 688, specific for VpreB, detects VpreB molecules on the surface of all CD34+ and a small fraction of CD34− cells that also expresses CD19 and CD10, but not surface (sIgM). These two precursor populations, the first TdT+ and the second TdT−, are analyzed in more detail below.
Age-related Changes in the Number of B Lineage Precursor Cells in Human Bone Marrow
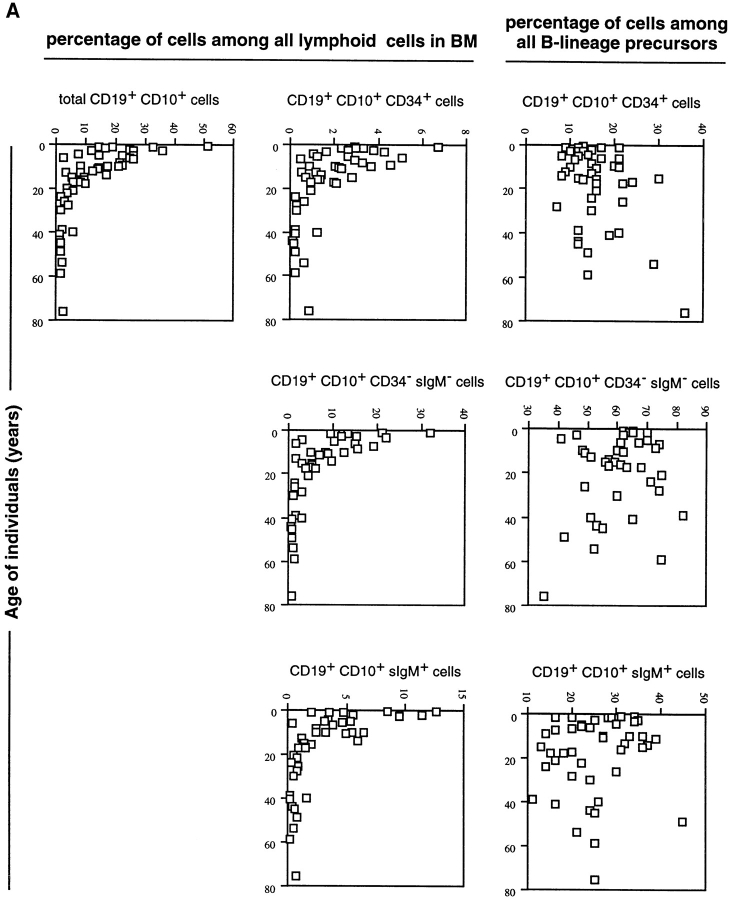
The representative of the CD19+CD10+ B lineage subpopulations of human bone marrow described above was analyzed in 53 donors who were 10 mo to 76 yr old. The results are presented in Fig. 2 a as the percentage values of cells within the lymphoid compartments, and as the percentage values of cells among the CD19+CD10+ B lineage precursor compartments in bone marrow.
Figure 2.
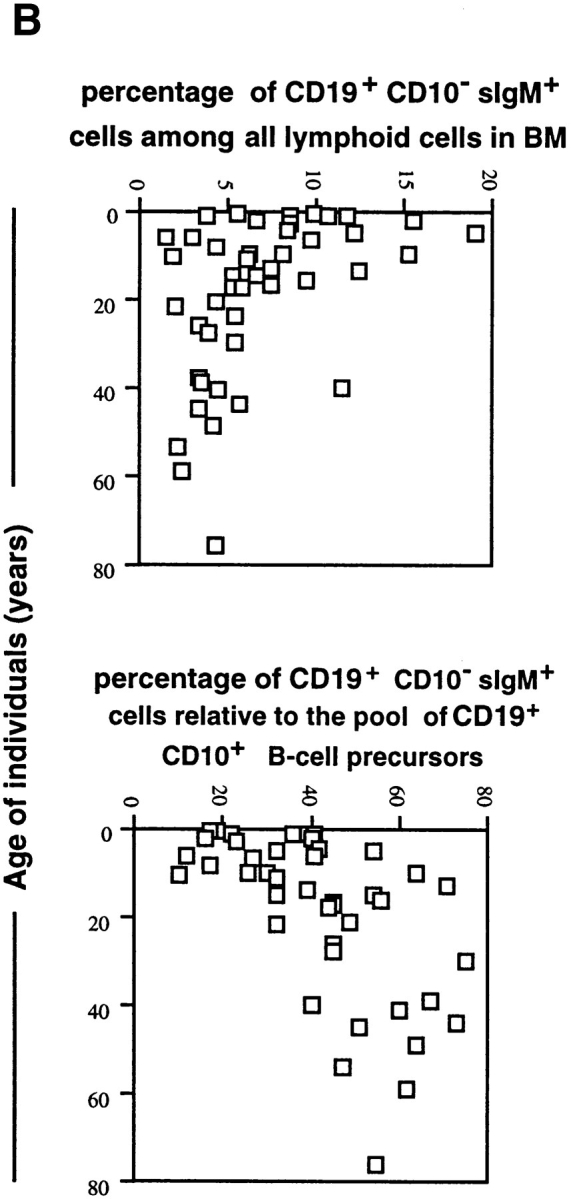
(a) Frequencies of CD19+ CD10+ B lineage cells and their subpopulations among total lymphoid cell compartment in human bone marrow of donors of different ages (one donor = one symbol). The frequencies of these subpopulations among all CD19+ CD10+ cells are also given. (b) Frequencies of CD19+CD10−sIgM+ mature B cells in the lymphoid compartments of human bone marrow, and their relative contribution to all CD19+ cells in human bone marrow of donors of different ages.
It is evident that the frequency of CD19+CD10+ B lineage cells and the frequency of cells in all identified B precursor compartments, hence also of the VpreB-expressing B lineage precursors, decrease with age. They are highest in young donors who are up to the age of 10 yr old. It appears, however, that the proportions of different B lineage precursors relative to each other remain the same throughout life.
The frequencies of CD19+CD10−sIgM+ mature B cells also decrease within the lymphoid compartment (Fig. 2 b). Their relative contribution to all CD19+ B lineage cells in bone marrow, however, increase with age.
RNA Expression of the VpreB, TdT, and RAG-1 Genes in Single Cells of Different B Lineage cells in Human Bone Marrow
Single cells from the subpopulations of CD19+CD10+ CD34+VpreB +, CD19+CD10+CD34−VpreB +, the large cycling CD19+CD10+CD34−sIgM−, the small resting CD19+ CD10+CD34−VpreB −sIgM− B lineage cells of bone marrow from three young donors were separated by FACS®, and were analyzed by RT-PCR developed for human VpreB, TdT, and RAG-1. As a control for cDNA integrity, B29 cDNA was amplified (Table 2b). The results of these experiments are summarized in Table 3.
Table 2b.
Approximate RT-PCR Product Lengths
| RT-PCR products of RNA of: | Product lengths |
|---|---|
| bp | |
| B29 | 409 and 97 |
| RAG-1 | 468 |
| TdT | 482 |
| VpreB | 428 |
Table 3.
RT-PCR Analyses of RAG-1, TdT, and VpreB mRNA Expression in Single Cells of the CD10+CD19+sIgM− Subpopulations of B Lineage Precursors in the Bone Marrow of Three Donors*
| B lineage precursors | CD34+VpreB + | CD34−VpreB + | Large CD34− VpreB − | Small CD34− VpreB − |
|---|---|---|---|---|
| Donor a (Male, 13-yr old) | ||||
| RAG-1 | 88% (21/24) | 0% (0/23) | 14% (4/24) | 44% (10/23) |
| TdT | 83% (20/24) | 17% (4/23) | 0% (0/24) | 0% (0/23) |
| VpreB | 75% (18/24) | 87% (20/23) | 80% (19/24) | 17% (4/23) |
| Donor b (female, 3-yr old) | ||||
| RAG-1 | 71% (17/24) | 9% (2/21) | 8% (2/24) | 25% (6/24) |
| TdT | 92% (22/24) | 9% (2/21) | 0% (0/24) | 4% (1/24) |
| VpreB | 88% (21/24) | 60% (12/21) | 17% (4/24) | 25% (6/24) |
| Donor c (female, 9-mo old) | ||||
| RAG-1 | 78% (25/32) | 0% (0/28) | 8% (2/24) | 49% (19/39) |
| TdT | 100% (32/32) | 0% (0/28) | 8% (2/24) | 7% (3/39) |
| VpreB | 71% (23/32) | 93% (12/28) | 50% (12/24) | 33% (13/39) |
TdT-specific RNA expression followed the pattern of protein expression that was detected in the earlier experiments (see Fig. 1). It was only expressed in the large majority of all CD34+ B lineage cells.
VpreB expression on the RNA level did not completely follow the pattern of expression of the VpreB protein. Hence, while CD34+ and some of the large cycling CD34− B lineage cells expressed the protein, and while the rest of the large and all the small resting CD34− cells did not, RNA was not only expressed in the two subpopulations with VpreB protein, but was expressed in the majority of CD34− large cells, as well as in up to 25% of all the small CD34− CD10+ pre-B cells. The VpreB-specific mRNA expressed in these cells was reverse transcribed, sequenced, and found to contain the expected VpreB characteristic sequences. It remains to be determined whether the VpreB RNA in the VpreB protein–negative cells is functional mRNA.
RAG-1 expression was detectable in most of the CD34+ B-lineage precursors, was suppressed in the majority of CD34− large cells, and was again detectable in half of the small CD34− cells. This modulated expression of RAG-1 RNA will be discussed below. We have shown previously that RAG-1 mRNA is expressed in immature CD34− CD10+ sIgM+ B cells, while it is not expressed in CD34− CD10− sIgM+ mature B cells (21).
PCR Analysis of DHJH and VκJκ Rearrangements in Single CD19+CD10+ B Lineage Cells of Different Bone Marrow Subpopulations
For the amplification of DHJH-rearranged IgH sequences of VκJκ-rearranged (κ-de) L chain gene loci, several primers were designed (see Materials and Methods and Table 1a). Practically the whole human κL chain gene locus is known, and that includes orphan gene segments (24). Genes in Vκ families I, II, III, and IV are expected to contain members that encode L chain proteins, while those in families V, VI, and VII are all nonfunctional. The VκJκ primer sets used in our studies are expected to detect at least 90% of the 32 potentially functional Vκ genes, as well as the 16 nonfunctional genes with minor sequence defects.
In contrast to the κL chain locus, the DH segments of the IgH chain locus are not yet completely known. A major locus of seven families have been detected within the IgH chain locus on chromosome 14 (22), while another six have been described on a minor locus on the same chromosome (23). With the primer sets shown in Table 1a, we expect to detect approximately two thirds of these DH segments in DHJH-rearranged configurations.
Eventual sequencing of many of the PCR products detected from single cells will be able to evaluate the validity of these expectations.
The results of the PCR analyses are discussed below:
DHJH Rearrangements.
A total of 36 single CD19+CD10+ CD34+ B cell precursors from three individuals were analyzed for their status of DHJH rearrangements within the IgH locus. The results, shown in Table 4, show that the majority of these cells have at least one of their IgH alleles in DHJH-rearranged conformation (see also legend to Table 4). In many cells, both alleles should be rearranged, since in most cases, no germline H chain locus was detectable in the PCR analyses. It remains to be analyzed how many of the rearranged H chain loci are VHDHJH rather than DHJH rearranged. If they were VHDHJH rearranged, the observed lack of μH chain expression in this cell population would make it likely that they would be nonproductively rearranged.
Table 4.
DHJH −, VκJκ −, VκJκκ(de)− and Vκκ(de)− Rearrangements in Single Cells of CD19+B Lineage Precursors in Human Bone Marrow*
| CD10+ CD34+‡ VpreB+ sIgM− | CD10+ CD34− VpreB+ sIgM− | Large CD10+ CD34− VpreB− sIgM− | Small CD10+CD34− VpreB− sIgM− | CD10+ sIgM+ immature | CD10−sIgM+sIgD+ mature | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | κLchain+ | λLchain+ | ||||||||||
| Donor | a | b | c | |||||||||
| Number of cells tested | 12 | 12 | 12 | |||||||||
| Configuration of H chain alleles | ||||||||||||
| GH | 4 | 0 | 1 | |||||||||
| DHJH | 5 | 7 | 8 | |||||||||
| Donor | a | b | a | b | a | b | a | b | a | a | a | a |
| Number of cells tested | 22 | 20 | 22 | 21 | 24 | 22 | 24 | 21 | 25 | 24 | 21 | 22 |
| Configuration of κL chain alleles | ||||||||||||
| Gκ | 21 | 17 | 22 | 21 | 22 | 22 | 14 | 13 | 11 | 9 | 6 | 3 |
| VκJκ | 1 | 0 | 2 | 0 | 7 | 1 | 15 | 13 | 22 | 23 | 18 | 20 |
| VκJκκ(de) | 0 | 2 | 1 | 1 | 0 | 1 | 8 | 8 | 8 | 9 | 1 | 22 |
| Vκκ(de) | 0 | 0 | 2 | 0 | 0 | 1 | 4 | 6 | 4 | 3 | 3 | 6 |
We conclude that the majority of CD19+CD10+CD34+ B lineage precursors appear to have their IgH chain alleles no longer in germline configuration, since many of them have at least one allele DHJH rearranged.
The vast majority of the CD19+CD10+CD34− cells express μH chains in their cytoplasm (Fig. 2). Hence, they are expected to be productively VHDJH rearranged on at least one allele. Therefore, we did not analyze their IgH gene loci by PCR.
Our results confirm previous suggestions of the order of human B cell subpopulations during development in bone marrow. The analyses of the conformations of the IgH gene loci show that CD19+CD10+CD34+TdT+ cells are DHJH-rearranged precursors of the CD19+CD10+CD34− TdT− cells.
VκJκ Rearrangements.
CD19+CD10+ B lineage cells of bone marrow were separated by FACS® into single cells of the subpopulations of CD34+VpreB +, CD34−VpreB +, large CD34−VpreB −, small CD34−VpreB −, sIgM+ immature, sIgM+ sIgD+, and sIg+ κL+ or λL+ mature cells. Ig κL chain gene loci of between 20 and 25 single cells of the different populations from the bone marrow of two donors were analyzed for their conformations (Table 4b).
It is evident that the vast majority of the DHJH-rearranged CD19+CD10+CD34+ precursors have their κL chain loci in germline configuration. κL chain gene rearrangements are detectable in a large part of all cells, i.e., in at least 60% of all CD34− small cells, and from there on, as expected, in 80–95% of all immature and mature sIg+ B cells. Interestingly, λL+ sIg+ B cells appear to have less κL chain gene loci in germline configuration, and have more of them deleted.
We conclude from these analyses that the majority of all κL chain gene rearrangements occur at the transition from VpreB − large CD34− to VpreB − small CD34− cells. Hence, the largest subpopulation of B lineage cells in the human bone marrow, the CD19+ CD10+CD34−VpreB −TdT− RAG-l+, cyto-plasmic μH chain+, small, resting sIg− precursor B cell has, to a large part, its Ig κL chain gene loci in VκJκrearranged conformation, but does not yet express them as proteins so that the cells would become sIg+. Collectively, these PCR analyses allow us to order CD19+ CD10+ B lineage subpopulations in human bone marrow in the following sequence: CD34+TdT+RAG-l+VpreB + cells are DHJHrearranged precursors of large, VpreB + and VpreB −, CD34−, TdT−, RAG-1−, productively VHDJH-rearranged cells which, in turn, are precursors of small, VpreB −, CD34−, TdT−, RAG-1+, VHDJH-rearranged cells, which, finally, are followed by sIgM+ immature and sIgM+ sIgD+ mature B cells.
Discussion
The analysis of the status of the IgH and L chain gene loci in single cells of B lineage subpopulations in the bone marrow of mice has been instrumental in ordering these subpopulations along a pathway of B lymphocyte development (17, 27). This ordering has been guided by the fact that rearrangements in the Ig loci occur in stepwise fashion, so that DH segments are rearranged first to JH segments, then VH segments are rearranged to DHJH segments, and finally, Vκ and Vλ segments are rearranged to Jκ and Jλ segments, respectively (28). Rearrangements in the κL chain locus appear often to precede those in the λL chain locus, and can be terminated by rearrangements into κL chain gene–deleting elements (29, 30). Stepwise, ordered rearrangements in the Ig gene loci are also characteristic for human B lymphocyte development.
Single-cell PCR analyses of the status of the Ig H and κL chain loci of B lineage subpopulations in human bone marrow performed in this paper allow a similar ordering of human B lineage subpopulations, characterized by the expression of CD19, CD10, CD34, VpreB, TdT, RAG-1, and by their cell cycle status. Comparisons with B cell development in the bone marrow of mice allow the following conclusions:
(a) Human CD19+CD10+CD34+TdT+RAG-l+ VpreB + μH chain− cycling cells are comparable to murine B220+ CD19+TdT+RAG-1/2+VpreB +/λ5 +c-kit +, μH chain− CD25− cycling pre-B I cells. Both have DHJH-rearranged Ig H chain loci. Mouse pre-B I cells often have both alleles DHJH rearranged. Their Ig L chain loci remain in germline configuration. In analogy to murine pre-B I cells, we will call them human pre-B I cells.
(b) Human CD19+CD10+CD34−TdT−RAG-1−VpreB +, μH chain+, pre-B receptor+ cycling cells are comparable to murine B220+CD19+TdT−RAG-1−/2−c-kit − CD25+ VpreB +/λ5 + μH chain+ pre-B receptor+ large pre-B II cells. They are productively VHDJH rearranged in the Ig H chain locus, while their L chain gene loci remain in germline configuration. In analogy with mouse pre-B cells, we will call them large, pre-B receptor+ pre-B II cells.
(c) Human CD19+CD10+CD34−TdT−RAG-1−VpreB −, μH chain+ pre-B receptor− cycling cells are comparable to murine B220+CD19+TdT−RAG-1−/2−c-kit −CD25+ VpreB −/λ5 −, pre-B receptor−, μH chain+ sIgM− large pre-B II cells. Their H chain locus is productively VHDJH rearranged, while their Ig L chain loci are largely in germline configuration. In analogy with mouse pre-B II cells, we will call them large pre-B receptor− pre-B II cells.
(d ) Human CD19+CD10+CD34−TdT−RAG-1+VpreB −, μH chain+ sIgM− small resting cells are comparable to murine B220+CD19+TdT−RAG-1+/2+c-kit −CD25+VpreB −/ λ5 −, μH chain+ small resting pre-B II cells. Both have a majority of their IgL chain gene loci in VLJL-rearranged configurations, but do not yet express L chains on their surface. In analogy with mouse pre-B cells, we will call them human small resting pre-B II cells.
(e) Human CD19+CD10+sIgM+ immature B cells are comparable to murine immature B cells, and human mature CD19+CD10−sIgM+sIgD+ B cells are comparable to murine mature B cells. The RT-PCR analyses of the expression of the RAG-1 genes in different B lineage subpopulations of human bone marrow show the same striking changes that have been observed in mouse B lineage subpopulations (31). It indicates that the same up- and downregulation of the rearrangement machinery that might be part of the mechanisms effecting allelic exclusion of the H chain alleles appear to be operative in the human. It appears that human and mouse B cell subpopulations in the bone marrow resemble each other, not only in their status of Ig gene rearrangements of TdT and RAG protein expression and pre-B receptor expression, but their relative amounts in the different compartments are also comparable.
The relative proportions of the CD19+CD10+ B lineage compartments remain the same throughout life, although they decrease in frequencies. This suggests that while the flow from early to late B cell precursors decreases with age, it continues from the same precursors through the same intermediates during life. The relative proportion of the mature, CD19+CD10−sIgM+ B cells increases over the CD19+ CD10+ B lineage cells with age, suggesting that the contribution of recirculating B cells in bone marrow increases with age. These results confirm earlier findings by Nuñez et al. (32), which showed that B cells are continuously generated through life though the relative frequencies of precursor B cells in human bone marrow decrease with age.
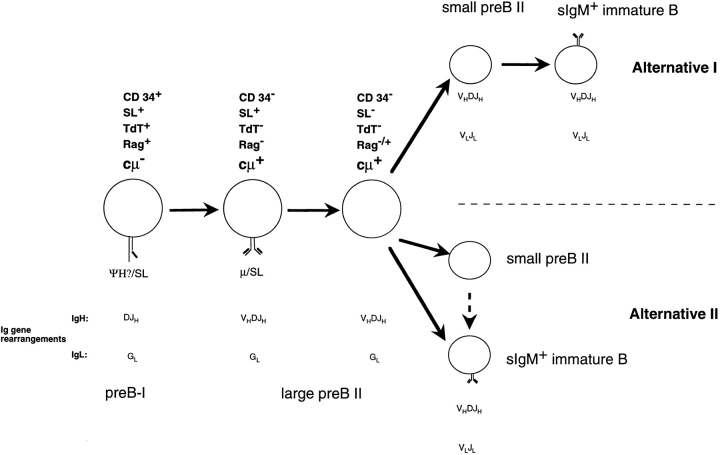
A few differences between human and mouse B lineage cells are apparent. First, a small but significant portion of the large, pre-B receptor expressing and pre-B receptor– negative human pre-B II cells have κL chain gene loci in VLJL-rearranged configurations. It cannot be excluded that this difference is a consequence of the way the bone marrow samples are collected. Murine bone marrow cells are prepared on ice within 15 min after the death of the animals, while human marrow is aspirated but then kept for considerable periods of time before staining with specific antibodies and FACS® separation procedures are initiated. This longer time ex vivo may allow the progression along the pathways of differentiation of a human precursor B cell and, hence, allow occasionally L chain rearrangements in large pre-B II cells, after ex vivo isolation.
Second, VpreB gene expression on mRNA level persists into a part of the large, pre-B receptor–negative and small, resting human pre-B II cells, while that is not so in the murine counterparts. Again, a longer period ex vivo before staining and FACS® might allow cells to change their surface phenotype, downregulate VpreB protein expression but keep VpreB mRNA intact. Also, it has been observed in human pre-B cells that stability of surrogate L chain protein differs in different precursor subpopulations (33). Different laboratories have developed different mAbs specific for different determinants on surrogate L chain expressed on either human or mouse B lineage cells (11, 12, 15). Three patterns of expressions on different B cell subpopulations of the marrow have been detected. Lassoued et al. (11) detected surrogate L chain only on CD19+CD10+TdT−, pre-B receptor+ (presumably large) cells, which we would call pre-B II cells. They have concluded that these cells are the immediate precursors of sIgM+ immature B cells, although they have not offered a placement of the pre-B receptor− cytoplasmic μH chain+ precursors, which, according to their studies, comprise as many as 80% of all CD19+CD10+ B lineage precursors in bone marrow. We would propose that these pre-B receptor− cytoplasmic μH chain+TdT− precursors are, in majority, VLJL-rearranged small pre-B II–like cells. They might be the nonproductively VLJL-rearranged portion of the immature cells and, hence, one of two direct products of the large, pre-B receptor–positive and –negative pre-B II cells (Fig. 3, Alternative II). This remains to be elucidated by sequencing of individual VκJκ rearrangements in these cells.
Figure 3.
Scheme of human B cell differentiation in bone marrow. Two alternative pathways of immature B cell generation are shown (see Discussion). The dotted arrow between small pre-B II and sIgM+ immature B cells in alternative II indicates that the RAG-expressing pre-B II cells continue to rearrange κL and λL chain loci (21). G, germline configuration. For a comparison to mouse B cell development, see Rolink et al. (36).
It is also possible that they are, at least in part, productively VLJL rearranged, but not (yet) capable of expressing κL chain protein. Such VLJL-rearranged, κL chain–negative cells have been found in the mouse in the case of the 70Z/3 cell line, which is inducible to κL chain expression by stimulation with lipopolysaccharides (34). If this is the case, they are precursors of the sIgM+ immature B cells and, hence, in between large pre-B II cells and immature sIgM+ B cells (Fig. 3, Alternative I).
For murine precursors B cells, Winkler et al. (15). have detected two subpopulations in bone marrow that express surrogate L chain on the surface (15). One is the pro-/preB I cell population, which expresses surrogate L chain together with a complex of glycoproteins (35), and the other is a population of large, cycling pre-B II cells, which is the progeny of the pre-B I cells and the precursors of large, pre-B receptor–negative and small, resting pre-B II cells. Guelpa-Fonlupt et al. (12) have observed the same pattern of surface expression of surrogate L chain in human bone marrow. Their antibody, however, also stained sIgM+ immature B cells. The VpreB-specific mAb 688 (16) used in this paper detects surrogate L chains on human pre-B I and large pre-B II, but not on small pre-B II and not on immature sIgM+ B cells, and hence follows the pattern of murine surrogate L chain expression on the surface. Since our RT-PCR analyses did not detect VpreB mRNA in immature sIgM+ B cells, and since our VpreB mRNA–positive small pre-B II cells did not express detectable levels of VpreB protein, we see no reason to expect that VpreB expression could be carried as far as into the sIgM+ immature B cells population.
The differential expression of CD34, TdT, RAG-1, and VpreB and differences in cell-cycle status, as well as single cell PCR analyses of the status of rearrangements in the IgH and κL chain loci, have allowed us to order cellular development of B cell precursors in human bone marrow as it has been done in the mouse. These analyses are expected to be helpful in an understanding of the cellular targets of dysregulation leading to autoimmune diseases, immunodeficiencies, and neoplasia in the human B lymphocyte lineage.
Acknowledgments
We thank Drs. Rod Ceredig and Thomas Winkler for critical reading of this manuscript. We are grateful to Marcus Dessing for his outstanding skill at the FACS® sorter and his extraordinary help during long, unusual hours. We thank Prof. A. Gratwohl, Dr. E. Signer, and Dr. U. Ramenghi for providing the bone marrow samples and Prof. F. Caligaris Cappio for continuous encouragement and discussions. We gratefully acknowledge Ms. Nadia Straube's technical experience in DNA sequencing.
Footnotes
The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche Ltd., Basel, Switzerland. E. Sanz was supported by contracts from the CSIC and grant CAM92/126, and A. de la Hera was supported by grants SAF-93-0925 and SAF-96-0201 from the CICY.
1 Abbreviations used in this paper: BIO, biotin; H, heavy; κde, κ-deleting element; L, light; RAG, recombination activating gene; RT, reverse transcription; s, surface; Tdt, terminal deoxynucleotidy transferase.
References
- 1.Osmond DG. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr Opin Immunol. 1991;3:179–185. doi: 10.1016/0952-7915(91)90047-5. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR. Variable gene usage, physiology and development of Ly1+ (CD5+) B cells. Curr Opin Immunol. 1992;4:181–185. doi: 10.1016/0952-7915(92)90010-c. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K. Early and late B-cell development in the mouse. Curr Opin Immunol. 1992;4:171–176. doi: 10.1016/0952-7915(92)90008-3. [DOI] [PubMed] [Google Scholar]
- 4.Tsubata T, Nishikawa S. Molecular and cellular aspects of early B-cell development. Curr Opin Immunol. 1991;3:186–192. doi: 10.1016/0952-7915(91)90048-6. [DOI] [PubMed] [Google Scholar]
- 5.Rolink A, Melchers F. Generation and regeneration of cells of the B-lymphocyte lineage. Curr Opin Immunol. 1993;5:207–217. doi: 10.1016/0952-7915(93)90006-e. [DOI] [PubMed] [Google Scholar]
- 6.Melchers F, Rolink A, Grawunder U, Winkler TH, Karasuyama H, Ghia P, Andersson J. Positive and negative selection events during B lymphopoiesis. Curr Opin Immunol. 1995;7:214–227. doi: 10.1016/0952-7915(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 7.Burrows PD, Cooper MD. B-cell development in man. Curr Opin Immunol. 1993;5:201–206. doi: 10.1016/0952-7915(93)90005-d. [DOI] [PubMed] [Google Scholar]
- 8.LeBien TW, Wormann B, Villablanca JG, Law CL, Steinberg LM, Shah VO, Loken MR. Multiparameter flow cytometric analysis of human fetal bone marrow B cells. Leukemia. 1990;4:354–358. [PubMed] [Google Scholar]
- 9.Loken MR, Shah VO, Dattilio KL, Civin CI. Flow cytometric analysis of human bone marrow. II. Normal B lymphocyte development. Blood. 1987;70:1316–1324. [PubMed] [Google Scholar]
- 10.Pontvert-Delucq S, Breton-Gorius J, Schmitt C, Baillou C, Guichard J, Najman A, Lemoine FM. Characterization and functional analysis of adult human bone marrow cell subsets in relation to B-lymphoid development. Blood. 1993;82:417–429. [PubMed] [Google Scholar]
- 11.Lassoued K, Nuñez CA, Billips L, Kubagawa H, Monteiro RC, Le Bien TW, Cooper MD. Expression of surrogate light chain receptors is restricted to a late stage in pre-B cell differentiation. Cell. 1993;73:73–86. doi: 10.1016/0092-8674(93)90161-i. [DOI] [PubMed] [Google Scholar]
- 12.Guelpa-Fonlupt V, Tonnelle C, Blaise D, Fougereau M, Fumoux F. Discrete early pro-B and pre-B stages in normal human bone marrow as defined by surface pseudolight chain expression. Eur J Immunol. 1994;24:257–264. doi: 10.1002/eji.1830240140. [DOI] [PubMed] [Google Scholar]
- 13.Melchers F, Karasuyama H, Haasner D, Bauer S, Kudo A, Sakaguchi N, Jameson B, Rolink A. The surrogate light chain in B-cell development. Immunol Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]
- 14.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt FW, Melchers F. The expression of VpreB/λ5surrogate light chain in early bone marrow precursor B cells of normal and B-cell deficient mutant mice. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 15.Winkler TH, Rolink AG, Melchers F, Karasuyama H. Precursor B cells of mouse bone marrow express two different complexes with the surrogate light chain on the surface. Eur J Immunol. 1995;25:446–450. doi: 10.1002/eji.1830250221. [DOI] [PubMed] [Google Scholar]
- 16.Sanz E, de la Hera A. A novel anti-VpreBantibody identifies immunoglobulin-surrogate receptors on the surface of human pre-B cells. J Exp Med. 1996;183:2693–2698. doi: 10.1084/jem.183.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ten Boekel, E., F. Melchers, and A. Rolink. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- 18.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinman RM. The dendritic cell system and its role in immunogenicity. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 20.Rolink A, Grawunder U, Haasner D, Strasser A, Melchers F. Immature surface Ig+B cells can continue to rearrange κ and λ L chain gene loci. J Exp Med. 1993;178:1263–1270. doi: 10.1084/jem.178.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghia P, Gratwohl A, Signer E, Winkler TH, Melchers F, Rolink AG. Immature B cells from human and mouse bone marrow can change their surface light chain expression. Eur J Immunol. 1995;25:3108–3114. doi: 10.1002/eji.1830251118. [DOI] [PubMed] [Google Scholar]
- 22.Ichihara Y, Matsuoka H, Kurosawa Y. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO (Eur Mol Biol Organ) J. 1988;7:4141–4150. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buluwela L, Albertson DG, Sherrington P, Rabbitts PH, Spurr N, Rabbitts TH. The use of chromosomal translocations to study human immunoglobulin gene organization: mapping DH segments within 35kb of the Cμ gene and identification of a new DHlocus. EMBO (Eur Mol Biol Organ) J. 1988;7:2003–2010. doi: 10.1002/j.1460-2075.1988.tb03039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schable KF, Zachau HG. The variable genes of the human immunoglobulin kappa locus. Biol Chem Hoppe Seyler. 1993;374:1001–1022. [PubMed] [Google Scholar]
- 25.Koyama M, Nakamura T, Higashihara M, Herren B, Kuwata S, Shibata Y, Okumura K. The novel variants of mb-1 and B29 transcripts generated by alternative mRNA splicing. Immunol Lett. 1995;47:151–156. doi: 10.1016/0165-2478(95)00071-x. [DOI] [PubMed] [Google Scholar]
- 26.Drach J, Gattringer C, Huber H. Combined flow cytometric assessment of cell surface antigens and nuclear TdT for the detection of minimal residual disease in acute leukemia. Br J Haematol. 1991;77:37–42. doi: 10.1111/j.1365-2141.1991.tb07945.x. [DOI] [PubMed] [Google Scholar]
- 27.Ehlich A, Matin V, Müller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 28.Tonegawa S. Somatic generation of antibody diversity. Nature (Lond) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 29.Graninger WB, Goldman PL, Morton CC, O'Brien SJ, Korsmeyer SJ. The κ-deleting element. J Exp Med. 1988;167:488–501. doi: 10.1084/jem.167.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siminovitch KA, Bakhshi A, Goldman P, Korsmeyer SJ. A uniform deleting element mediates the loss of κ genes in human B cells. Nature (Lond) 1985;316:260–262. doi: 10.1038/316260a0. [DOI] [PubMed] [Google Scholar]
- 31.Grawunder U, Leu TMJ, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG-1 and RAG-2 gene expression in pre-B cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 32.Nuñez C, Nishimoto N, Gastland GL, Billips LG, Burrows PD, Kubagawa H, Cooper MD. B cells are generated throughout life in humans. J Immunol. 1996;156:866–872. [PubMed] [Google Scholar]
- 33.Lassoued K, Illges H, Benlagha K, Cooper MD. Fate of surrogate light chains in B lineage cells. J Exp Med. 1996;183:421–429. doi: 10.1084/jem.183.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978;121:641–647. [PubMed] [Google Scholar]
- 35.Karasuyama H, Melchers F, Rolink A. A complex of glycoproteins is associated with VpreB/λ5surrogate light chain on the surface of μ heavy chain-negative early precursor B cell lines. J Exp Med. 1993;178:469–478. doi: 10.1084/jem.178.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F. IL-2 receptor α chain (CD25, TAC) expression defines a crucial stage in preB cell development. Int Immunol. 1994;6:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- 37.Küppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO (Eur Mol Biol Organ) J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]