Proteolytic Activation of Protein Kinase C δ by an ICE/CED 3-like Protease Induces Characteristics of Apoptosis (original) (raw)
Abstract
Recent studies have shown that protein kinase C (PKC) δ is proteolytically activated at the onset of apoptosis induced by DNA-damaging agents, tumor necrosis factor, and anti-Fas antibody. However, the relationship of PKCδ cleavage to induction of apoptosis is unknown. The present studies demonstrate that full-length PKCδ is cleaved at DMQD330N to a catalytically active fragment by the cysteine protease CPP32. The results also demonstrate that overexpression of the catalytic kinase fragment in cells is associated with chromatin condensation, nuclear fragmentation, induction of sub-G1 phase DNA and lethality. By contrast, overexpression of full-length PKCδ or a kinase inactive PKCδ fragment had no detectable effect. The findings suggest that proteolytic activation of PKCδ by a CPP32-like protease contributes to phenotypic changes associated with apoptosis.
The protein kinase C (PKC) family consists of multiple subspecies that possess a conserved catalytic domain. The classic or group A isoforms (α, β, and γ) require Ca2+ for activity and contain cysteine-rich motifs that confer phospholipid-dependent binding of diacylglycerol (1). The group A PKCs are cleaved at the third variable region (V3) by the neutral proteases, calpains I and II, to catalytically active fragments (2). Recent studies have demonstrated that the Ca2+-independent δ isoform, and not the group A PKCs, is selectively cleaved at V3 to a catalytically active fragment in cells induced to undergo apoptosis (3, 4). Inhibition of apoptosis by overexpression of Bcl-2 or Bcl-xL is associated with a block of PKCδ cleavage (3, 4). The finding that PKCδ is cleaved at a site (DMQD/N) adjacent to aspartic acid has supported the potential involvement of aspartate-specific cysteine proteases which are known to be activated during apoptosis.
The nematode Ced-3 cysteine protease is related to the mammalian interleukin-1β converting enzyme (ICE) (5, 6). The demonstration that overexpression of Ced-3 or ICE induces apoptosis has provided support for involvement of these cysteine proteases in cell death pathways (7). ICE/ Ced-3 family members include Nedd2/Ich-1, CPP32/ YAMA/apopain, Tx/Ich-2/ICErelII, ICErelIII, Mch2, Mch3/ ICE-LAP3/CMH-1 (reviewed in reference 8), ICE-LAP6 (9), FLICE/Mch5 (10, 11), and Mch4 (11). ICE cleaves the precursor of IL-1β to the active cytokine (6, 12, 13). Other known substrates of the ICE/Ced-3 family include: (a) poly (ADP-ribose) polymerase (PARP) which is cleaved by CPP32, Mch3 and Ced-3, but not ICE (14–16); and (b) DNA-dependent protein kinase (DNA-PK), the U1 small nuclear ribonucleoprotein and D4-GDP dissociation inhibitor for the Rho family GTPases (D4-GDI), which are cleaved by CPP32 (17, 18). However, the functional role of these cleavage products in the induction of apoptosis is unclear.
The present results demonstrate that PKCδ is cleaved by CPP32 and not certain other ICE/Ced-3 family members. We also demonstrate that overexpression of the PKCδ catalytic fragment is involved in the induction of phenotypic changes that are characteristic of apoptosis.
Materials and Methods
In Vitro Cleavage of PKCδ and PARP.
The full-length PKCδ cDNA was cloned into the SpeI and BamH1 sites of a modified pSVβ plasmid (Clontech, Palo Alto, CA). PKCδ(D327A/D330A) was generated in two steps by overlapping primer extension. PARP cDNA was generated by PCR cloning. The proteins were labeled with [35S]methionine by coupled transcription and translation reactions (Promega, Madison, WI). Labeled proteins were incubated with 5 μg/ml _Escherichia coli_-derived CPP32β in 50 mM Hepes (pH 7.5), 10% glycerol, 2.5 mM DTT, and 0.25 mM EDTA at room temperature for 30 min. The reaction products were analyzed by electrophoresis in 10-20% SDS–polyacrylamide gels and then autoradiography. For the kinase assays, full-length PKCδ, PKCδ(D327A/D330A), PKCδ catalytic fragment (CF), and PKCδCF(K-R) were prepared by coupled transcription and translation. PKCδ and PKCδ(D327A/D330A) were incubated with 5 μg/ml CPP32β at room temperature for 30 min. Protein kinase assays using MBP as a substrate were performed as described (PKC Assay Kit; GIBCO BRL, Gaithersburg, MD).
Analysis of Peptide Proteolysis.
Peptides were synthesized and purified to ∼95% by standard methods and confirmed by mass spectrometry. Reaction mixtures (810 μl) contained: 100 mM Hepes (pH 7.5), 20% (vol/vol) glycerol, 5 μM dithiothreitol, 0.5 mM EDTA, and 380 ng _N_-His CPP32 (19). Peptide substrates were added to final concentrations of 10 μM. The reaction mixtures were incubated at 30°C. Aliquots were removed at 10 min intervals for 60 min and added to vials containing 3 M HCl to stop the reactions. The amount of substrate remaining at each time was quantitated by reverse phase HPLC. Data were fit to the equation (St/So) = e−kt, where k is the decay rate constant equal to Vmax/Km. Observed Vmax/Km values were normalized to 1.00 for the PARP peptide.
Cell Transfections.
Cells were seeded at a density of 1.7 × 105 in each well of 6-well dishes 24 h before transfection. For each well, 2 μg DNA construct and 0.5 μg pSvβ plasmid containing β-gal were coprecipitated with calcium phosphate. Cells were incubated with the coprecipitate for 30 h at 37°C and then analyzed by X-gal staining. Cells (1.7 × 105/well) were also transfected with 2 μg DNA construct for 30 h at 37°C, fixed with 4% paraformaldehyde, postfixed with 5% acetic acid in ethanol and then stained with 5 μg/ml Hoechst dye. For sub-G1 DNA content, cells transfected by lipofectamine were stained with propidium iodide and monitored by FACScan®. Chromatin condensation was assessed by staining with acridine orange and ethidium bromide (20).
Results and Discussion
To determine whether PKCδ is cleaved by one of the known ICE-like proteases, full-length 78-kD PKCδ labeled with [35S]methionine was incubated with purified recombinant proteases. Cleavage of PKCδ to a 40-kD fragment was observed with purified CPP32β (14) (Fig. 1 A). In contrast, ICE failed to cleave PKCδ at concentrations up to 600 U/μl (3). The related Ich-1, Ich-2, Mch2, Mch3, and ICErelIII proteases also failed to cleave PKCδ (data not shown). Because PKCδ is cleaved at DMQD330N in vivo (3, 4), we asked whether this site is responsible for CPP32mediated cleavage in vitro. CPP32 may prefer peptidic substrates with aspartic acid at the P1 and P4 positions (15). Consequently, we prepared a PKCδ mutant with substitution of D327A and D330A. Incubation with CPP32 resulted in no detectable CPP32-mediated cleavage of this mutant to the 40-kD catalytic fragment, while there was partial digestion to a species of ∼55 kD (not observed with wild-type substrate) (Fig. 1 A). Recombinant CPP32 also cleaved the 116-kD full-length PARP to the predicted 85-kD fragment (14, 15) (Fig. 1 A). Using peptides derived from the cleavage sites of PARP and PKCδ in proteolytic assays, we found that CPP32 cleaves both substrates and not a peptide spanning the IL-1β maturation site (Table 1). These findings confirm that PKCδ, like PARP, is a substrate for CPP32.
Figure 1.

PKCδ is proteolytically activated by CPP32 in vitro. (A) PKCδ (full-length: FL), PKCδ(D327A/D330A) and PARP were labeled with [35S]methionine and incubated with recombinant CPP32β. The reaction products were analyzed by SDS-PAGE and autoradiography. The kinase active PKCδ catalytic fragment (CF) and the kinase inactive PKCδCF(K-R) were labeled with [35S]methionine and analyzed under similar conditions. (B) Recombinant PKCδ and PKCδ (D327A/D330A) were incubated with CPP32 and then assayed for protein kinase activity using MBP as substrate.
Table 1.
CPP32 Proteolysis of Peptides Spanning the PARP, PKCδ, and IL-1β Cleavage Sites
| Substrate | Sequence | Relative Vmax/Km |
|---|---|---|
| PARP | Ac-WGDEVD216-GVDEVW-NH2 | 1.00 |
| PKCδ | Ac-GEDMQD330NSGTYW-NH2 | 0.42 |
| IL-1β | Ac-NEAYVHD116APVRSLY-NH2 | 0.00 |
We also asked whether cleavage of PKCδ by CPP32 is associated with activation of the kinase function. Fulllength PKCδ exhibited a low level of myelin basic protein (MBP) phosphorylation, while incubation with CPP32 resulted in a greater than sixfold increase in kinase activity (Fig. 1 B). In contrast, CPP32 had no detectable effect on kinase function of the PKCδ(D327A/D330A) mutant (Fig. 1 B). A recombinant 40-kD CF of PKCδ (amino acids 331676) exhibited constitutive kinase activity, while a mutant of the fragment with K-378 in the ATP binding site mutated to R (K378R; designated K-R) yielded background levels of MBP phosphorylation found with control bacterial lysates (Figs. 1, A and B). These findings collectively demonstrate that CPP32-mediated cleavage of the DMQD330N site activates PKCδ.
To study the role of PKCδ in apoptosis, we used the transient HeLa cell transfection system previously found to demonstrate induction of apoptosis by ICE-like proteases (7). Cotransfection of the kinase inactive PKCδCF(K-R) mutant with the β-galactosidase (β-gal) marker gene had little effect on HeLa cell morphology (Fig. 2 A). Most of the blue X-gal positive cells remained flat and attached to the dish (Fig. 2 A). Cotransfection of the kinase active PKCδCF and β-gal resulted in condensed, small blue cells (Fig. 2 B), consistent with the induction of apoptosis (7). Similar findings were obtained with NIH3T3 cells (Figs. 2, C and D). Overexpression of PKCδ in both cell types also resulted in detachment of non-viable cells into the culture medium.
Figure 2.
Transfection of the PKCδ catalytic fragment (CF) induces morphologic changes characteristic of apoptosis. HeLa (upper panels) and NIH3T3 (lower panels) cells were cotransfected with pSvβ-β-gal and vectors expressing: (A and C) kinase inactive PKCδCF(K-R) and (B and D) kinase active PKCδCF. Transfection was determined by X-gal staining and apoptotic cells were identified by their condensed morphology.
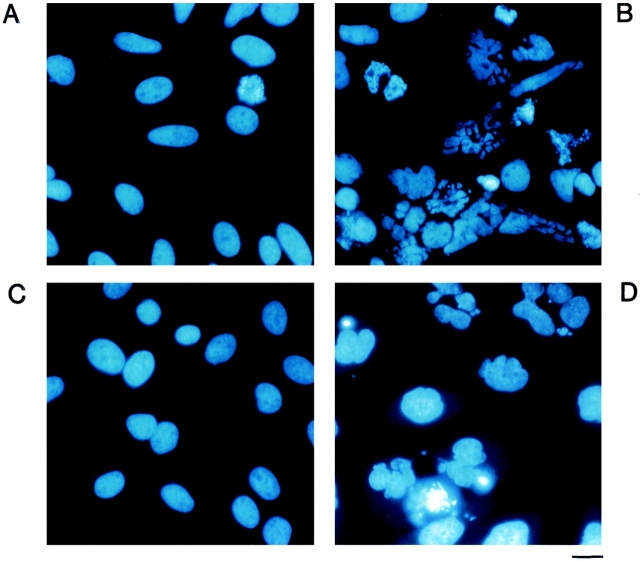
Hoechst staining of HeLa cells transfected with a vector that expresses full-length PKCδ had no detectable changes in nuclear morphology (Fig. 3 A), but overexpression of PKCδCF resulted in fragmented nuclei (Fig. 3 B). Transfection of kinase inactive PKCδCF(K-R) was associated with a normal nuclear morphology (Fig. 3 C). The changes observed with expression of the PKCδ CF were also compared to those found upon exposure to 1-β-d-arabinofuranosylcytosine (ara-C), a DNA-damaging agent that induces proteolytic cleavage of PKCδ and apoptosis (4). Treatment of HeLa cells with ara-C resulted in a similar pattern of nuclear fragmentation (Fig. 3 D).
Figure 3.
Expression of PKCδCF results in nuclear fragmentation. HeLa cells were transfected with vectors that express: (A) full-length PKCδ; (B) kinase active PKCδCF; and (C) kinase inactive PKCδCF(K-R). (D) Cells were exposed to 2 μM ara-C. The cells were fixed with paraformaldehyde and then stained with Hoechst dye. Cotransfection of PKCδFL, PKCδCF and PKCδCF(K-R) with pSvβ-β-gal demonstrated transfection efficiencies of 50, 42, and 43%, respectively. Percentage of apoptotic cells for the PKCδFL, PKCδCF, and PKCδCF(K-R) transfected populations was 2, 26, and 4%, respectively. Bar, 15 μM.
To confirm that the nuclear changes induced by PKCδCF are associated with induction of apoptosis, we assessed the effects of transfection on the appearance of HeLa cells with sub-G1 DNA content. Transfection of the empty vector, full-length PKCδ or PKCδCF(K-R) resulted in 10-15% of cells with sub-G1 DNA (Fig. 4 A and data not shown). By contrast, transfection of PKCδCF was associated with 30– 35% of cells with sub-G1 DNA (Fig. 4 A). Cells were also stained with acridine orange and ethidium bromide to assess chromatin condensation (20). Transfection of PKCδCF, but not PKCδCF(K-R), resulted in the appearance of bright yellow-green nuclear staining of condensed chromatin (Fig. 4 B).
Figure 4.
Overexpression of PKCδCF induces sub-G1 DNA and chromatin condensation. (A) HeLa cells were transfected with PKCδFL, PKCδCF, or PKCδCF(K-R). Cells were assessed for DNA content by flow cytometry at 48 h after transfection. The small triangle denotes G0/G1 DNA. (B) HeLa cells transfected with PKCδCF(K-R) (left) and PKCδCF (right) were assessed for chromatin condensation after staining with acridine orange and ethidium bromide.
To quantify the effects of PKCδCF expression on cell viability, we cotransfected PKCδCF or PKCδCF(K-R) and the green fluorescence gene (Clontech) into HeLa cells. Positive transfectants were selected by flow cytometry, reseeded in culture medium and assayed at 24 h for viability by trypan blue exclusion. Less than 5% of the PKCδCF transfectants were viable, while over 90% of the kinase inactive PKCδCF(K-R) transfectants were viable and attached to the dish. Viability of 90–95% was observed after transfection of the null vector and sorting. We conclude that the kinase active catalytic domain of PKCδ induces characteristics typical of cells undergoing apoptosis: (a) size reduction and round morphology; (b) nuclear fragmentation; (c) chromatin condensation; (d) sub-G1 DNA content; and (e) detachment and loss of viability.
Multiple events that lead to destruction of nuclear and cytoplasmic integrity are probably required for apoptosis. Activation of ICE-family proteases may be a central trigger, resulting in the cleavage of substrates such as PARP (21), lamin B1 (22, 23), topoisomerase 1 (23), D4-GDI (18), DNA-PK, and the U1 small nuclear ribonucleoprotein (17). PKCδ, but not PKCα, β, ε, or ζ, is also cleaved at the onset of apoptosis (3, 4). Little is known about the physiological function of PKCδ (24, 25). We demonstrate that PKCδ is cleaved by CPP32 and not other ICE/Ced-3 family members in vitro. The results also demonstrate that expression of the PKCδ catalytic fragment induces morphologic changes characteristic of apoptosis. We propose that the proteolytic cleavage of PKCδ is a key mediator of nuclear fragmentation and cell death, and not a bystander effect of protease activation. Moreover, the finding that proteolytic activation of PKCδ is blocked by Bcl-2 and Bcl-xL suggests that these anti-apoptotic proteins act upstream to this event (3). Elucidation of the substrates phosphorylated as a consequence of PKCδ cleavage should provide insights into the pathways activated by the catalytic fragment.
Footnotes
This investigation was supported by Public Health Service grants CA66996, CA55241, and CA29431 awarded by the National Cancer Institute, DHHS.
References
- 1.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science (Wash DC) 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto, A. 1990. Limited proteolysis of protein kinase C by calpain, its possible implication. In The Biology and Medicine of Signal Transduction. Y. Nishizuka, M. Endo, and C. Tanaka, editors. Raven Press, New York. 472–477.
- 3.Emoto Y, Manome G, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong WW, Kamen R, Weichselbaum R, Kufe D. Proteolytic activation of protein kinase C δ by an ICE-like protease in apoptotic cells. EMBO (Eur Mol Biol Organ) J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emoto Y, Kisaki H, Manome Y, Kharbanda S, Kufe D. Activation of protein kinase C δ in human myeloid leukemia cells treated with 1-β-d-arabinofuranosylcytosine. Blood. 1996;87:1990–1996. [PubMed] [Google Scholar]
- 5.Hugunin M, Quintal LJ, Mankovich JA, Ghayur T. Protease activity of in vitro transcribed and translated Caenorhabditis elegans cell death gene (ced-3) product. J Biol Chem. 1996;271:3517–3522. doi: 10.1074/jbc.271.7.3517. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. eleganscell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell. 1993;75:641–752. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 7.Miura M, Zhu H, Rotello R, Hartweig EA, Yuan J. Induction of apoptosis in fibroblasts by IL-1β-converting enzyme, a mammalian homolog of the C. eleganscell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson DW. ICE/CED3-like proteases as therapeutic targets for the control of inappropriate apoptosis. Nat Biotechnol. 1996;14:297–301. doi: 10.1038/nbt0396-297. [DOI] [PubMed] [Google Scholar]
- 9.Duan H, Orth K, Chinnaiyan AM, Poirier GG, Froelich CJ, He W-W, Dixit VM. ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B. J Biol Chem. 1996;271:16720–16724. doi: 10.1074/jbc.271.28.16720. [DOI] [PubMed] [Google Scholar]
- 10.Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, et al. FLICE, a novel FADD-homologous ICE/ CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F, Fritz L, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Ness KV, Greenstreet TA, March CJ, Kronheim SR, Druck T, Canizzaro LA, et al. Molecular cloning of the interleukin-1β-converting enzyme. Science (Wash DC) 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 13.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature (Lond) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 14.Tewari M, Quan LT, O'Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Yama/CPP32β, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and apoptosis. Nature (Lond) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, Takahashi A, Armstrong R, Krebs J, Fritz L, Tomaselli KJ, Wang L, Yu Z, Croce CM, Salveson G, Earnshaw WC, Litwack G, Alnemri ES. Mch3, a novel human apoptotic cysteine protease highly related to CPP32. Cancer Res. 1995;55:6045–6052. [PubMed] [Google Scholar]
- 17.Casciola-Rosen L, Nicholson DW, Chong T, Rowan KR, Thornberry NA, Miller DK, Rosen A. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Na S, Chuang T-H, Cunningham A, Turi TG, Hanke JH, Bokoch GM, Danley DE. D4-GDI, a substrate of CPP32, is proteolyzed during Fas-induced apoptosis. J Biol Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- 19.Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science (Wash DC) 1995;269:18851888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 20.McGahon AJ, Martin SJ, Bissonnette RP, Mahboubi A, Shi Y, Mogli RJ, Nishioka WK, Green DR. The end of the (cell) line: methods for the study of apoptosis in vitro. Methods Cell Biol. 1995;46:153–185. doi: 10.1016/s0091-679x(08)61929-9. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 22.Neamati N, Fernandez A, Wright S, Kiefer J, McConkey DJ. Degradation of lamin B1 precedes oligonucleosomal DNA fragmentation in apoptotic thymocytes and isolated thymocyte nuclei. J Immunol. 1995;154:3788–3795. [PubMed] [Google Scholar]
- 23.Voelkel-Johnson C, Entingh AJ, Wold WSM, Gooding LR, Laster SM. Activation of intracellular proteases is an early event in TNF-induced apoptosis. J Immunol. 1995;154:1707–1716. [PubMed] [Google Scholar]
- 24.Watanabe T, Ono Y, Taniyama Y, Hazama K, Igarashi K, Ogita K, Kikkawa U, Nishizuka Y. Cell division arrest induced by phorbol ester in CHO cells overexpressing protein kinase C-δ subspecies. Proc Natl Acad Sci USA. 1992;89:10159–10163. doi: 10.1073/pnas.89.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mischak H, Goodnight J, Kolch W, Martiny-Baron G, Schaechtle C, Kazanietz MG, Blumberg PM, Pierce JH, Mushinski JF. Overexpression of protein kinase c-δ and -ε in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence, and tumorigenicity. J Biol Chem. 1993;268:6090–6096. [PubMed] [Google Scholar]