Regulation of the Receptor Specificity and Function of the Chemokine RANTES (Regulated on Activation, Normal T Cell Expressed and Secreted) by Dipeptidyl Peptidase IV (CD26)-mediated Cleavage (original) (raw)
Abstract
CD26 is a leukocyte activation marker that possesses dipeptidyl peptidase IV activity but whose natural substrates and immunological functions have not been clearly defined. Several chemo-kines, including RANTES (regulated on activation, normal T cell expressed and secreted), have now been shown to be substrates for recombinant soluble human CD26. The truncated RANTES(3–68) lacked the ability of native RANTES(1–68) to increase the cytosolic calcium concentration in human monocytes, but still induced this response in macrophages activated with macrophage colony-stimulating factor. Analysis of chemokine receptor messenger RNAs and patterns of desensitization of chemokine responses showed that the differential activity of the truncated molecule results from an altered receptor specificity. RANTES(3–68) showed a reduced activity, relative to that of RANTES(1–68), with cells expressing the recombinant CCR1 chemokine receptor, but retained the ability to stimulate CCR5 receptors and to inhibit the cytopathic effects of HIV-1. Our results indicate that CD26-mediated processing together with cell activation–induced changes in receptor expression provides an integrated mechanism for differential cell recruitment and for the regulation of target cell specificity of RANTES, and possibly other chemokines.
Monocytes differentiate into macrophages as they migrate from the blood to tissues during immune surveillance. At sites of inflammation, monocyte infiltration and macrophage accumulation are coordinated, in part, by chemokines (1). The mechanisms that control the recruitment of monocytes and macrophages by chemoattractants have not been clearly defined, but they may include regulation of the expression of chemokines and their receptors (2) as well as the modification of chemokine activity by posttranslational processing (3–5). Several chemokines share a conserved NH2-X-Pro sequence (where X is any amino acid) at the NH2 terminus (6), which conforms to the substrate specificity of dipeptidyl exopeptidase IV (DPPIV; reference 7).1 DPPIV cleaves the first two amino acids from peptides with penultimate proline or alanine residues, although no natural substrate with immune function has been identified. This enzyme is also a leukocyte differentiation antigen, known as CD26 (8–10), that is expressed on the cell surface mostly by T lymphocytes and macrophages. Expression of CD26 has been associated with T cell activation (8–10) and with susceptibility of a T cell line to infection with macrophage-tropic HIV-1 (11).
In this study, we identify the chemokines RANTES (regulated on activation, normal T cell expressed and secreted), interferon-γ-inducible protein monocyte chemotactic protein (MCP)–2, eotaxin, and IP-10 as the first natural CD26 substrates with immune function. It is shown that the cleavage product of RANTES is a chemokine agonist with altered receptor specificity. We also describe, for the first time, differential changes in the expression pattern of chemokine receptors after activation of monocytes by M-CSF. Therefore, target cell recruitment into inflammatory sites may depend both on the extent of CD26 activity on chemokines and on the maturational status of the responding cells.
Materials and Methods
Cell Cultures and Transfections.
Monocytes were isolated from human PBMCs of healthy donors by counter-current centrifugal elutriation. Monocyte-derived macrophages were prepared by culturing monocytes for 6 d at a density of 106 cells/ml in serum-free macrophage medium (GIBCO BRL, Gaithersburg, MD) supplemented with recombinant human (rh) M-CSF (10 ng/ml; R&D Systems, Inc., Minneapolis, MN).
Human embryonic kidney (HEK)-293 cells grown to confluence in DMEM supplemented with 10% heat-inactivated FCS, penicillin, streptomycin, 2 mM glutamine, and 10 mM hepes (pH 7.4) were transfected with plasmid DNA encoding CCR5 (12). CD4-positive human osteosarcoma (HOS-CD4) cell lines transfected with individual chemokine receptor cDNAs were obtained from N. Landau (Aaron Diamond AIDS Research Center, New York), and were grown in the above culture medium supplemented with puromycin.
The derivative of the PM1 cell line chronically infected with the recombinant HIV-1 clone MV3-HXB2 has been previously described (11).
sCD26 Cleavage and Electrospray Mass Spectrometry.
To create the recombinant soluble (s) human CD26 construct, a signal peptidase cleavage consensus sequence was introduced in the pTZ-CD26.11 cDNA (13) by a Leu to Ala substitution at residue 28. To obtain enzyme negative construct, the Ser at residue 630 was further replaced by Ala. The two constructs were cloned into the pEE14.HCMV expression vector and transfected into CHO-K1 cells (14). The enzymatically active (E+) and enzymatically deficient (E−) sCD26 proteins were purified from cell culture supernatants of stable transfectants, and were tested in Western blotting and DPPIV enzyme assays (15). Both proteins had a relative molecular weight of 110 kD and bound equally well to several CD26 mAbs, but only the E+ sCD26 showed detectable DPPIV activity.
rhRANTES, MCP-1, MCP-2, eotaxin, and IP-10 (100 nM; all from PeproTech, Rocky Hill, NJ) were incubated overnight at 37°C with different amounts of E+ or E− sCD26 in 50 μl of PBS. Samples were desalted and concentrated by using a peptide trap (Michrom BioResources, Inc., Auburn, CA), or a reversed-phase–HPLC interface. Electrospray mass spectrometry (ES-MS) analysis of samples was performed in 50% acetonitrile, supplemented with 0.1% (vol/vol) glacial acetic acid, using a Finnigan MAT (San Jose, CA) TSQ 7000 triple-stage quadrupole mass spectrometer. Several scans were summed to obtain the final spectrum.
Peptide Synthesis.
Full-length and truncated RANTES were synthesized with an Applied Biosystems, Inc. (Foster City, CA) peptide synthesizer according to fluorenyl methoxycarbonyl (FMOC) chemistry. FMOC-protected amino acids were added stepwise with ninhydrin monitoring at each cycle. The peptides were folded by air oxidation and purified by reversed-phase– HPLC. Peptide sequences were confirmed by amino acid analysis and Edman sequence analysis, and the molecular masses were confirmed by ES-MS analysis. There was no substantial difference in the activities of chemically synthesized full-length RANTES and rhRANTES(1-68) as judged by the Ca2+ influx and anti– HIV-1 assays used in this study.
Colorimetric DPPIV Enzyme Assay.
The _p_-nitroanilide (_p_NA)– conjugated Gly-Pro dipeptide substrate and test competitors were mixed and added to human placental DPPIV (Enzyme Systems Products, Dublin, CA), and the resulting mixture was incubated at room temperature in a final volume of 150 μl containing 50 mM tris-HCl (pH 8.0) and 0.15 M NaCl. The final concentrations of DPPIV and Gly-Pro-_p_NA were 1.25 mU/ml and 400 μM, respectively. The kinetics of the enzyme reaction were monitored by measuring absorbance at 405 nm with a Vmax kinetic microplate reader (Molecular Devices Corp., Menlo Park, CA). The percentage of inhibition of enzyme activity was calculated from the maximal velocity for each sample and from that apparent in the absence of competitor (100% activity).
Reverse Transcriptase–PCR Analysis.
Isolated total cellular RNA of monocytes was subjected to first-strand cDNA synthesis. PCR amplification of cDNA was performed for 30 cycles (92°C for 1 min, 40°C for 1 min, 72°C for 1 min) with primers specific for CCR1, CCR2b, CCR3, CCR5, CXCR4, and glyceraldehyde phosphate dehydrogenase (GAPDH). Separated products were stained with SYBR Green I (Molecular Probes, Eugene, OR).
Cytosolic Calcium Measurements.
Cells (107/ml) were washed and incubated in the dark at 37°C for 45 min in Ca2+ buffer (136 mM NaCl, 4.8 mM KCl, 5 mM glucose, 1 mM CaCl2, 20 mM Hepes, pH 7.4) supplemented with 5 μM Fura-2 acetoxymethyl ester that had been premixed with 10% Pluronic® F-127 (Molecular Probes). The cells were then washed and resuspended at 2 × 106 cells/ml in Ca2+ buffer containing BSA (1 mg/ml), and portions (2 ml) of the cell suspension were exposed at different time points in a stirred cuvette at 37°C to chemokines. Fluorescence was monitored with a δ scan (Photon Technology Intl., Monmouth Junction, NJ), and data were recorded as the relative ratio of fluorescence at excitation wavelengths of 340 and 380 nm, with emission measured at 510 nm. After each measurement, maximal and minimal fluorescence were assessed by addition of 20 μM ionomycin followed by 5 mM MnCl2.
Assay for HIV-1–induced Cytopathicity.
HOS-CD4.CCR5 cells (2 × 104) were incubated for 1 h at 37°C with RANTES variants in 150 μl of culture medium containing 20% FCS, and were then mixed with 50 μl (2 × 105 cells/ml) of uninfected PM1 cells or PM1 cells chronically infected with MV3-HXB2 virus. After 3 d, photomicrographs of cultures were taken and cell viability was measured by adding of 50 μl of 1 mg/ml 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2_H_-tetrazolium-5-carboxanilide solution containing 20 μM phenazine methosulfate and recording the OD at 450 nm. Data are expressed as the percentage of inhibition of cytopathicity [calculated as 100% × (R − V)/(U − V), where U, V, and R represent OD values obtained for HOS-CD4.CCR5 cells cultured with uninfected PM1 cells, or with HIV-1–infected cells in the absence or presence of chemokine, respectively].
Results
RANTES, MCP-2, eotaxin, and IP-10 Are Substrates of CD26.
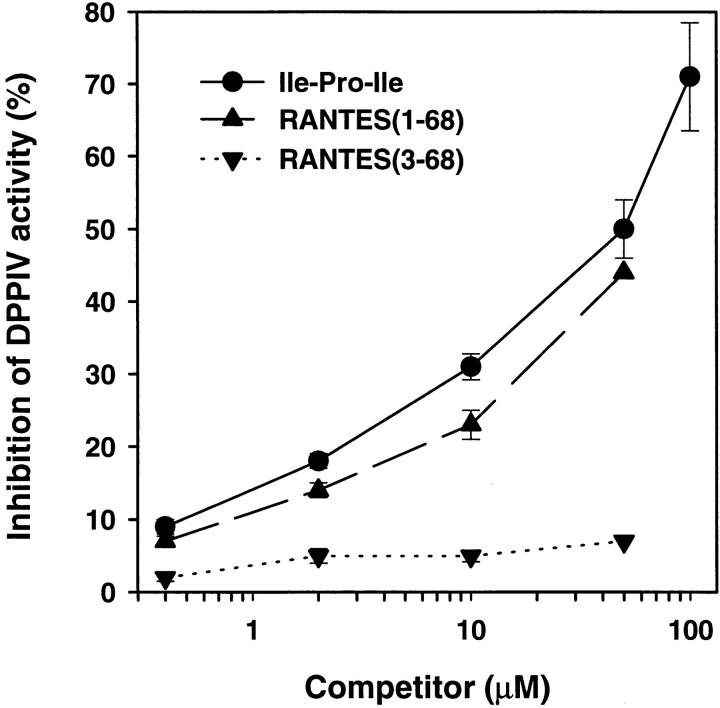
ES-MS analysis revealed that 100 nM rhRANTES underwent partial to complete hydrolysis when incubated overnight at 37°C with increasing amounts (25–250 μU) of sCD26 (Fig. 1). Taking into account cationization (K+) of the multiply charged ions, the measured molecular masses of the native and degraded polypeptides corresponded to the theoretical masses of full-length (residues 1–68) and truncated (residues 3–68) forms of RANTES, respectively. The calculated difference between the molecular masses of the native and the truncated forms ranged from 183 to 185 daltons, which is consistent with the expected mass (184 daltons) of a released Ser-Pro dipeptide, the predicted NH2 terminus of RANTES (16). In contrast to the effect of enzymatically active sCD26, shortened RANTES was not generated by incubation of the chemokine with a mutant sCD26 deficient in enzyme activity (Fig. 1). RANTES also inhibited, possibly in a competitive manner, the rapid hydrolysis of a _p_NA-conjugated Gly-Pro dipeptide by human placental DPPIV, as measured in a colorimetric enzyme assay (Fig. 2). The efficacy of inhibition by chemically synthesized RANTES(1–68) was similar to that observed with the DPPIV substrate and competitive inhibitor Ile-Pro-Ile (Diprotin A; reference 17), whereas RANTES(3–68) did not inhibit the reaction.
Figure 1.
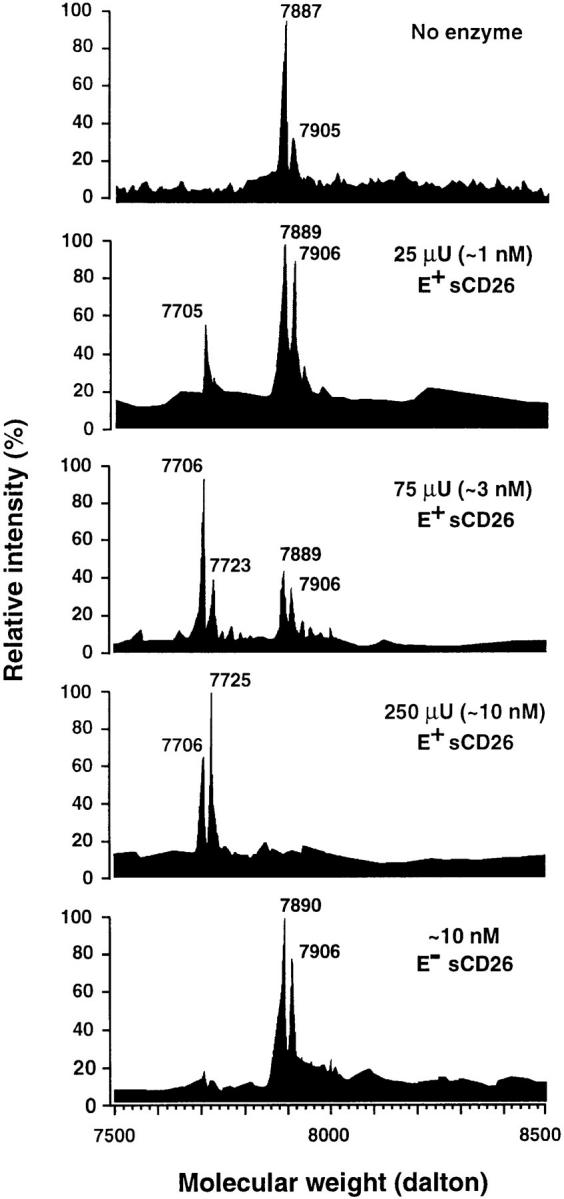
RANTES cleavage products after digestion with sCD26. RANTES was incubated overnight with the indicated amounts of E+ or sCD26 and samples were subjected to ES-MS analysis. The peaks in the spectrum at masses (m) of 7,905 to 7,906 and 7,887 to 7,890 are tentatively identified as [M + K+]+ of RANTES with (7,904 daltons) and without (7,886 daltons) a molecule of H2O, respectively; the labeled peaks at the left of the spectrum correspond to each of these molecular ions minus a Ser-Pro dipeptide (184 daltons).
Figure 2.
Competitive inhibition of DPPIV by RANTES(1–68). Colorimetric DPPIV enzyme assay was performed using human placental DPPIV and the Gly-Pro-_p_NA substrate, in the presence or absence of the test competitors Ile-Pro-Ile, RANTES(1–68), or RANTES(3–68); the competitor concentration is indicated on the horizontal axis. Data are means ± SEM (n = 3), except for the highest concentration of RANTES(1–68) and RANTES(3–68), for which only one sample was assayed in order to conserve material. Similar results were obtained in a repeat experiment.
Sensitivity to CD26-mediated cleavage was not a unique property of RANTES (Table 1). Cleavage products with the predicted molecular masses were also evident in samples of MCP-2, eotaxin, and IP-10 after incubation with sCD26. In contrast, MCP-1, which has a 62% sequence similarity with MCP-2 including the NH2-terminal Gln-Pro dipeptides, was not cleaved by the enzyme under the same experimental conditions.
Table 1.
Chemokine Cleavage Products after Digestion with sCD26
| Chemokine | NH2-terminal dipeptide | CD26 cleavage | Molecular masses by mass spectrometry (Da) | |||
|---|---|---|---|---|---|---|
| Full length | Truncated | |||||
| Theoretical | Observed | Theoretical | Observed | |||
| Eotaxin | GP | Yes | 8,361 | 8,361 | 8,207 | 8,207 |
| IP-10 | VP | Yes | 8,633 | 8,637/8,751* | 8,437 | 8,440/8,555* |
| MCP-1 | QP | No | 8,681 | 8,678 | 8,456 | ND§ |
| MCP-2 | QP | Yes | 8,910 | 8,909 | 8,685 | 8,686/8,703‡ |
CD26-specific Truncation of RANTES Modifies its Target Cell Specificity.
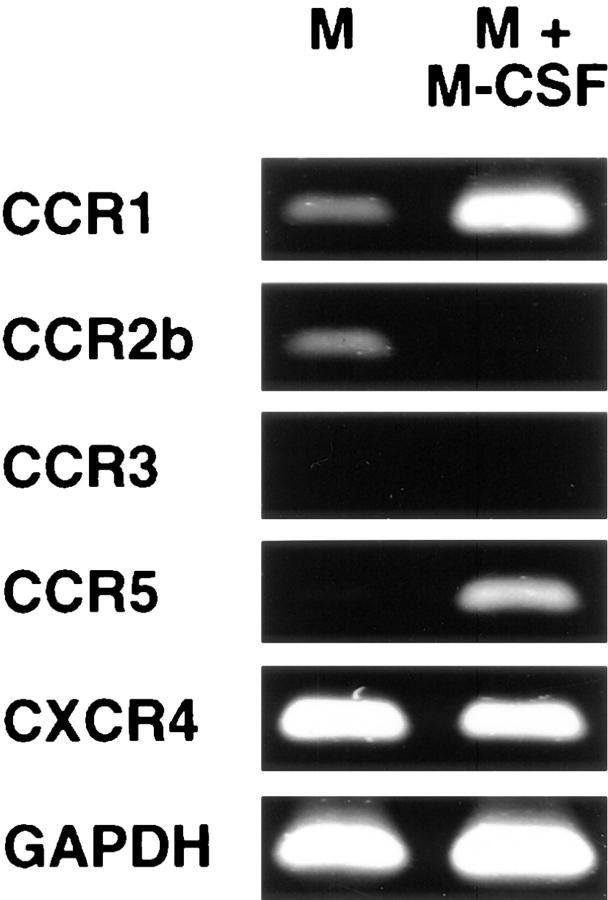
To investigate the functional significance of DPPIV-mediated truncation of RANTES, we compared the effects of chemically synthesized RANTES(1–68) and RANTES(3–68) on monocytes and monocyte-derived macrophages. Both resting cells and cells activated with M-CSF were analyzed because reverse transcriptase–PCR revealed marked changes in the abundance of chemokine receptor transcripts in response to M-CSF activation (Fig. 3). In resting cells, transcripts encoding the chemokine receptors CCR1, CCR2b, or CXCR4, as well as control GAPDH mRNA, were readily detectable, whereas CCR5 receptor transcripts were virtually absent. After differentiation to macrophages, the intensity of the CXCR4 and GAPDH signals remained virtually unchanged, whereas the abundance of CCR1 and CCR5 mRNAs increased substantially and the CCR2b transcript virtually disappeared. CCR3 mRNA was not detected in either cell type.
Figure 3.
Reverse transcriptase– PCR analysis of chemokine receptor transcripts in monocytes cultured in the absence (M) or presence (M + M-CSF) of M-CSF. Total cellular RNA was subjected to reverse transcriptase– PCR analysis as described in Materials and Methods. Control reactions performed without reverse transcriptase were negative for each PCR product.
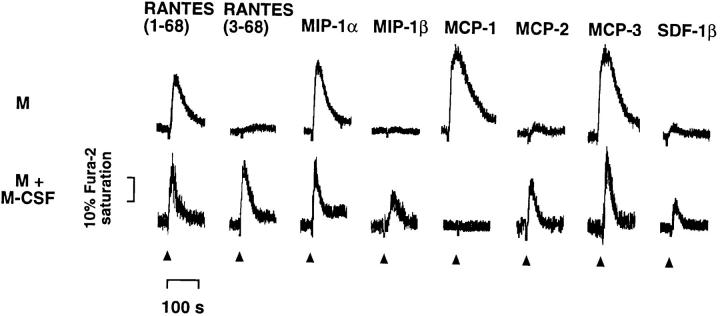
Transient changes in the cytosolic free Ca2+ concentration ([Ca2+]i) were recorded after stimulation of monocytes or macrophages with an optimal concentration of RANTES- (1–68) or RANTES(3–68), and the effects were compared with those of other chemokines (Fig. 4). Addition of 100 nM RANTES(1–68) to cells loaded with the fluorescent Ca2+ probe Fura-2 induced a rapid increase in [Ca2+]i in both monocytes and macrophages. In contrast, the same concentration of RANTES(3–68) increased [Ca2+]i in macrophages but not in monocytes. Among the other chemo-kines tested, macrophage inflammatory protein (MIP)–1α, MCP-1, MCP-3 (1, 6), and stromal-derived factor–1β (SDF-1β; references 18–20) also increased [Ca2+]i in resting monocytes, whereas MCP-2 (21) induced a barely detectable response and MIP-1β (1, 6) was inactive. On the basis of the previously described receptor specificities of these chemokines (1, 6, 19, 20), the obtained activity pattern is consistent with expression of CCR1, CCR2b, and CXCR4 receptors on monocytes (Fig. 3). Macrophages showed marked Ca2+ responses to MIP-1α, MIP-1β, MCP-2, MCP-3, and SDF-1β, but were resistant to MCP-1, consistent with the presence of transcripts encoding CCR1, CCR5, and CXCR4, and the absence of those encoding CCR2b, in these cells (Fig. 3).
Figure 4.
Effects of chemokines on [Ca2+]i in monocytes cultured in the absence (M) or presence (M + M-CSF) of M-CSF. Fura-2–labeled cells were exposed (at the times indicated by arrowheads) to chemically synthesized RANTES variants (100 nM) or other indicated rh chemokines (30 nM; R & D Systems), and Ca2+ responses were measured. The final concentrations of chemokines in this and subsequent experiments were sufficient to induce a maximal increase in [Ca2+]i in the responding cells, and further challenge with the same dose produced little or no detectable change in [Ca2+]i. The duration (∼100 s) and amplitude (∼20– 30% of Fura-2 saturation) of Ca2+ responses were similar to those obtained for chemokines with human monocytes (36). Similar results were obtained in two additional experiments.
RANTES(3–68) Is a Chemokine Agonist, with Altered Receptor Specificity.
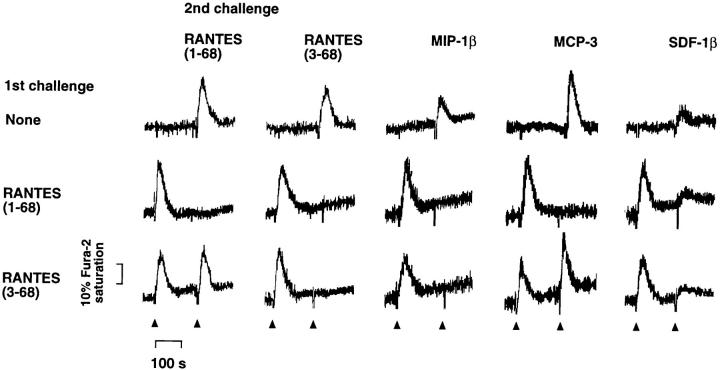
Agonists that act at common chemokine receptors block each other's activity as a result of receptor desensitization, whereas responses to chemokines that act at different receptors generally are not affected (1, 6). Therefore, we performed comparative desensitization experiments to define the types of receptors that mediate the effects of native versus truncated RANTES in macrophages (Fig. 5). Macrophages that were stimulated first with 100 nM RANTES(1–68) did not exhibit a second Ca2+ response when challenged with the same dose of either full-length or truncated RANTES. In contrast, cells stimulated with 100 nM RANTES(3–68) fully retained their ability to respond to a subsequent challenge with full-length RANTES, but were desensitized to the effect of the truncated form. These results suggest that the receptor repertoire available for truncated RANTES is more restricted than that available for the native chemokine. To further characterize the receptor usage of the different forms of RANTES and other chemokines, we also studied the sensitivity of MIP-1β–, MCP-3–, and SDF-1β–induced Ca2+ responses to RANTES-mediated receptor desensitization (Fig. 5). Of the known receptors, RANTES signals via CCR1, CCR4, and CCR5, whereas MIP-1β acts at CCR5 exclusively and MCP-3 binds only to CCR1 and CCR2b at the concentrations used in our experiments (1, 6). The only receptor known to bind SDF-1β is CXCR4 (19, 20). Pretreatment of macrophages with full-length RANTES blocked the ability of MIP-1β and MCP-3, but not that of SDF-1β, to increase [Ca2+]i. In contrast, RANTES(3–68) desensitized cells to the effect of MIP-1β but did not affect the response to MCP-3 or SDF-1β. These results are consistent with previous data on RANTES-induced receptor desensitization (1) and with our data on chemokine receptor mRNA abundance (Fig. 3). They suggest that, in M-CSF–activated macrophages, full-length RANTES shares CCR1 and CCR5 receptors with MCP-3 and MIP-1β, respectively. Our results also indicate that, without its two NH2-terminal residues, RANTES is still able to signal via CCR5 but can no longer act at the CCR1 receptor.
Figure 5.
Desensitization of chemokine-induced Ca2+ responses by full-length or truncated RANTES. Fura-2–labeled cells were stimulated first with 100 nM RANTES(1–68) or RANTES(3–68), or were left unstimulated. After ∼150 s, the cells were challenged with the RANTES variants (100 nM) or other chemokines (30 nM) as indicated, and Ca2+ responses were measured.
Removal of the NH2-Ser-Pro Residues Affects the CCR1-, but not the CCR5-mediated Signaling of RANTES.
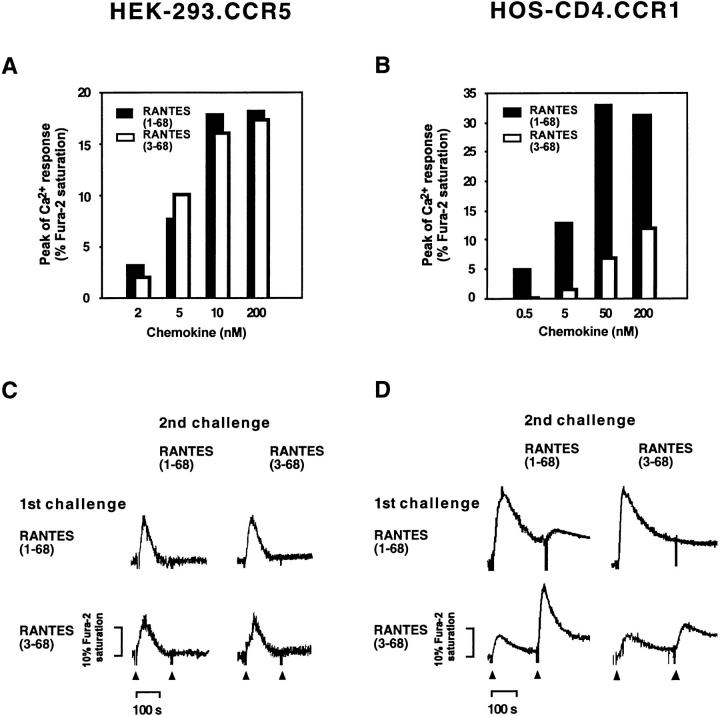
HEK-293 cells expressing CCR5 and HOS-CD4 cells expressing CCR1 were loaded with Fura-2 and exposed to various concentrations of RANTES(1–68) or RANTES(3–68). The two RANTES variants showed similar abilities to increase [Ca2+]i in the CCR5 transfectant (Fig. 6 A); the responses were dose dependent, with 10 nM of each variant sufficient to induce a maximal Ca2+ response. In contrast, in the cells expressing CCR1, the amount of RANTES(3–68) required to produce a detectable Ca2+ response was ∼100 times that for RANTES(1–68) (Fig. 6 B); the effect of RANTES(1–68) saturated at 50 nM, whereas that of RANTES(3–68) appeared not to have achieved saturation at 200 nM. Furthermore, bidirectional cross-desensitization between the two RANTES variants was evident only with the cells expressing CCR5 (Fig. 6 C); in the CCR1 transfectant, cross-desensitization was induced by full-length RANTES but not by the truncated form, which also did not exhibit self-desensitization (Fig. 6 D). Control cells transfected with vector alone or with vectors encoding CCR2b, CCR3, or CXCR4 did not respond to these ligands (data not shown). These results thus confirm that the native and CD26-truncated RANTES variants exhibit markedly different activities at the CCR1 receptor.
Figure 6.
Activity of full-length and truncated RANTES in cells expressing recombinant CCR5 or CCR1 receptors. The [Ca2+]i was measured in HEK-293 cells expressing CCR5 (A and C) and HOS-CD4 cells expressing CCR1 (B and D). (A and B) Cells were stimulated with various concentrations of the two RANTES variants as indicated and maximal fluorescence values were calculated from the peaks of the Ca2+ response curves. (C and D) Homologous and heterologous desensitization of the responses induced by RANTES(1–68) and RANTES(3–68) was measured in transfectants as described in Fig. 5.
RANTES(3–68) Is a Potent Inhibitor of HIV-1.
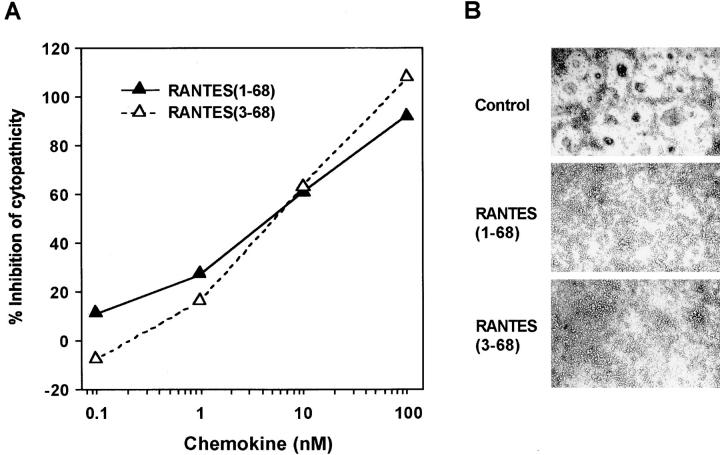
In addition to their function in chemotaxis, RANTES, MIP-1α, and MIP-1β each inhibit HIV-1 infection by competitive binding to CCR5 (22–27), and this inhibition does not require receptor-mediated cell signaling (27, 28). To examine whether removal of the two NH2-terminal residues affects the antiviral activity of RANTES, we mixed HOS-CD4 cells expressing recombinant CCR5 and PM1 cells chronically infected with the macrophage-tropic recombinant MV3-HXB2 virus and cocultured them in the absence or presence of various concentrations of RANTES(1–68) or RANTES(3–68). Both RANTES variants inhibited HIV-1-induced syncytium formation and cytopathicity (Fig. 7). Thus, similar to signaling activity through CCR5, competitive inhibition of HIV-1 infection does not require the NH2-terminal Ser-Pro residues of RANTES.
Figure 7.
Effects of full-length and truncated RANTES on HIV-1–induced cytopathicity. (A) HOS-CD4.CCR5 cells were incubated with uninfected PM1 cells or PM1 cells chronically infected with MV3-HXB2 virus in the presence or absence of the indicated concentrations of RANTES variants. After 3 d, cell viability was measured by the XTT (2,3-bis[2-methoxy- _h_-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide) method. Data are means of triplicate samples (SEM, <20% of mean). (B) Representative photomicrographs of HOS-CD4.CCR5 cells cultured with HIV-1–infected PM1 cells in the absence or presence of RANTES(1–68) or RANTES(3–68), as indicated.
Discussion
Chemical modifications at the NH2 terminus of chemo-kines have been previously suggested to produce polypeptides that are antagonists of the native chemoattractants (3, 4). However, these alterations did not correspond to the physiological specificities of known enzymes and no natural equivalents of the modified chemokines were identified. In contrast, the CD26 cleavage product of RANTES, RANTES (3–68), acts as a chemokine agonist with altered receptor specificity. Hydrolysis by CD26 might explain why RANTES (3–68) has been isolated as a second component in addition to intact RANTES from culture supernatants of stimulated human fibroblasts, skin samples, and platelet preparations (29, 30). The CC-chemokines RANTES, MCP-2, and eotaxin, and the CXC-chemokine IP-10 are the first immune modulators and the longest polypeptides identified as natural substrates for CD26.
CD26 exists in both soluble and membrane-expressed forms. Secreted forms of CD26 have been identified in cell cultures and in human serum (31, 32), although CD26 may be more active when expressed as an ectoenzyme at high concentrations on endothelial cells, hepatocytes, kidney brush border membranes, and leukocytes (10). Upregulation of CD26 expression on T lymphocytes and macrophages has been linked to cell activation and development of immunological memory (10). Thus, activation-induced changes in CD26 expression could affect the course of an inflammatory response by modifying the target cell specificity of RANTES or other chemokines, and by regulating the equilibrium between the migrating cell subsets. We are currently addressing whether cells with different levels of CD26 expression (e.g., naive versus memory T cells) secrete truncated forms of RANTES or other chemoattrac-tants, or are capable of modifying exogenous chemokines.
The differential effects of CD26-truncated RANTES on monocytes versus macrophages illustrate a role for cell differentiation in regulating chemokine sensitivity through altered receptor expression. Our functional and receptor transcript data indicate that CCR1 and CCR2b may be the two principal CC chemokine receptors in resting monocytes, although other unidentified and functionally overlapping receptors may also contribute to chemokine function. Cell differentiation markedly changes the pattern of chemokine sensitivity by reducing CCR2b expression, thereby rendering the cells resistant to MCP-1, while increasing CCR5 expression, thereby augmenting the responses to CD26-truncated RANTES and MIP-1β. An increase in CCR5 expression may also render macrophages more susceptible to infection by macrophage-tropic variants of HIV-1. We have shown that macrophages also express CXCR4, the coreceptor for T cell line–tropic HIV-1 variants (33, 34), as assessed by receptor transcript abundance and functional activity of the CXCR4 ligand SDF-1β. Nevertheless, activated macrophages are relatively resistant to infection by T cell line–tropic HIV-1 variants (35), which suggests that factors other than CXCR4 may also be required for efficient infection of macrophages by these types of viruses.
Removal of two NH2-terminal residues by CD26 abolishes the interaction of RANTES with CCR1, but does not affect the anti–HIV-1 activity or the CCR5 signaling properties of the chemokine. Proline residues also influence the susceptibility of proximal peptide bonds to proteolytic enzymes (6), and so the removal of such residues by CD26 may also reduce the half-life of RANTES and other chemo-kines during an inflammatory response. It will be important to determine whether CD26-mediated cleavage is a general mechanism for changing the receptor specificity and functional activity of other chemokines, including those examined in this study (MCP-2, eotaxin, and IP-10).
Many, but not all CC- and CXC-chemokines contain the X-Pro– or X-Ala–NH2-terminal sequence and are potential substrates of DPPIV. We are currently exploring whether the inability of CD26 to cleave MCP-1 is due to aggregation of this chemokine under these experimental conditions or to a conformational requirement of the enzyme that is not fulfilled by MCP-1. Selectivity of CD26 activity on chemokines may function to reduce redundancy in chemokine target cell specificity as illustrated by the different activity of full-length and truncated RANTES on monocytes versus macrophages. Finally, truncated analogs of chemokines with selective activity on distinct functional receptors, or analogs that resist CD26 cleavage, may prove therapeutically beneficial in blocking or inducing the infiltration of specific subsets of effector cells mediating inflammation, allergy, and antitumor responses.
Acknowledgments
We thank K. Faust and V. Calvert for help in separation of mononuclear cells, C. Abbott for mutant CD26 cDNAs, and M. Samson for the CCR5 vector. The HOS-CD4 cell lines were obtained from N. Landau through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases).
This study was supported in part by funds from the National Institutes of Health AIDS Targeted Antiviral Program and from the National Health and Medical Research Council of Australia (M.D. Gorrell).
Footnotes
Note added in proof. We note that a CD8+ T cell–derived HIV-1 suppressor activity has been recently identified as a truncated form of macrophage-derived chemokine (MDC), missing a glycine–proline dipeptide from the NH2 terminus (Pal, R., A. Garzino-Demo, P.D. Markham, J. Burns, M. Brown, R.C. Gallo, and A.L. DeVico. 1997. Science. 278:695–698). Based on our results, we suggest that truncation of MDC is a consequence of CD26-mediated cleavage that may have resulted in enhanced MDC antiviral activity.
1
Abbreviations used in this paper: [Ca2+]i, cytosolic free Ca2+ concentration; DPPIV, dipeptidyl peptidase IV; E+, enzymatically active; E−, enzymatically deficient; ES-MS, electrospray mass spectrometry; GAPDH, glyceraldehyde phosphate dehydrogenase; HEK, human embryonic kidney; HOS, human osteosarcoma; IP, interferon-γ-inducible protein; MCP, monocyte chemotactic protein; MIP, macrophage inflammatory protein; _p_NA, _p_-nitroanilide; RANTES, regulated on activation, normal T cell expressed and secreted; rh, recombinant human; s, soluble; SDF, stromal-derived factor.
References
- 1.Murphy PM. Chemokine receptors: structure, function and role in microbial pathogenesis. Cytokine Growth Factor Rev. 1996;7:47–64. doi: 10.1016/1359-6101(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 2.Sica A, Saccani A, Borsatti A, Power CA, Wells TN, Luini W, Polentarutti N, Sozzani S, Mantovani A. Bacterial lipopolysaccharide rapidly inhibits expression of C–C chemokine receptors in human monocytes. J Exp Med. 1997;185:969–974. doi: 10.1084/jem.185.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber M, Uguccioni M, Baggiolini M, Clark-Lewis I, Dahinden CA. Deletion of the NH2-terminal residue converts monocyte chemotactic protein 1 from an activator of basophil mediator release to an eosinophil chemoattractant. J Exp Med. 1996;183:681–685. doi: 10.1084/jem.183.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong J-H, Uguccioni M, Dewald B, Baggiolini M, Clark-Lewis I. RANTES and MCP-3 antagonists bind multiple chemokine receptors. J Biol Chem. 1996;271:10521–10527. doi: 10.1074/jbc.271.18.10521. [DOI] [PubMed] [Google Scholar]
- 5.Arenzana-Seisdedos F, Virelizier J-L, Rousset D, Clark-Lewis I, Loetscher P, Moser B, Baggiolini M. HIV blocked by chemokine antagonist. Nature. 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 6.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 7.Walter R, Simmons WH, Yoshimoto T. Proline specific endo- and exopeptidases. Mol Cell Biochem. 1980;30:111–127. doi: 10.1007/BF00227927. [DOI] [PubMed] [Google Scholar]
- 8.Fox DA, Hussey RE, Fitzgerald KA, Acuto O, Poole C, Palley L, Daley JF, Schlossman SF, Reinherz EL. Ta1, a novel 105 kD human T cell activation antigen defined by a monoclonal antibody. J Immunol. 1984;133:1250–1256. [PubMed] [Google Scholar]
- 9.Hegen M, Niedobitek G, Klein CE, Stein H, Fleischer B. The T cell triggering molecule Tp103 is associated with dipeptidyl aminopeptidase IV activity. J Immunol. 1990;144:2908–2914. [PubMed] [Google Scholar]
- 10.Fleischer B. CD26: a surface protease involved in T-cell activation. Immunol Today. 1994;15:180–184. doi: 10.1016/0167-5699(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 11.Oravecz T, Roderiquez G, Koffi J, Wang J, Ditto M, Bou-Habib DC, Lusso P, Norcross MA. CD26 expression correlates with entry, replication and cytopathicity of monocytotropic HIV-1 strains in a T-cell line. Nat Med. 1995;1:919–926. doi: 10.1038/nm0995-919. [DOI] [PubMed] [Google Scholar]
- 12.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Camerini D, Seed B, Torimoto Y, Dang NH, Kameoka J, Dahlberg HN, Schlossman SF, Morimoto C. Cloning and functional expression of the T cell activation antigen CD26. J Immunol. 1992;149:481–486. [PubMed] [Google Scholar]
- 14.Davis SJ, Ward HA, Puklavec MJ, Willis AC, Williams AF, Barclay AN. High level expression in Chinese hamster ovary cells of soluble forms of CD4 T lymphocyte glycoprotein including glycosylation variants. J Biol Chem. 1990;265:10410–10418. [PubMed] [Google Scholar]
- 15.McCaughan GW, Wickson JE, Creswick PF, Gorrell MD. Identification of the bile canalicular cell surface molecule GP110 as the ectopeptidase dipeptidyl peptidase IV: an analysis by tissue distribution, purification and N-terminal amino acid sequence. Hepatology. 1990;11:534–544. doi: 10.1002/hep.1840110403. [DOI] [PubMed] [Google Scholar]
- 16.Schall TJ, Jongstra J, Dyer BJ, Jorgensen J, Clayberger C, Davis MM, Krensky AM. A human T cell–specific molecule is a member of a new gene family. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 17.Rahfeld J, Schierhorn M, Hartrodt B, Neubert K, Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? . Biochim Biophys Acta. 1991;1076:314–316. doi: 10.1016/0167-4838(91)90284-7. [DOI] [PubMed] [Google Scholar]
- 18.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth–stimulating factor. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 20.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler DF, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line–adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 21.Van Damme J, Proost P, Lenaerts J-P, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkathib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: A RANTES, MIP-1α, MIP-1β, receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 23.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 24.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 25.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–673. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 26.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 27.Oravecz T, Pall M, Norcross MA. β-chemo-kine inhibition of monocytotropic HIV-1 infection: interference with a postbinding fusion step. J Immunol. 1996;157:1329–1332. [PubMed] [Google Scholar]
- 28.Farzan M, Choe H, Martin KA, Sun Y, Sidelko M, Mackay CR, Gerard NP, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1beta–mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 29.Mallet AI, Kay I. Characterization of chemokine proinflammatory proteins by combined liquid chromatography–mass spectrometry. Biochem Soc Trans. 1995;23:911–913. doi: 10.1042/bst0230911. [DOI] [PubMed] [Google Scholar]
- 30.Noso NM, Sticherling M, Bartels J, Mallet AI, Christophers E, Schröder J-M. Identification of an N-terminally truncated form of the chemokine RANTES and granulocyte-macrophage colony-stimulating factor as major eosinophil attractants released by cytokine-stimulated dermal fibroblasts. J Immunol. 1996;156:1946–1953. [PubMed] [Google Scholar]
- 31.Tanaka T, Duke-Cohan JS, Kameoka J, Yaron A, Lee I, Schlossman FF, Morimoto C. Enhancement of antigen-induced T-cell proliferation by soluble CD26/dipeptidyl peptidase IV. Proc Natl Acad Sci USA. 1994;91:3082–3086. doi: 10.1073/pnas.91.8.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duke-Cohan JS, Morimoto C, Rocker JA, Schlossman S. Serum high molecular weight dipeptidyl peptidase IV (CD26) is similar to a novel antigen DPPT-L released from activated T cells. J Immunol. 1996;156:1714–1721. [PubMed] [Google Scholar]
- 33.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein–coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 34.Berson JF, Long D, Doranz BJ, Rucker J, Jirik FR, Doms RW. A seven-transmembrane domain receptor involved in fusion and entry of T-cell–tropic human immu-nodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng-Mayer C, Quiroga M, Tung JW, Dina D, Levy JA. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JM, McVicar DW, Oppenheim JJ, Kelvin DJ. Identification of RANTES receptors on human monocytic cells: competition for binding and desensitization by homologous chemotactic cytokines. J Exp Med. 1993;177:699–705. doi: 10.1084/jem.177.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]