Structure of the Murray Valley encephalitis virus RNA helicase at 1.9 Å resolution (original) (raw)
Abstract
Murray Valley encephalitis virus (MVEV), a mosquito-borne flavivirus endemic to Australia, is closely related to Japanese encephalitis virus and West Nile virus. Nonstructural protein 3 (NS3) is a multifunctional enzyme with serine protease and DEXH/D-box helicase domains, whose activity is central to flavivirus replication and is therefore a possible target for anti-flaviviral compounds. Cloning, purification, and crystal structure determination to 1.9 Å resolution of the NS3 helicase of MVEV and characterization of its enzymatic activity is reported. Comparison with the structures of helicases from related viruses supports a possible mechanism of ATP hydrolysis-driven strand separation.
Keywords: Flaviviridae, Murray Valley encephalitis virus, helicases, viral enzymes
Arthropod-borne viruses of the genus Flavivirus (family Flaviviridae) cause a range of serious diseases in humans, the prevention and treatment of which are global public health priorities. Unfortunately no suitable antivirals are available and there are commercial vaccines for only three flaviviruses (Ray and Shi 2006). Murray Valley encephalitis virus (MVEV) has been isolated from mosquitoes in Australia and Papua New Guinea and, like West Nile virus (WNV), is grouped in the Japanese encephalitis (JE) serocomplex (Burrow et al. 1998). Clinical encephalitis due to MVEV has a mortality rate of ∼30%, with a similar proportion of patients left with significant neurological deficits (Burrow et al. 1998). Hepatitis C virus (HCV) is only 20% identical in sequence to Flaviviruses but has a similar replication strategy. The single-stranded positive sense RNA flavivirus genome (11 kb) encodes a large polyprotein precursor that is processed into three structural (C, prM/M, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Fields et al. 2001). NS3 is a multifunctional protein that displays several distinct enzymatic functions (Lain et al. 1989). The N-terminal portion of the protein, which possesses a serine protease activity, is involved in the proteolytic cleavage of the viral polyprotein precursor, while the C-terminal portion displays nucleoside triphosphatase (NTPase) and 3′ to 5′ helicase activities. Mutagenesis studies in Dengue virus (DENV) (Matusan et al. 2001) have demonstrated that NS3 helicase activity is crucial for pathogen virulence. NS3 is a DEAH/D box helicase within superfamily 2 (SF2) and as such possesses seven conserved sequence motifs (including motifs I and II, the Walker A and Walker B motifs), functions as a monomer, and provides a model system for studying oligonucleotide translocation and strand separation. Several crystal structures of SF2 DEAH/D box helicases have been reported (Singleton et al. 2007) that reveal a triangular-shaped molecule, composed of three distinct domains, with clefts formed at the interface between the domains. The helicase motifs form tandem α/β structures (domains 1 and 2: d1, d2), with the NTPase active site formed by their interface. The binding site for single-stranded nucleic acid is believed to be in a cleft between a third domain (d3) and d1 and d2. A generic model for the NTPase coupled translocation/strand separation for SF2 helicases has been proposed, named “inchworming,” in which the RNA strand is translocated through the cleft by NTPase-induced conformational changes in the hinges between the d1 and d2 (Singleton et al. 2007).
Recently, the structures of DENV NS3 (DENVh) (Xu et al. 2005) and YFV NS3 helicase (YFVh) (Wu et al. 2005) have revealed the structural organization of flavivirus helicases. Nevertheless, several issues remain unanswered regarding the unwinding mechanism of SF2 helicases, in particular the mechanism of coupling of NTP hydrolysis with oligonucleotide-translocation and strand-separation activities. The NS3 helicase is an attractive target for antiviral drug discovery, and several compounds have been shown to effectively inhibit helicase activity in vitro (Borowski et al. 2002; Zhang et al. 2003). Understanding NS3 helicase–substrate interactions should lead to improved compounds and perhaps novel inhibitors. Here we report the cloning, expression, purification, and structure determination of an ATPase-active recombinant NS3 helicase from MVEV, performed as part of the Vizier project, which aims to solve structures for replication proteins of RNA viruses (http://www.vizier-europe.org/).
Results
Crystallization of the NS3 helicase
The high-throughput pipeline of the Oxford protein production facility (OPPF) was used to test three constructs to establish the N-terminal boundary of the helicase domain of NS3. The constructs, comprising amino acid residues 159–619, 178–619, and 184–619, were designed using sequence alignment with flavivirus NS3 homologs and RONN disorder predictions (Yang et al. 2005). All three could be expressed in a soluble form using either N- or C-terminal His-6 tags; however, only the N-terminally tagged NS3178–619 crystallized after removal of the His-6 tag. This construct is referred to below as MVEVh.
Structure determination
The structure was determined by molecular replacement with the DENV NS3171–618 (DENVh) structure. The crystallographic asymmetric unit contains one monomer, and, with the exception of residues 247–250 and a disordered loop (582–592), the refined (at 1.9 Å resolution) model is complete and has good stereochemistry (see Table 1), with more than 90% of residues in the most favored region of the Ramachandran plot (Laskowski et al. 1993). Attempts to obtain structures of complexes with ADP, ATP, or nonhydrolysable ATP analogs (by cocrystallization and soaking) yielded no evidence of bound nucleotides.
Table 1.
X-ray data collection and refinement statistics for MVEV NS3 helicase
Overall structure
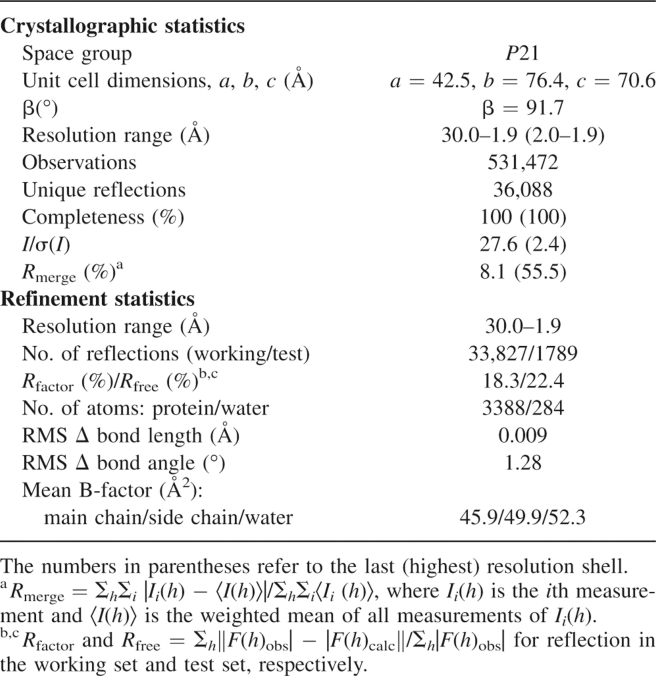
MVEVh (Fig. 1A) comprises three domains (d1–d3) of about 150 residues each, separated by clefts (Fig. 1B). d1 (residues 176–332) and d2 (333–484) have similar RecA-like folds (but no significant sequence identity) and form the catalytic core of the molecule, as observed in other SF2 helicases. The NS3 helicase of MVEV shares 64% sequence identity with its counterpart in DENV and 47% with YFV, and this is reflected in the structural similarity (RMSD of 1.6 Å for 421 equivalent Cα atoms for DENVh [PDB 2BMF] and 2.4 Å for 396 equivalent Cα atoms for YFVh [PDB 1YMF]). Domain 3 (residues 485–618) has a fold distinctive to flavivirus NS3 but retains strong topological similarities to the related domain in HCV NS3 (residues 491–624) (Fig. 1D). Despite no significant sequence identity (10%), we were able to superimpose 41 out of 123 residues with a RMSD of 4.3 Å (light pink in Fig. 1D); indeed, excluding the insertion of an α-helix in the N terminus of HCV, the major structural difference is repositioning of the C-terminal helix (α12). These differences have implications for both polyprotein processing at the NS3–NS4A junction and for interactions of NS3 with the RNA-dependent RNA polymerase, NS5. Thus while the N and C termini of HCV NS3 are on the same side of the protein, in MVEV they are on opposite sides, removing the NS3–NS4a junction from the protease active site. Secondly, flavivirus NS5 is thought to interact specifically with d3 of NS3 helicase, while HCV NS5 binds to the protease region of NS3, presumably reflecting differences in the biology of these proteins. Superposition of the individual domains of flavivirus NS3 helicase reveals that d3 is structurally the most conserved, with RMSD of 0.9 Å (123 [100%] equivalent Cα atoms) for DENVh and 1.2Å (118 [96%] equivalent Cα atoms) for YFVh. Superimpositions based on d3 reveal significant deviations in d2 and set YFVh apart from MVEVh (Fig. 2A) and DENVh. The hinge between d2 and d1 is closed by about 15° in YFVh compared to the structures of MVEV, DENV, and HCV helicases. These structural differences are presumably due to nucleotide binding (YFVh is the only structure of a Flaviviridae helicase to be observed with bound nucleotide [ADP-Mg2+]), in line with the domains hinging relative to one another during the catalytic cycle. The most significant changes are at the d1–d2 boundary, where specific contacts between motif H6 in d2 and motifs H1 and H2 in d1 of YFVh are lost in MVEVh and DENVh. Similarly, arginine finger Arg464 (motif H6) in MVEVh, which is thought to facilitate hydrolysis by inserting in the nucleotide binding site, is not in a position to contact the nucleotide γ-phosphate (Fig. 2A, bottom panel), whereas Arg467 in YFVh is. The groove between d2 and d3 is also affected and is tighter in MVEVh (where the d2–d3 interface is 980 Å2 compared 630 Å2 in YFV), with a salt bridge formed between Arg388 and Asp542 in MVEVh that is absent in YFVh (Fig. 2A, top panel, B). As a result a pocket-like cavity is produced in YFVh (volume 5400 Å3 calculated with CASTp; Fig. 2B; Binkowski et al. 2003).
Figure 1.
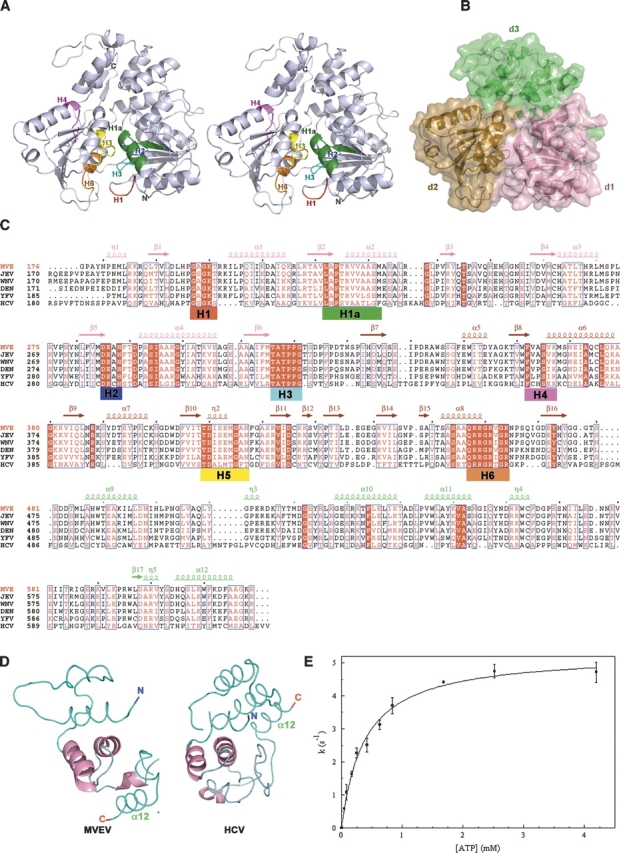
(A) Stereo ribbon diagram of MVEVh. The seven helicase motifs are colored using the same scheme as in panel C. (B) Surface representation of MVEVh, d1 (pink) and d2 (brown) are at the bottom and d3 (lime) at the top (the color scheme matches that of the secondary elements of MVE displayed above the sequence alignments in panel C. (C) Structure-based alignment (produced using T-Coffee) (O'Sullivan et al. 2004) of Flaviviridae NS3 helicases. The sequences of Japanese encephalitis virus (JEV, strain JaOArS982, NP_059434), West Nile virus (WNV, strain B956, AAT02759), Yellow Fever virus (YFV, strain 17D, NP_041726), and Hepatitis C virus (HCV, strain H77, NP_671491) were obtained from GenBank. Secondary structure elements of MVEVh are displayed above the sequence alignment. The conserved SF2 motifs are marked red (H1), green (H1a), blue (H2), cyan (H3), magenta (H4), yellow (H5), and orange (H6). (D) d3 of MVEVh (left) structurally compared to d3 of the HCV helicase (right). The structural superimposition was performed using SHP (Stuart et al. 1979). The secondary structural elements are shown in a loop representation (cyan) except for the regions that were best structurally aligned (pink), which are shown in a cartoon representation. (E) The release of inorganic phosphate in the presence of the indicated concentrations of ATP. The solid line corresponds to the fit to the Michaelis–Menten equation. Results of two protein batches were averaged and standard deviations are shown.
Figure 2.
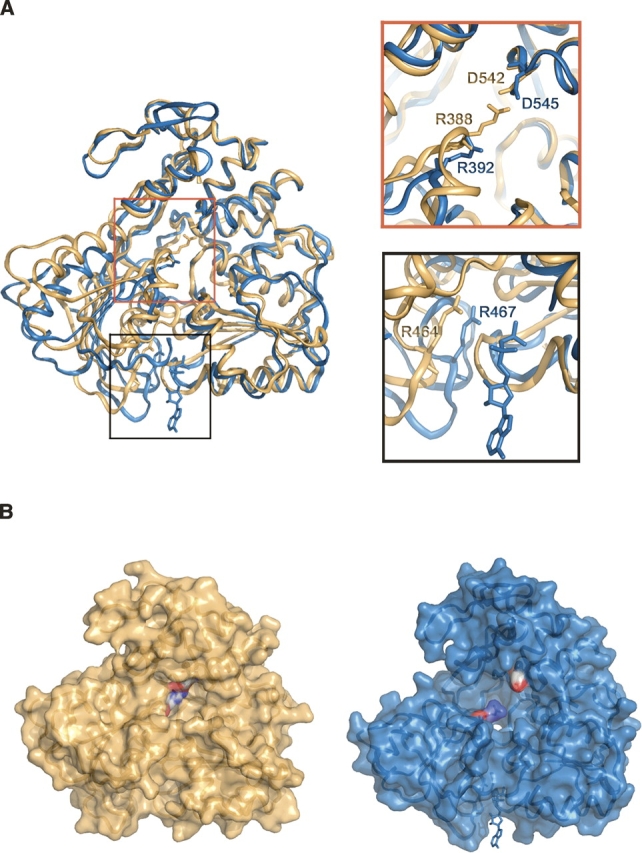
(A) The structures of MVEVh (yellow) and YFVh (blue) superimposed via domain 3 (superimposition performed using SHP) (Stuart et al. 1979). Two regions of the superimposition (top panel: groove between d2 and d3; bottom panel: nucleotide binding groove) are enlarged to highlight the differences between the two structures. (B) Surface representation of MVEVh and YFVh illustrating the opening and closure of the groove between domains 2 and 3. The color scheme matches that of panel A.
Nucleotide binding and helicase motifs
The nucleotide binding site between d1 and d2 is lined with residues of helicase motifs H1–H6 (defined in Fig. 1A) involved in ATP binding/hydrolysis and the chemo-mechanical coupling of ATP hydrolysis to RNA unwinding and translocation. In the absence of substrate (MVEVh and DENVh), the phosphate binding loop (P-loop) (H1 motif, red in Fig. 1A,C) is more flexible than in the presence of ADP-Mg2+ (YFVh) (average B-factors: MVEVh = 52, DENVh = 57, YFVh = 30). In the DENVh:SO4 (PDB 2BHR) and in YFVh:ADP-Mg2+complexes, residues belonging to the P-loop (H1, red in Fig. 1A) bind to each other and additionally to the residues of the conserved TAT sequence in motif H3 (cyan in Fig. 1A). Motif H3 is part of the flexible “switch” region connecting d1 to d2, which is thought to transmit conformational changes coupled to ATP hydrolysis. In our structure the P-loop does not contact the switch motif, suggesting that, in the absence of substrate, there is no communication between d1 and d2.
ATPase activity of the NS3 helicase
ATPase activity was measured at enzyme concentrations ranging from 0.003 to 0.04 μM. Activity increased linearly with the amount of enzyme at 0.3 and 3mM ATP concentrations (not shown), ruling out ATP induced self-association and cooperativity. The steady-state rate dependence on ATP concentration followed the standard Michaelis–Menten model (Michaelis constant Km = 0.38 ± 0.03 mM, turnover number kcat = 5.3 s−1, Fig. 1D). The turnover number and Km are similar to those of DENV full-length NS3 (kcat = 5.8 s−1, Km = 0.30 mM); (Xu et al. 2005). Only slight (∼10%) ATPase stimulation was observed upon addition of ssRNA (poly[C]; data not shown). These data suggest that MVEVh retains the enzymatic properties of the full-length protein.
Oligomeric state
It has been proposed recently that a dimeric form of Kunjin virus NS3 helicase is the biologically relevant oligomer (Mastrangelo et al. 2006), providing the helicase with multiple nucleic acid binding sites and aiding processive translocation along RNA. Different crystallographic dimers have also been observed for HCV (Cho et al. 1998) and DENV (Xu et al. 2005), although DENVh (Xu et al. 2005) and YFVh (Wu et al. 2005) are monomeric in solution. Since MVEVh is monomeric in the crystal and solution and lacks cooperativity (see Fig. 1D), it is likely that the monomer constitutes the active form of the enzyme.
Discussion
The inchworm model explains the mechanism of action of SF2 helicases by postulating that d1 and d2 switch alternatively between tight and weak DNA binding states and move relative to each other as the helicase binds and hydrolyzes ATP, conformational changes causing the power stroke that drives directional movement along RNA. Since the interface between d1 and d3 is very extensive, it is likely d2 moves relative to these domains. The conformation differences between the MVEVh structure presented here and that of ADP-Mg2+-bound YFVh might correspond to two of these conformational states. These snapshots might provide a route to novel therapeutics since the obvious target for inhibitors, the ATP binding site, lacks nucleotide specificity, and the presence of similar binding sites in many cellular proteins may make it difficult to achieve selectivity. In contrast d3 is unique to flavivirus helicases, and the relative movement of d2 and d3 might be blocked by binding a compound in the pocket-like cavity at the interface between the domains. Specifically, an inhibitor blocking the formation of the conserved salt bridge in this region might arrest translocation and stall the enzyme.
Material and Methods
Cloning and expression
Three constructs of the MVEV NS3 helicase were designed with different N-terminal domain boundaries, cloned into pOPINE (adds a C-terminal KHHHHHH tag) and pOPINF (adds an N-terminal MAHHHHHH tag and a rhinovirus 3C protease cleavage site) vectors (Berrow et al. 2006) and expressed in Escherichia coli Rosetta (DE3) pLysS in 1 L of Overnight Express instant TB medium (Novagen) containing 50 μg/mL carbenicillin, 35 μg/mL chloramphenicol, and 1% glucose. Cultures were grown for 6 h at 37°C, 220 rpm, and then for 20 h at 25°C. Cells were harvested by centrifugation and stored at −80°C.
Protein purification
Ni-NTA affinity chromatography was followed by overnight His-tag cleavage by 3C tagged protease. The cleavage mixtures were then passed over Ni-NTA columns, followed by gel filtration in 20 mM Tris (pH 7.5), 200 mM NaCl, 1 mM TCEP, and 1 mM EDTA buffer. Purity was assessed by SDS-PAGE and mass spectrometry.
Crystallization
Protein was concentrated to 30 mg/mL and crystallization experiments were performed using a Cartesian Microsys MIC4000 pipetting robot (Genomic Solutions) to set up 200 nL drops (100 nL protein + 100 nL reagent) (Walter et al. 2003, 2005) and monitored using the OPPF plate storage and imaging system (Mayo et al. 2005). Initial crystals grew as stacked plates within 2 d in 5% (w/v) PEG6000, 0.1 M HEPES (pH 7.0) and diffracted anisotropically to 3.5 Å. Single crystals of good diffraction quality (resolution limit 1.9 Å) grew only from drops seeded according to OPPF protocols (T.S. Walter, unpubl.).
Data collection and processing
A crystal was cryoprotected by brief immersion in mother liquor containing 25% glycerol and flash-frozen in liquid nitrogen. Four hundred degrees of data were collected at the ESRF beamline ID23-1 (Grenoble, France) as a series of 1.0° oscillations. Data were acquired with a Q315R CCD detector (ADSC) using an exposure time of 1 s per frame and processed using the programs DENZO and SCALEPACK (Otwinowski 1997).
Structure solution and refinement
The structure was solved by molecular replacement (program CaspR) (Claude et al. 2004) with the DENVh structure (PDB ID 2BMF) as a search model. One molecule was located in the asymmetric unit (solvent content 47%). The model was subjected to rigid body refinement and simulated annealing using CNS (Brünger et al. 1998), automatic rebuilding with ArpWarp (Perrakis et al. 1999), manual model building with Coot (Emsley and Cowtan 2004), and refinement with REFMAC (Collaborative Computational Project, Number 4 1994; see Table 1). Translation-libration-screw (TLS) parameters were refined, treating residues 1–550 and 551–618 as independent rigid bodies (Winn et al. 2001).
ATPase activity
The steady-state rate of ATP hydrolysis was determined using the phosphate release assay EnzCheck (Molecular Probes) in a standard buffer (20 mM Tris at pH 7.5, 75 mM NaCl, 7.5 mM MgCl2) at 28°C as described (Lisal and Tuma 2005). Absorbance at 360 nm was converted to inorganic phosphate concentration using KH2PO4 standards. AMP was used as a background control in all experiments. Km and kcat were determined using the GraphPad Prism Software. Protein concentrations were determined from absorbance at 280 nm (molar extinction coefficient, 71,850 L cm−1 mol−1 calculated from the amino acid composition).
Data deposition
Coordinates and structure factors are deposited with the Protein Data Bank; 2v8o.
Acknowledgments
We thank the staff at ID23-1 ESRF Grenoble for support with data collection and R. Hurrelbrink and S. Fuller for useful discussions. E.J.M. and J.M.G. are supported by the Royal Society, D.I.S. and the Oxford Protein Production Facility by the UK Medical Research Council and European Commission grant number QLG2-CT-2002-00988 (SPINE) and LSHG-CT-2004-511960 (VIZIER). R.T. is supported by the Finnish Centre of Excellence in Virus Research 2006-11.
Footnotes
Reprint requests to: David I. Stuart, Division of Structural Biology, The Henry Wellcome Building for Genomic Medicine, Oxford University, Roosevelt Drive, Oxford, OX3 7BN, United Kingdom; e-mail: dave@strubi.ox.ac.uk; fax: 44-1865-56-287547.
References
- Berrow N.S., Bussow, K., Coutard, B., Diprose, J., Ekberg, M., Folkers, G.E., Levy, N., Lieu, V., Owens, R.J., Peleg, Y., et al. 2006. Recombinant protein expression and solubility screening in Escherichia coli: A comparative study. Acta Crystallogr. D Biol. Crystallogr. 62**:** 1218–1226. [DOI] [PubMed] [Google Scholar]
- Binkowski T.A., Naghibzadeh, S., and Liang, J. 2003. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 31**:** 3352–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski P., Lang, M., Haag, A., Schmitz, H., Choe, J., Chen, H.M., and Hosmane, R.S. 2002. Characterization of imidazo[4,5-d]pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: Evidence for activity on the level of substrate and/or enzyme. Antimicrob. Agents Chemother. 46**:** 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54**:** 905–921. [DOI] [PubMed] [Google Scholar]
- Burrow J.N., Whelan, P.I., Kilburn, C.J., Fisher, D.A., Currie, B.J., and Smith, D.W. 1998. Australian encephalitis in the Northern Territory: Clinical and epidemiological features, 1987–1996. Aust. N. Z. J. Med. 28**:** 590–596. [DOI] [PubMed] [Google Scholar]
- Cho H.S., Ha, N.C., Kang, L.W., Chung, K.M., Back, S.H., Jang, S.K., and Oh, B.H. 1998. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273**:** 15045–15052. [DOI] [PubMed] [Google Scholar]
- Claude J.B., Suhre, K., Notredame, C., Claverie, J.M., and Abergel, C. 2004. CaspR: A Web server for automated molecular replacement using homology modelling. Nucleic Acids Res. 32**:** W606–W609. doi: 10.1093/nar/gkh400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Cryst. D. 50**:** 760–763. [DOI] [PubMed] [Google Scholar]
- Emsley P. and Cowtan, K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60**:** 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fields B.N., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Straus, S.E., and Knipe, D.M. 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- Lain S., Riechmann, J.L., Martin, M.T., and Garcia, J.A. 1989. Homologous potyvirus and flavivirus proteins belonging to a superfamily of helicase-like proteins. Gene 82**:** 357–362. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Moss, D.S., and Thornton, J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231**:** 1049–1067. [DOI] [PubMed] [Google Scholar]
- Lisal J. and Tuma, R. 2005. Cooperative mechanism of RNA packaging motor. J. Biol. Chem. 280**:** 23157–23164. [DOI] [PubMed] [Google Scholar]
- Mastrangelo E., Bollati, M., Milani, M., Brisbarre, N., de Lamballerie, X., Coutard, B., Canard, B., Khromykh, A., and Bolognesi, M. 2006. Preliminary crystallographic characterization of an RNA helicase from Kunjin virus. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62**:** 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusan A.E., Pryor, M.J., Davidson, A.D., and Wright, P.J. 2001. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: Effects on enzyme activity and virus replication. J. Virol. 75**:** 9633–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo C.J., Diprose, J.M., Walter, T.S., Berry, I.M., Wilson, J., Owens, R.J., Jones, E.Y., Harlos, K., Stuart, D.I., and Esnouf, R.M. 2005. Benefits of automated crystallization plate tracking, imaging, and analysis. Structure 13**:** 175–182. [DOI] [PubMed] [Google Scholar]
- O'Sullivan O., Suhre, K., Abergel, C., Higgins, D.G., and Notredame, C. 2004. 3DCoffee: Combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 340**:** 385–395. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.A.M.W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. 276**:** 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6**:** 458–463. [DOI] [PubMed] [Google Scholar]
- Ray D. and Shi, P.Y. 2006. Recent advances in flavivirus antiviral drug discovery and vaccine development. Recent Pat. Anti-Infect. Drug Discov. 1**:** 45–55. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham, M.S., and Wigley, D.B. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76**:** 23–50. [DOI] [PubMed] [Google Scholar]
- Stuart D.I., Levine, M., Muirhead, H., and Stammers, D.K. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 134**:** 109–142. [DOI] [PubMed] [Google Scholar]
- Walter T.S., Diprose, J., Brown, J., Pickford, M., Owens, R.J., Stuart, D.I., and Harlos, K. 2003. A procedure for setting up high-throughput nanolitre crystallization experiments. I. Protocol design and validation. J. Appl. Crystallogr. 36**:** 308–314. [Google Scholar]
- Walter T.S., Diprose, J.M., Mayo, C.J., Siebold, C., Pickford, M.G., Carter, L., Sutton, G.C., Berrow, N.S., Brown, J., Berry, I.M., et al. 2005. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61**:** 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Isupov, M.N., and Murshudov, G.N. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57**:** 122–133. [DOI] [PubMed] [Google Scholar]
- Wu J., Bera, A.K., Kuhn, R.J., and Smith, J.L. 2005. Structure of the Flavivirus helicase: Implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 79**:** 10268–10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Sampath, A., Chao, A., Wen, D., Nanao, M., Chene, P., Vasudevan, S.G., and Lescar, J. 2005. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J. Virol. 79**:** 10278–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.R., Thomson, R., McNeil, P., and Esnouf, R.M. 2005. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21**:** 3369–3376. [DOI] [PubMed] [Google Scholar]
- Zhang N., Chen, H.M., Koch, V., Schmitz, H., Liao, C.L., Bretner, M., Bhadti, V.S., Fattom, A.I., Naso, R.B., Hosmane, R.S., et al. 2003. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 46**:** 4149–4164. [DOI] [PubMed] [Google Scholar]