Investigations of CHD1 Function in Transcription and Development of Drosophila melanogaster (original) (raw)
Abstract
In this report we describe chd1 mutant alleles and show that the CHD1 chromatin-remodeling factor is important for wing development and fertility. While CHD1 colocalizes with elongating RNA polymerase II (Pol II) on polytene chromosomes, elongating Pol II can persist on chromatin in the absence of CHD1. These results clarify the roles of chromatin remodelers in transcription and provide novel insights into CHD1 function.
IN eukaryotic cells, RNA polymerase II (Pol II) encounters nucleosomes at each step in transcription. To deal with these obstacles, Pol II requires the action of a plethora of proteins including chromatin-remodeling factors, which function as DNA-translocating machines that draw waves of DNA around the histone octamer to slide, perturb, or disassemble the nucleosomes (Cairns 2005; Smith and Peterson 2005; Saha et al. 2006). While a number of chromatin-remodeling factors have been identified, the relative roles of these proteins in Pol II transcription are unclear.
The stages of transcription are largely defined by the phosphorylation status of the C-terminal domain (CTD) of the largest subunit of Pol II. During initiation, the CTD is unphosphorylated (Pol IIa); as Pol II clears the promoter, it is phosphorylated at serine 5 within the heptad repeats of the CTD; and later in elongation, Pol II is phosphorylated on serine 2 (Pol IIoser2) (Phatnani and Greenleaf 2006). Recent studies utilizing transcriptionally active polytene chromosomes derived from Drosophila melanogaster larval salivary glands suggest that three distinct chromatin-remodeling factors may be required for a successful round of transcription. Pol IIa levels are reduced on polytene chromosomes derived from larvae expressing a dominant-negative allele of brahma (brm), suggesting that the SWI2-like BRM chromatin remodeler is critical for transcription initiation (Armstrong et al. 2002). Polytene chromosomes derived from larvae lacking the Kismet (KIS) chromatin-remodeling protein retain Pol IIoser5, but lose Pol IIoser2, suggesting that KIS is required for an early step in the transition to transcription elongation (Srinivasan et al. 2005). The CHD1 (_c_hromodomain, _h_elicase, _D_NA-binding protein 1) chromatin-remodeling protein localizes to active genes of polytenes in a pattern nearly identical to that of Pol IIoser2, suggesting a role for CHD1 in facilitating elongation (Stokes et al. 1996; Srinivasan et al. 2005).
Taken together, these results lead to a model in which three Drosophila chromatin-remodeling factors function sequentially to allow transcription: (1) BRM is required for initiation, (2) KIS is required for the transition from promoter clearance to the elongation phase, and (3) CHD1 is required for continued elongation by Pol II. A place for CHD1 in elongation is also supported by data in yeast. Saccharomyces cerevisiae CHD1 is localized to a transcriptionally active gene and physically interacts with transcription elongation factors (Simic et al. 2003). To determine whether Drosophila CHD1 is required for elongation, we generated loss-of-function alleles of chd1 and examined the consequences of the loss of CHD1 on global chromosome structure and transcription.
chd1 is not an essential gene:
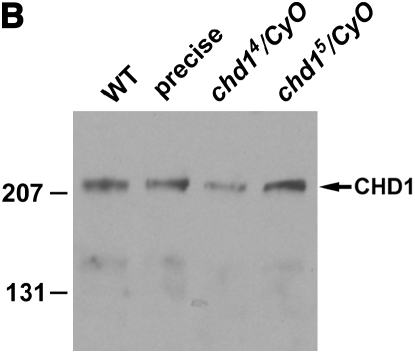
To investigate the function of CHD1, we generated two deletion alleles (chd14 and chd15) by imprecise excision of an EP element inserted into position -2 of the chd1 promoter (GenExel stock G2213) (Figure 1A). We observed no differences in the behavior of the two alleles. As described below, chd14 and chd15 homozygous and chd14/chd15 heteroallelic individuals display phenotypes that are less severe than those seen in hemizygous mutants [using _Df(2L)Exel7014_]. These genetic data would suggest that chd14 and chd15 are hypomorphic alleles. We investigated the possibility that the two chd1 alleles could generate proteins with N-terminal truncations. Given the location of the earliest in-frame start codon, we predict that both chd14 and chd15 alleles would generate a 166-kDa protein. However, Western blot analysis of heterozygous embryo extracts failed to detect an N-terminal truncated protein (Figure 1B). While it is formally possible that chd14 and chd15 express an unstable protein that is not detectable by Western blot, we propose that our alleles are protein nulls and conclude that chd1 is not an essential gene. chd14 and chd15 homozygous, heteroallelic, and hemizygous mutant individuals are viable, although they display a 1- to 2-day developmental delay, and some marker combinations reduce viability of chd1 mutants. Given the genetic data described above, we propose that 1 of the other 18 genes uncovered by Df(2L)Exel7014 may dominantly enhance chd1 mutant phenotypes. For example, okra (the RAD54 homolog, a SNF2-like helicase) is located 20 kb away from chd1. okra mutant phenotypes include female sterility (Kooistra et al. 1997), one of the chd1 phenotypes that may be dominantly enhanced by the deficiency.
Figure 1.—
The generation of two chd1 alleles. (A) The chd14 allele carries a deletion from −1 to +1994 with the additional sequence CATGATGAAATAACATATAGTTAGATATGAAATAA. The chd15 allele carries a deletion from −1 to +1871 with the additional sequence CATGATGAAATAACATCATCATAACATGAAATAAC. Much of the additional sequence in both alleles is derived from the P element. (B) Western blot analysis of embryo extracts derived from Oregon-R (WT), flies in which the P element was precisely excised (precise), and chd14/CyO and chd15/CyO heterozygotes. Full-length CHD1 is observed in each lane, and a truncated protein (predicted to be 166 kDa) is not observed in the mutant heterozygous embryo extracts. The CHD1 rabbit polyclonal antibody was raised and affinity purified against CHD1 amino acids 1706–1721 (CRLNMDRHEDRKKHHRG) (Covance). This peptide antibody recognizes CHD1 on polytenes in a pattern identical to that observed with the antibody raised by Robert Perry (Stokes et al. 1996) (data not shown).
chd1 mutants reveal unexpected defects in fertility and wing development:
We examined the distribution of chd1 mRNA by in situ hybridization and found that chd1 is broadly expressed throughout embryogenesis and in imaginal discs (data not shown). This broad expression pattern is similar to that of brm and kis (Elfring et al. 1998; Daubresse et al. 1999) and suggests that, like BRM and KIS, CHD1 could function globally to regulate transcription.
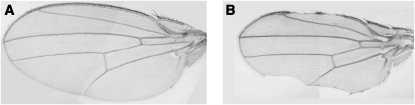
While chd1 is broadly expressed, we observed specific phenotypes in mutant animals, suggesting that CHD1 may function as a tissue-specific chromatin-remodeling factor. Wing margins in chd14 and chd15 homozygous, heteroallelic, and hemizygous mutant individuals displayed notching (Figure 2B), with 3.8–36% of heteroallelic individuals and 75–94% of hemizygous individuals showing notched wing margins (Table 1). Several control individuals are presented in Table 1 to ensure that the cut-in wing margins were not a consequence of the chromosomal markers (which are a result of meiotic mapping of the chd1 alleles), although the markers may affect how often the phenotype is seen (Table 1). The variability of the wing-notching phenotype was not correlated with developmental delay or viability. Individuals homozygous for the precise excision did not show cut-in wing margins, indicating that the phenotype is due to lack of CHD1. This specific wing phenotype is not observed in animals lacking BRM or KIS and suggests that genes critical for wing-margin formation are especially sensitive to loss of CHD1.
Figure 2.—
chd1 mutant individuals display wing defects that include notched wing margins. In contrast to a wild-type Oregon-R wing (A), wings from chd14 b pr c px sp/chd5 b individuals show notched wing margins (B). Wings from chd1 mutant animals are ∼80% the size of wild-type wings, consistent with their overall smaller size.
TABLE 1.
Wing defects in chd1 mutant flies
| Genotype | Total no. of flies | % of flies displaying cut-in wing marginsa |
|---|---|---|
| chd14 b pr c px sp/chd15 b | 285 | 3.8 |
| chd14 b pr c px sp/chd15 b c sp | 115 | 36 |
| chd14 b pr c px sp/b | 269 | 0 |
| chd14 b pr c px sp/al b c sp | 294 | 0 |
| b pr c px sp/chd15 b c sp | 280 | 0 |
| chd14 b pr c px sp/Df(2L)Exel7014 | 159 | 78 |
| chd15 b/Df(2L)Exel7014 | 48 | 94 |
| In(2LR) Gla, Bc/Df(2L)Exel7014 | 209 | 0 |
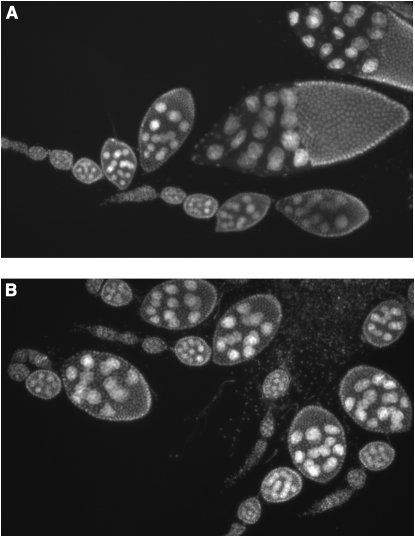
chd14 and chd15 homozygous, heteroallelic, and hemizygous males are sterile; CHD1 is therefore required for male fertility (Table 2). chd1 mutant males displayed normal mating behaviors, and there were no obvious defects in the general morphology of testes of chd14/chd15 mutant males (data not shown). While our mutant males produced zero progeny, control males produced an average of 101 progeny per single male under the same conditions. CHD1 is also important for female fertility as chd1 mutant females produced few offspring (Table 2) (by comparison, a single wild-type female produced 108 progeny under the same conditions). We propose that the majority of the fertilized eggs cannot develop due to an inability to repackage the sperm pronuclear DNA into H3.3-containing chromatin (Konev et al. 2007). Examination of egg chambers from hemizygous mutant females [_chd14/Df(2L)Exel7014_] reveals that, while eggs are occasionally formed, oogenesis often fails at stage 8, the start of yolk production (Figure 3B). An 8.5-kb genomic chd1 transgene (−456 to + 8019 relative to the chd1 start site) fully rescued all mutant phenotypes: male sterility, reduced female fertility, and notching of wing margins.
TABLE 2.
chd1 mutant males and females have reduced fertility
| Genotype | Gender | No. of chd1 mutant parents | No. of adult offspring |
|---|---|---|---|
| chd14 b pr c px sp/chd15 b | Male | 120 | 0 |
| chd14 b pr c px sp/chd15 b | Female | 69 | 39 |
| chd14 b pr c px sp/Df(2L)Exel7014 | Female | 47 | 2 |
Figure 3.—
chd1 mutant individuals display defects in oogenesis. Ovarioles derived from wild-type (A) and chd14/Df(2L)Exel7014 hemizygous mutant females (B) were stained with DAPI and prepared as described (Verheyen and Cooley 1994). While eggs are occasionally produced, oogenesis of hemizygous mutant females often fails at stage 8.
CHD1 and transcriptional elongation:
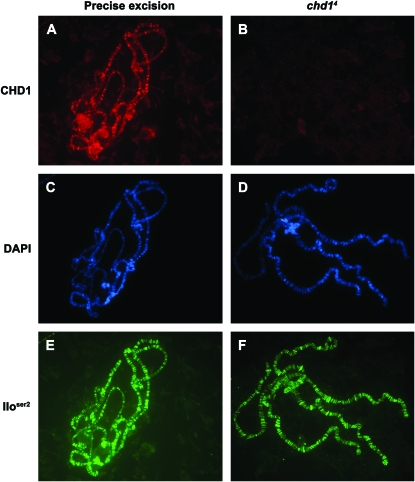
CHD1 protein persisted into early third instar larval stages in chd1 mutant larvae (likely a consequence of maternal perdurance). However, in contrast to CHD1 levels in control chromosomes (Figure 4A), CHD1 levels were greatly reduced on chromosomes derived from chd1 mutant individuals in mid-to-late third instar larval development (Figure 4B), providing us with an excellent system in which to dissect the role of CHD1 on chromosomes. Unlike the chromatin-remodeling factor ISWI (Deuring et al. 2000), CHD1 is not essential for global chromosome structure (Figure 4D).
Figure 4.—
CHD1 binding is not required for the structure of interphase chromosomes or for elongation by Pol II (IIoser2). Polytene chromosomes were derived from control individuals that underwent a precise excision of the P element (A, C, and E) or from homozygous chd14 b pr c px sp individuals (B, D, and F). Chromosomes were immunostained with CHD1 (A and B) or Pol IIoser2 (E and F) or were stained with DAPI (C and D). Despite lack of the CHD1 protein on chromosomes derived from chd1 mutant individuals (B), the chromosomes display an overall normal morphology (D) and contain normal levels of elongating Pol II (F). Control and mutant larvae were immunostained and imaged at the same time under identical conditions. Immunostains were carried out as described (Armstrong et al. 2002; Corona et al. 2004) using an antibody directed against the C-terminal region of CHD1 (Stokes et al. 1996) and the commercially available H5 antibody (Covance). Secondary antibodies (Jackson ImmunoResearch Laboratories) were tested with individual primary antibody to ensure specificity. Images were obtained on a Zeiss Axioskop 2 plus microscope with an Axioplan HRm camera and Axiovision 4 software. Control and mutant chromosomes were photographed using identical exposure times, and images were processed identically in Photoshop 7.0.
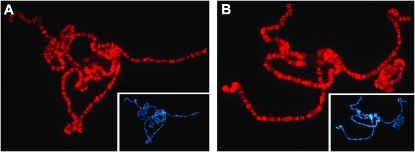
Given the observation that chromosomes lacking KIS are not bound by CHD1 (Srinivasan et al. 2005), we proposed that CHD1 is functionally downstream of Kismet. Alternatively, Kismet and CHD1 could be mutually dependent upon each other for chromosome binding. To distinguish between these two possibilities, we asked whether KIS was found on chromosomes lacking CHD1. KIS was present at wild-type levels in chd1 mutant individuals (Figure 5), indicating that, while CHD1 localization depends upon KIS, KIS localization is not dependent upon CHD1.
Figure 5.—
CHD1 is not required for KIS binding. Polytene chromosomes derived from control individuals that underwent a precise excision of the P element (A) or homozygous chd14 b pr c px sp individuals (B) were immunostained for KIS. DAPI stains of the same chromosomes are shown in insets. Immunostains were carried out as described (Armstrong et al. 2002; Corona et al. 2004) using a previously described KIS antibody (Srinivasan et al. 2005). Control and mutant chromosomes were processed in parallel and photographed using identical exposure times.
To test our hypothesis that CHD1 is required for the continued elongation of Pol II, we examined the levels of Pol IIoser2 on chromosomes derived from mutant chd1 individuals. We observed chromosomes that lack CHD1 protein that still possessed normal levels of Pol IIoser2 (Figure 4F). An antibody recognizing all the forms of Pol II (4H8, Abcam) similarly showed normal levels and distribution of total Pol II on chd1 mutant chromosomes (data not shown). Given that we observed chromosomes lacking observable CHD1 protein that retain elongating Pol II, we conclude that while CHD1 colocalizes with elongating Pol II, it is not absolutely required for the association of Pol II with chromatin. We note that lower levels of CHD1 protein on polytenes from chd1 mutant larvae can correlate with reduced levels of Pol IIoser2 (data not shown). Reduced levels of transcription may be a secondary affect of healthy larvae; alternatively, loss of CHD1 may affect subsequent rounds of transcription, leading to a reduction of Pol IIoser2 levels over time.
In conclusion, we have generated two chd1 null alleles that have revealed roles for CHD1 in male fertility, oogenesis, and wing development. While it is formally possible that transcription is affected in a subtle way, our experiments allow us to conclude that CHD1 is not absolutely required for the association of elongating Pol II on chromosomes. However, since Pol IIoser2 levels can occasionally be reduced on chromosomes derived from chd1 mutant larvae, CHD1 activity may indirectly impact transcriptional elongation. Yeast Chd1 is implicated in Pol II elongation, termination, and the response to transcriptional stress (Alen et al. 2002; Simic et al. 2003; Zhang et al. 2005). Drosophila CHD1 participates in nucleosome assembly in vitro (Lusser et al. 2005) and was recently found to repackage the sperm pronuclear DNA into H3.3-containing chromatin in vivo (Konev et al. 2007). Whether CHD1 functions to facilitate chromatin disassembly or reassembly during transcription remains to be determined.
Acknowledgments
We thank John Tamkun for antibodies, discussions, and helpful comments on this manuscript; Kristel Dorighi, Giorgia Siriaco, Shrividhya Srinivasan, Nick Reeves, Grant Hartzog, and Joseph Schulz for helpful discussions and advice; Robert Perry for generously providing CHD1 antibody; Helen McNeill for the _chd1 P_-element insertion line; Laura Lee and Rebecca Zabinsky for fly assistance; and Rancho Santa Ana Botanic Garden for use of their sequencing facilities. This work was supported by a grant from the National Science Foundation (MCB-0641379).
References
- Alen, C., N. A. Kent, H. S. Jones, J. O'Sullivan, A. Aranda et al., 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10**:** 1441–1452. [DOI] [PubMed] [Google Scholar]
- Armstrong, J. A., O. Papoulas, G. Daubresse, A. S. Sperling, J. T. Lis et al., 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21**:** 5245–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, B. R., 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15**:** 185–190. [DOI] [PubMed] [Google Scholar]
- Corona, D. F., J. A. Armstrong and J. W. Tamkun, 2004. Genetic and cytological analysis of Drosophila chromatin-remodeling factors. Methods Enzymol. 377**:** 70–85. [DOI] [PubMed] [Google Scholar]
- Daubresse, G., R. Deuring, L. Moore, O. Papoulas, I. Zakrajsek et al., 1999. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126**:** 1175–1187. [DOI] [PubMed] [Google Scholar]
- Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas et al., 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5**:** 355–365. [DOI] [PubMed] [Google Scholar]
- Elfring, L. K., C. Daniel, O. Papoulas, R. Deuring, M. Sarte et al., 1998. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148**:** 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev, A. Y., M. Tribus, S. Y. Park, V. Podhraski, C. Y. Lim et al., 2007. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317**:** 1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra, R., K. Vreeken, J. B. M. Zonneveld, A. de Jong, J. C. J. Eeken et al., 1997. The Drosophila melanogaster RAD54 homolog, DmRAD54, is involved in the repair of radiation damage and recombination. Mol. Cell. Biol. 17**:** 6097–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser, A., D. L. Urwin and J. T. Kadonaga, 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12**:** 160–166. [DOI] [PubMed] [Google Scholar]
- Phatnani, H. P., and A. L. Greenleaf, 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20**:** 2922–2936. [DOI] [PubMed] [Google Scholar]
- Saha, A., J. Wittmeyer and B. R. Cairns, 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7**:** 437–447. [DOI] [PubMed] [Google Scholar]
- Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa et al., 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22**:** 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. L., and C. L. Peterson, 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65**:** 115–148. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., J. A. Armstrong, R. Deuring, I. K. Dahlsveen, H. McNeill et al., 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA polymerase II. Development 132**:** 1623–1635. [DOI] [PubMed] [Google Scholar]
- Stokes, D. G., K. D. Tartof and R. P. Perry, 1996. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 93**:** 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen, E., and L. Cooley, 1994. Looking at oogenesis. Methods Cell Biol. 44**:** 545–561. [PubMed] [Google Scholar]
- Zhang, L., S. Schroeder, N. Fong and D. L. Bentley, 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’ EMBO J. 24**:** 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]