2B4, the Natural Killer and T Cell Immunoglobulin Superfamily Surface Protein, Is a Ligand for CD48 (original) (raw)
Abstract
2B4 is a cell surface glycoprotein related to CD2 and implicated in the regulation of natural killer and T lymphocyte function. A recombinant protein containing the extracellular region of mouse (m)2B4 attached to avidin-coated fluorescent beads bound to rodent cells, and binding was completely blocked by CD48 monoclonal antibodies (mAbs). Using surface plasmon resonance, we showed that purified soluble mCD48 bound m2B4 with a six- to ninefold higher affinity (K d ≈ 16 μM at 37°C) than its other ligand, CD2. Human CD48 bound human 2B4 with a similar affinity (K d ≈ 8 μM). The finding of an additional ligand for CD48 provides an explanation for distinct functional effects observed on perturbing CD2 and CD48 with mAbs or by genetic manipulation.
Keywords: 2B4, CD48, natural killer cell, T cell, ligand
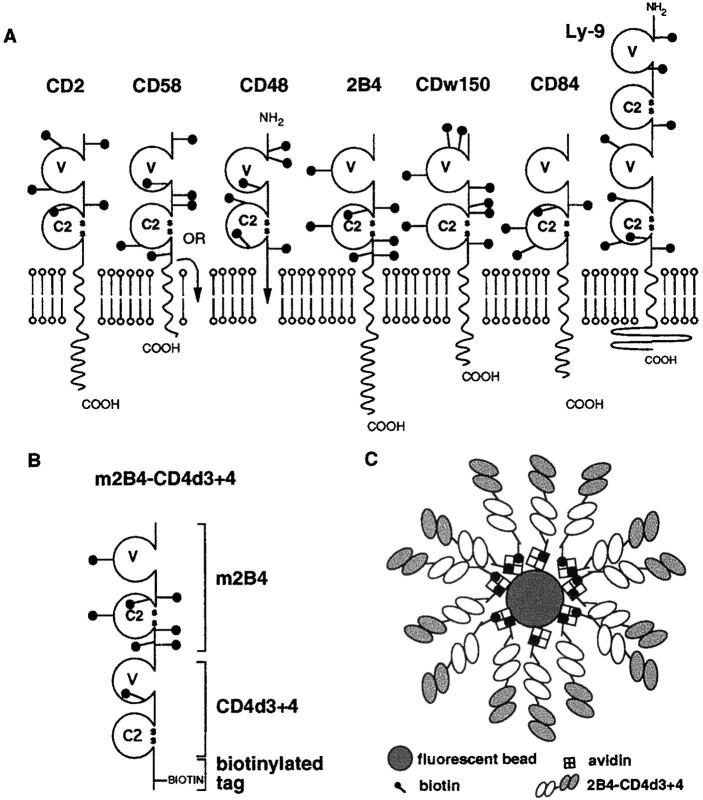
The majority of leukocyte surface proteins can be grouped according to the type of domain they contain (1). The immunoglobulin superfamily (IgSF)1 domain is the most highly represented domain type, found in 34% of leukocyte surface proteins. Of these, 45% have extracellular regions comprised of two IgSF domains in tandem, an arrangement that appears to be particularly favored for cell– cell recognition molecules, including the TCR. Other two-IgSF-domain leukocyte surface antigens known to mediate cell–cell interactions include CD2, CD58, CD48, CD80, CD86, CD102 (1), and OX2 (2). Among these, CD2 and its ligands, CD58 in humans (3, 4) and CD48 in rodents (5, 6) belong to the CD2 subset of the IgSF, which also includes 2B4, CDw150, CD84, and Ly-9 (Fig. 1), all of which are expressed on hematopoietic cells (1, 7, 8). With the exception of Ly-9, the extracellular regions of all of these molecules contain a membrane distal IgSF V-set domain and a membrane proximal C2-set domain (Fig. 1 A). In Ly-9, this arrangement is duplicated to give four IgSF domains. The related sequence and chromosomal location of this subset together with interactions among members suggest they evolved from one molecule which mediated homophilic recognition (1, 9).
Figure 1.
CD2 IgSF subfamily. (A) Schematic representation of mouse CD2, CD48, 2B4, and Ly-9 and human CD58, CDw150 (SLAM), and CD84. (B) Recombinant chimeric m2B4-CD4d3+4 protein. (C) The 2B4-CD4d3+4 fluorescent beads used for ligand identification.
The 2B4 antigen was identified as an antigen on NK cells and T cells capable of non-MHC–restricted killing (10). 2B4 is also expressed on murine dendritic epidermal γ/δ T cells (DETCs [11, 12]). A correlation in DETCs between 2B4 expression and capacity to lyse transformed keratinocytes suggests a role for 2B4 in killing skin tumors (11). Evidence for a role in regulation of NK and T cell activity by 2B4 is provided by the cross-linking effects of the 2B4 mAb, which enhances lytic function of 2B4-expressing cells, suggesting that 2B4 can transduce stimulatory signals (10, 12). Costimulation experiments with CD3 and 2B4 mAbs suggest 2B4, like CD2 (7), can enhance signals through the TCR (12). However, the mechanisms by which 2B4 and CD2 regulate cell activation are presumably different, as there are marked differences in their cytoplasmic regions. For example, the 2B4 cytoplasmic region contains tyrosine motifs that are potential targets for phosphorylation (13), whereas the CD2 cytoplasmic region contains proline-rich motifs and no tyrosine residues (14).
Since CD2 binds to CD58 and CD48, it seemed reasonable to predict that 2B4 would also bind another molecule within the CD2 subfamily. Interactions between leukocyte cell surface molecules are weak and transient, often making them difficult to identify (15). For use as ligand-binding tools, recombinant forms of extracellular regions must be made sufficiently multivalent to achieve the necessary avidity to bind to cells. In a modification of a previously described assay (16), an enzymatically biotinylated peptide (17, 18) engineered onto CD4d3+4 (domains 3 and 4 of rat CD4) fusion proteins was used to attach the recombinant molecules in an oriented manner to fluorescent beads. We used this assay to explore the ligand-binding properties of 2B4, and discovered it is a ligand for CD48 in rodents and humans.
Materials and Methods
mAbs.
mAbs used were 2B4 (10); rat (r)CD4d3+4 mAb, OX68; mouse (m)CD2 mAb, RM2.1 (19); mCD48 mAb, OX78; rCD48 mAb, OX45; and rCD147 mAb, OX47. The OX mAbs are all referenced in the European Collection of Animal Cell Cultures (Porton Down, Wiltshire, UK). Purified HM48-1 (hamster anti–mouse CD48 [5]) was provided by Dr. H. Yagita (Juntendo University School of Medicine, Tokyo, Japan), and control 2C11 (hamster anti–mouse CD3) by Dr. E. Adams (Sir William Dunn School of Pathology). Human (h)CD48 mAbs were MEM 102 (20) and 6.28 (21), provided by Drs. V. Horejsi (Institute of Molecular Genetics, Prague, Czech Republic) and D. Thorley-Lawson (Department of Pathology, Tufts University, Boston, MA), respectively.
Vector Construction for Recombinant Proteins.
The constructs for CD4d3+4 chimeric proteins used previously (22) were modified to contain at the COOH terminus a consensus peptide sequence recognized by the Escherichia coli biotin holoenzyme synthetase, BirA (17, 18). DNA encoding the amino acid sequence NSGSLHHILDAQKMVWNHR* was amplified by PCR using 5′ (agggttgaattccggatcactgcatcatattctgg) and 3′ (ctactaggatccttaacgatgattccacacc) oligonucleotides (restriction enzyme sites are underlined) and an HLA A2 construct as template (17), provided by Dr. Paul Moss (Institute of Molecular Medicine, Oxford, UK). This fragment was inserted via a 5′ EcoRI site and a 3′ BamHI site into a modified Bluescript (BS KS+) vector containing a rat CD4 leader and rat CD4d3+4 (CD4Ld3+4RI [23]). An XbaI-BamHI fragment encoding XbaI-CD4L-SalI-CD4d3+4-EcoRI-biotin peptide-stop-BamHI was transferred to a modified pEF-BOS vector (24), pEF-BOS-XB. To construct fusion proteins, the XbaI-SalI fragment encoding CD4L was replaced with DNA encoding the extracellular region of m2B4 or h2B4 amplified from plasmid DNA (13; Boles, K., and P.A. Mathew, manuscript in preparation) or domains 1–3 of rCD5 (25). XbaI sites were introduced 42 bases (m2B4) and 52 bases (h2B4) upstream of the initiation Met. The join at the SalI (g tcg acc) junction with rCD4d3 was m2B4:VPSNFRST (the residues containing the SalI site are underlined) and at the homologous region for h2B4. Confirmatory sequence analysis of all constructs (model 373A DNA sequencing system; Applied Biosystems, Inc., Foster City, CA) revealed for m2B4-CD4d3+4 a 3-base deletion, the correct sequence reading at base 619: gctttgtac encoding ALY in the C strand of domain 2. To construct soluble His-tagged hCD48, DNA (sequence data available from EMBL/GenBank/ DDBJ under accession no. M37766) encoding the extracellular region with an XbaI site 13 bases upstream of the initiation Met and a His-tag:TLARSTHHHHHH* contained in the 3′ oligonucleotide (ccactaggatcctaatgatggtggtgatgatgggtcgaccgggccagggtacagggtggactgag) was amplified by PCR from mouse macrophage cDNA (provided by Dr. J. Mahoney, Sir William Dunn School of Pathology) and cloned into pEF-BOS-XB.
Expression of Recombinant Biotinylated Chimeric Proteins.
Recombinant proteins were expressed by transfection of 293T cells with 40 μg plasmid DNA/5 × 106 cells/175-cm2 flask using calcium phosphate. After removal of precipitate at 16 h, cells were incubated for 48 h in 7.5–10 ml X-Vivo-10 serum-free medium (BioWhittaker, Walkersville, MD). Levels of four Ig domain fusion proteins as assayed in a CD4d3+4 inhibition ELISA (22) were routinely in the region of 50–150 μg/175-cm2 flask. To enzymatically biotinylate CD4d3+4 fusion proteins, supernatant was exchanged (∼1:20) into 10 mM Tris-HCl, pH 8, and concentrated to 0.5–0.8 ml using a 10,000 mol wt cutoff 15-ml centricon (Amicon, Inc., Beverly, MA) and incubated with 1 μl BIR enzyme (Avidity, Denver, CO). Biotinylation was complete after 2–4 h at 30°C or overnight incubation at room temperature as assessed by binding of streptavidin to chimeras immobilized via a CD4d3+4 mAb in a BIAcore™ (reference 26, and see below). Excess biotin was removed by dialysis (twice with 800 ml PBS for 20 h), and biotinylated material was aliquoted and stored at −20°C.
Expression of His-tagged Recombinant Proteins.
Purified monomeric mCD48 containing a COOH-terminal His-tag was prepared as described (27). Soluble hCD48 was expressed as for CD4d3+4 fusion proteins in DMEM containing 10% FCS. Soluble hCD48 was purified using an FPLC (Pharmacia Biotech Ltd., St. Albans, UK) first on a 1-ml HiTrap® column (Pharmacia Biotech Ltd.). Monomeric soluble hCD48 was then eluted in 0.1-ml fractions from Superdex 75 (Pharmacia Biotech Ltd.) and used within 4 h. Concentration was measured at 280 nm using an extinction coefficient of 1.6 cm2/mg (28), and purity was checked by SDS-PAGE. Purified hCD48 was shown to be antigenically active by binding stoichiometric amounts of the 6.28 mAb when immobilized via the His-tag on an NTA chip (Biacore AB, Uppsala, Sweden). In experiments with h2B4-CD4d3+4 and control CD4d3+4 immobilized on streptavidin-coated beads (Dynal A.S., Oslo, Norway), >70% hCD48 could be depleted as assessed by SDS-PAGE and densitometry.
Cell-binding Experiments.
Experimental procedures were essentially as described (16). Biotinylated proteins (1–2 μg/sample) were mixed with 10–20 μl avidin-coated fluorescent beads/sample (VFP-0552-5; Spherotech, Inc., Libertyville, IL). Small volumes were shaken in round-bottomed microtiter plates. Where volume adjustment was necessary, beads were centrifuged and resuspended in PBS containing 0.2% BSA (PBS/BSA). For coating beads with mAbs in blocking studies, beads were washed twice in PBS/BSA and resuspended in PBS/BSA containing 2B4 or OX68 mAb (1 μg/20 μl beads).
BIAcore™ Analysis of Affinity and Kinetics of the 2B4–CD48 Interaction.
Experiments were carried out on a BIAcore™ 2000 (Biacore AB) using HBS buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P20) supplied by the manufacturer. Streptavidin 0.2 mg/ml (Pierce Chemical Co., Rockford, IL) was coupled in 10 mM sodium acetate, pH 5, to a research grade CM5 chip (Biacore AB) using an amine coupling kit (Biacore AB) using an activation time of 5 min, resulting in immobilization of ∼2,000–3,000 response units (RU). Soluble mCD2 comprising the extracellular region of mCD2 (27) at 90 μg/ml was coupled similarly with an activation time of 1.8 min. The surface of the chip was washed with 0.1 M glycine-HCl, pH 2.5, after coupling. Biotinylated 2B4-CD4d3+4 and control rCD5-CD4d3+4 or CD4d3+4 were then injected over immobilized streptavidin. Equilibrium affinity measurements were performed as described (26). Increasing and decreasing concentrations of mCD48 (5-μl injections at 20 μl/min) were passed over m2B4-CD4d3+4, rCD5-CD4d3+4, and mCD2. Kinetic measurements were made at 37°C (28). mCD48 (23.4 μM) was injected for 3 s at 100 μl/min over high and low levels of m2B4-CD4d3+4, rCD5-CD4d3+4, and mCD2. Similarly, to measure affinity, hCD48 was passed over h2B4-CD4d3+4 and CD4d3+4, and for kinetics, hCD48 (14.4 μM) was passed over high and low levels of h2B4-CD4d3+4 and CD4d3+4. Dissociation rates were measured for mCD48 as described (28) using Origin software, version 5.0 (Microcal Software, Inc., Northampton, MA). The effect of 2B4 mAb on mCD48 (23.4 μM) binding was tested by first passing 2B4 mAb at 10 μg/ml at 5 μl/min over m2B4-CD4d3+4 until saturation was reached.
Results
m2B4 Binds to CD48.
A binding reagent for a m2B4 ligand was constructed by expressing the two IgSF domains of 2B4 as a recombinant chimeric protein with domains 3 and 4 of rCD4 (m2B4-CD4d3+4; Fig. 1 B). The recombinant protein differed from previously described CD4d3+4 chimeras (22) in that it contained a peptide sequence at its COOH terminus that can be specifically biotinylated by the E. coli enzyme, BirA (18). The expressed protein was biotinylated by the BirA enzyme in vitro and tested for reactivity with streptavidin and 2B4 mAb by surface plasmon resonance (not shown). The in vitro biotinylation of the recombinant protein simplified the previously described method (16) for detecting low-affinity interactions at the cell surface by enabling the biotinylated m2B4-CD4d3+4 to be directly immobilized on avidin-coated beads (Fig. 1 C) without the use of a CD4d3+4 mAb. The assay was shown to be at least as sensitive as coupling CD4d3+4 fusion proteins to beads using a CD4d3+4 mAb (data not shown).
m2B4-CD4d3+4 beads bound to LPS and Con A–activated mouse spleen cells (Fig. 2, A and B). The broad distribution of the ligand and our own expectation that it might be another member of the CD2 subset suggested CD48 as a likely candidate. In support of this, the binding of m2B4-CD4d3+4 beads was blocked by the CD48 mAb, OX78, but not by a control CD2 mAb (Fig. 2, A and B). A second CD48 mAb, HM48-1, which blocks the CD2–CD48 interaction (5) and is known to have functional effects in vivo (29, 30), also inhibited the binding of m2B4-CD4d3+4 beads (Fig. 2 C).
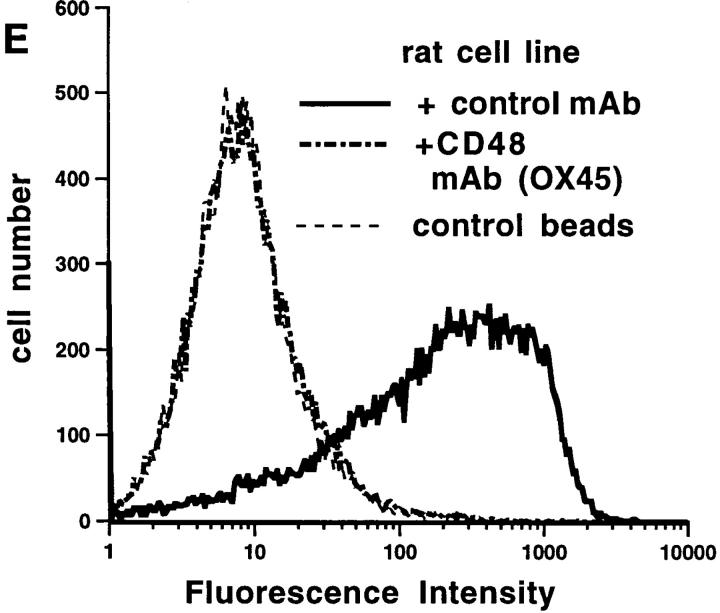
Figure 2.
m2B4-CD4d3+4 fluorescent beads bind to rodent CD48 on cells. m2B4-CD4d 3+4 fluorescent beads bind to LPS (A and D) and Con A (B)– activated and resting (C) mouse spleen cells and a rat basophilic leukemia cell line (E). Preincubation of cells with mCD48 mAbs OX78 (A and B) or HM48-1 (C) or rCD48 mAb (E) blocked binding to the level seen with the negative control, rCD5-CD4d3+4. Control mCD2 mAb (A and B), mCD3 mAb 2C11 (C), or rCD147 mAb (E) did not block binding. Preincubation of m2B4-CD4d3+4 fluorescent beads with 2B4 mAb but not a CD4d3+4 mAb partially blocks binding to cells (D).
Preincubation of m2B4-CD4d3+4 beads with 2B4 mAb before presentation to cells partially inhibited binding, whereas a control CD4d3+4 mAb had no effect (Fig. 2 D). The partial inhibition suggests that sites recognized by 2B4 mAb and CD48 may be close but do not overlap.
The m2B4-CD4d3+4 beads also bound to rat thymocytes (not shown) and a rat mast cell line (Fig. 2 E). This binding was completely inhibited by a rat CD48 mAb (Fig. 2 E), indicating that the 2B4–CD48 interaction is conserved between these species, as is the CD2–CD48 interaction (27).
h2B4 Binds to CD48.
The finding that rodent 2B4 and CD48 bind to each other suggests that 2B4 may be a ligand for CD48 in humans. The recent isolation of h2B4 cDNA (Boles, K., and P.A. Mathew, manuscript in preparation) made it possible to test this hypothesis. An h2B4 rCD4d3+4 fusion protein with a COOH-terminal biotinylation tag (h2B4-CD4d3+4) was expressed at levels similar to those obtained with the m2B4-CD4d3+4 construct, consistent with it being correctly folded. Biotinylated h2B4-CD4d3+4 immobilized on fluorescent beads bound to PBMCs (Fig. 3 A). Two hCD48 mAbs (6.28 and MEM 102) were tested for blocking effects on h2B4-CD4d3+4 cell binding. Preincubation of cells with the mAbs revealed blocking of h2B4-CD4d3+4 binding by 6.28 mAb but not by MEM 102 mAb (Fig. 3 A). Both 6.28 and MEM 102 mAbs bound to the cells, with the IgG3 6.28 mAb giving a lower signal as detected by the FITC-labeled secondary reagent (Fig. 3 B). The specificity of blocking by a CD48 mAb against a particular epitope provides convincing evidence that antigenically active h2B4 is binding to CD48 at the cell surface.
Figure 3.
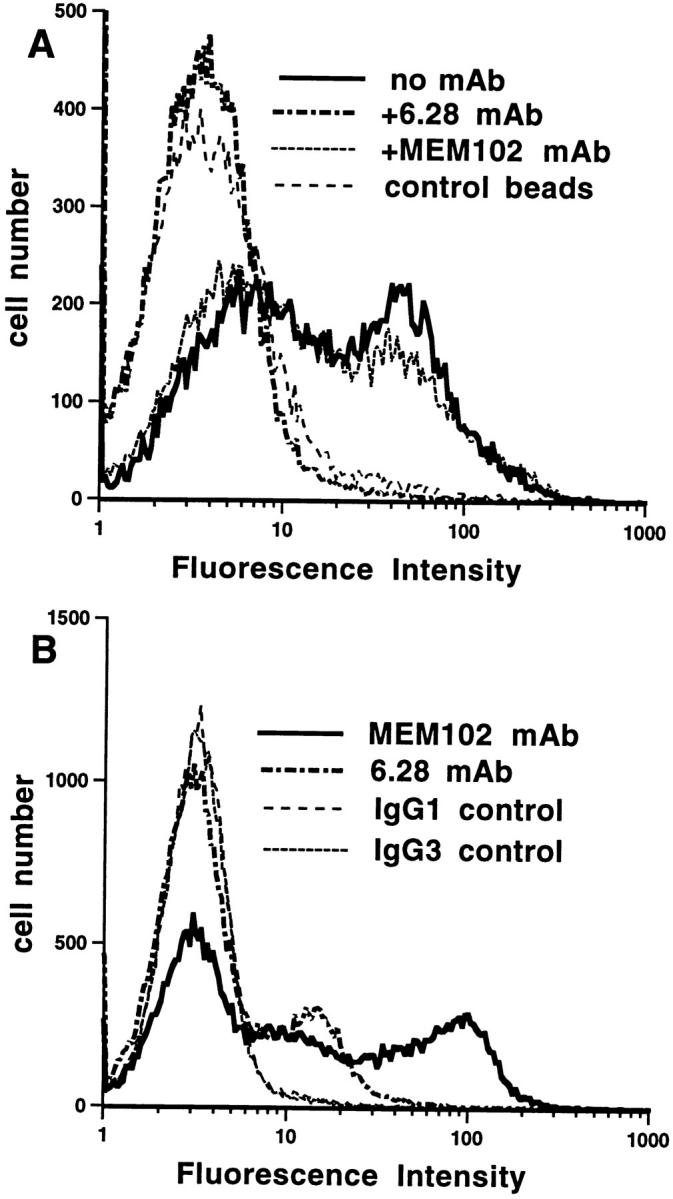
h2B4-CD4d3+4 fluorescent beads bind to hCD48 on cells. (A) h2B4-CD4d3+4 fluorescent beads bind to PBMCs. Preincubation of cells with hCD48 mAb 6.28 but not MEM 102 blocked binding to the level seen with the negative control, rCD4d3+4. (B) CD48 mAbs 6.28 and MEM 102 bind to the cells in A as detected with a fluorescent- labeled second mAb.
Affinity and Kinetic Analysis of the 2B4–CD48 Interaction.
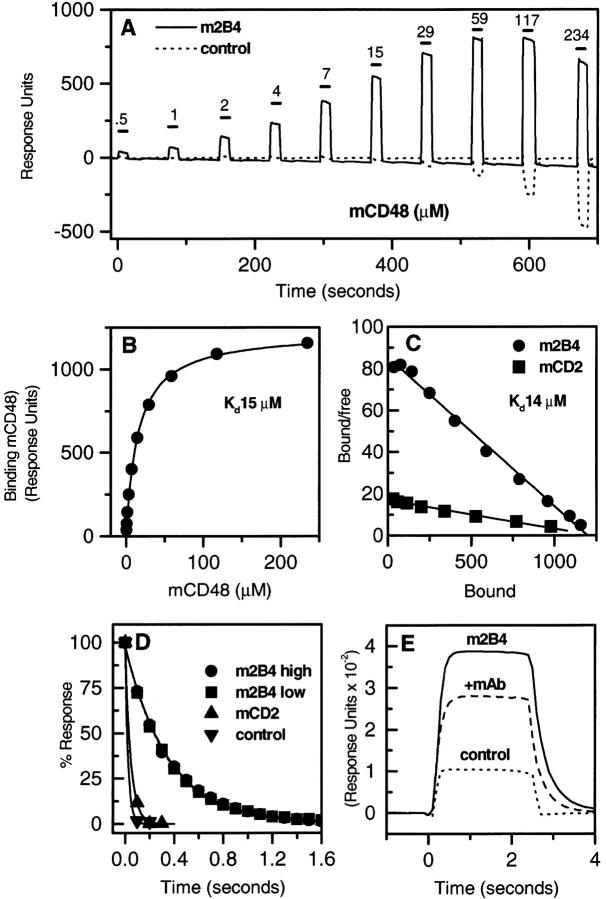
The finding that mCD48 has two ligands, 2B4 and CD2, raises the question of which interaction dominates in a particular response. This will depend on the distribution and level of expression of the proteins and the affinities of the two interactions. The latter was investigated by analysis on a BIAcore™, which permits protein interactions to be followed in real time using a detection system based on surface plasmon resonance. The affinity was measured by determining the level of binding at equilibrium after injection of a range of concentrations of mCD48 over immobilized m2B4-CD4d3+4 and mCD2. Immobilized rCD5-CD4d3+4 was used as a negative control in the reference flow cell. A representative experiment with increasing concentrations of mCD48 injected at 37°C is shown in Fig. 4 A. The difference between the response at equilibrium in the m2B4 and control flow cells represents binding (Fig. 4 B). A Scatchard transformation of the data is shown in Fig. 4 C. mCD48 bound to m2B4 with a K d ≈ 16 μM at 37°C (Fig. 4, B and C, and Table 1). This was significantly higher than the mCD48–mCD2 interaction (K d ≈ 90 μM) measured simultaneously (Fig. 4 C, and Table 1). Values determined for the mCD48–mCD2 interaction agree with published data (27; Table 1). At 25°C, mCD48 bound m2B4 and mCD2 with a K d of 7 and 49 μM, respectively (data not shown).
Figure 4.
Affinity and dissociation rate of soluble mCD48 binding to m2B4-CD4d3+4. (A) Soluble mCD48 was injected at the indicated concentration through flow cells with immobilized m2B4-CD4d3+4 (3,733 RU) or as a negative control, rCD5-CD4d3+4 (1,857 RU), at 37°C. (B) The difference between the response at equilibrium in the m2B4-CD4d3+4 and control flow cells is plotted against the mCD48 concentration. A K d = 15 μM and maximum binding of 1,197–1,225 RU were calculated by nonlinear curve fitting of the Langmuir binding isotherm (line) to data (circles) from A with negative control subtracted. (C) Scatchard plot of data in B plus data for mCD48 binding mCD2. (D) Soluble mCD48 (23.4 μM) was injected over immobilized m2B4-CD4d3+4 at high (1,060 RU) and low (572 RU) levels, mCD2 (557 RU), and rCD5-CD4d3+4 (1,083 RU). A k off = 3 s−1 for 2B4, 22 s−1 for mCD2, and 41 s−1 for rCD5-CD4d3+4 was calculated by exponential decay curve fitting (line) to dissociation data (symbols). (E) In the experiment shown in D, soluble mCD48 (23.4 μM) was injected over immobilized m2B4-CD4d3+4 (1,060 RU) before and after saturation with 2B4 mAb.
Table 1.
Comparison of Affinity and Dissociation Rates for Soluble CD48 Binding to Immobilized 2B4 and CD2 at 37°C
| Immobilized | Soluble | K d | k off |
|---|---|---|---|
| μM | s−1 | ||
| m2B4 | mCD48 | 16 | 3 |
| h2B4 | hCD48 | 8 | >7 |
| mCD2 | mCD48 | 90* | >10* |
| hCD2 | hCD58 | 9–22‡ | >4‡ |
Kinetic studies were carried out at 37°C to determine the dissociation rate constant (k off) of the mCD48–m2B4 interaction. mCD48 dissociated more slowly from m2B4 (k off = 3 s−1) than from mCD2 (k off >20 s−1) (Fig. 4 D). The k off was the same when the level of immobilized m2B4-CD4d3+4 was decreased, indicating that the dissociation rate measured was not affected by rebinding or mass transport limitations. This suggests that the higher affinity of the 2B4–CD48 interaction is the result of a decreased k off.
The partial blocking effect of 2B4 mAb on cell binding (Fig. 2 D) was reproduced in BIAcore™ analysis as shown in Fig. 4 E. Saturation of m2B4-CD4d3+4 with 2B4 mAb before injection of mCD48 reduced the level of mCD48 binding.
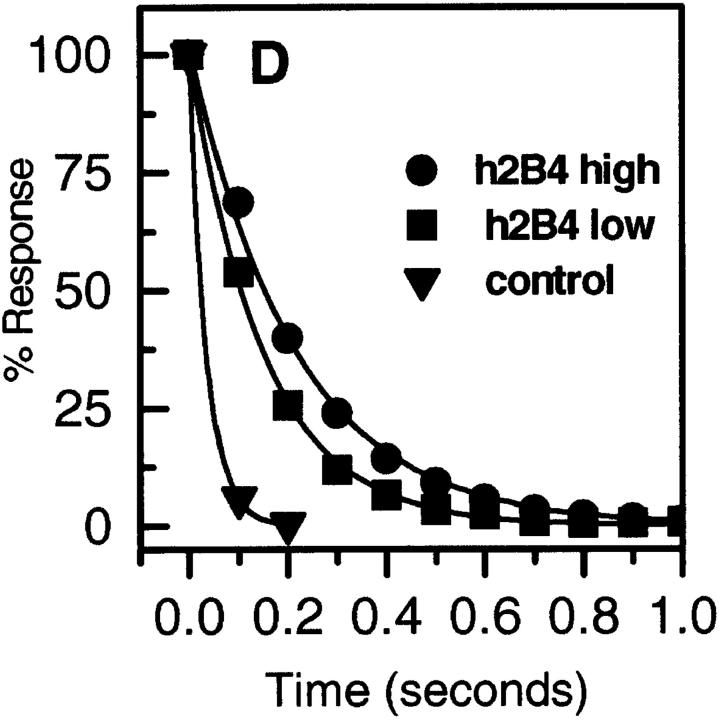
To measure the affinity of the interaction between h2B4 and hCD48, a soluble form of hCD48 was produced. In analyses similar to that carried out for the m2B4–mCD48 interaction, affinity measurements were made with soluble hCD48 and immobilized h2B4-CD4d3+4. Both nonlinear curve fitting (Fig. 5 B) and linear regression analysis of the Scatchard plot (Fig. 5 C) yielded a K d ≈ 8 μM (Table 1). The apparent k off was 5 and 7 s−1 when measured with high and low levels of immobilized h2B4 (Fig. 5 D). This indicates that dissociation is limited by mass transport and/or rebinding and that the true k off is ≥7 s−1.
Figure 5.
Affinity and dissociation rate of soluble hCD48 binding to h2B4-CD4d3+4. (A) Soluble hCD48 was injected at the indicated concentration through flow cells with immobilized h2B4-CD4d3+4 (2,166 RU) or as a negative control, CD4d3+4 (2,112 RU), at 37°C. (B) The difference between the response at equilibrium in the h2B4-CD4d3+4 and control flow cells is plotted against the hCD48 concentration. A K d = 8 μM and maximum binding of 945–959 RU were calculated by nonlinear curve fitting of the Langmuir binding isotherm (line) to data (circles) from A with negative control subtracted. (C) Scatchard plot of data in B. (D) Soluble hCD48 (14.4 μM) was injected over immobilized h2B4-CD4d3+4 at high (1,073 RU) and low levels (578 RU), and CD4d3+4 (1,782 RU). A k off = 5 s−1 for 2B4 high, 7 s−1 for h2B4 low, and 41 s−1 for CD4d3+4 was calculated by exponential decay curve fitting (line) to dissociation data (symbols).
Discussion
The interaction between 2B4 and CD48 identified in this study represents a new ligand pair that is conserved between rodents and humans within the CD2 subset of IgSF molecules. A previous study failed to demonstrate an additional ligand for CD48 in the rat (16). However, 2B4 is expressed on a limited range of cell types, and these were not examined (16). Interestingly, in preliminary experiments, the binding of biotinylated multivalent CD48-CD4d3+4 to lymphokine-activated killer (LAK) cells shown to express both 2B4 and CD2 was entirely blocked by a CD2 mAb (Brown, M.H., unpublished observations). It is possible that with the particular conditions used to generate the LAK cells, too few expressed 2B4 at a sufficiently high surface density and/or 2B4 has insufficient lateral mobility to support multivalent binding. Limitations of this type of assay have been demonstrated previously (16). Thus, our observation that CD48 mAbs completely block 2B4-CD4d3+4 binding to cells does not rule out the existence of another ligand for 2B4. Ianelli et al. have reported that an hCD48–IgM fusion protein binds to epithelial cells, which do not express CD2 (31). It is unclear whether 2B4 is the CD48 ligand on these cells.
Our finding that the m2B4–mCD48 interaction in the mouse has a significantly higher affinity than the mouse or rat CD2–CD48 interactions (26, 27; Table 1) suggests that it is biologically significant. Moreover, the strength of the 2B4–CD48 interaction is conserved across species, with similar affinities being observed for the mouse and human 2B4–CD48 interactions (Table 1). Although fast, the 2B4– CD48 dissociation rate constants are comparable with values measured for other cell–cell recognition molecules, including the CD2–CD48 and CD2–CD58 interactions (27, 28). An interaction has been reported between hCD48 and hCD2 (32, 33), but this appears to be exceptionally weak (K d >0.5 mM [28]). Since CD2 binds with a much higher affinity to CD58 and essentially all cells that express CD48 also express CD58, it is doubtful that the CD2–CD48 interaction is of physiological significance in humans. In contrast, the relatively high affinity of h2B4 for hCD48 suggests that this latter interaction is physiologically important.
The same CD48 mAbs block CD2 and 2B4 binding to CD48, suggesting that the 2B4 ligand binding site, like the CD2 binding site (34), lies on the GFCC′C′′ face of the V domain. Thus, a similar topology for the CD2–CD48 and 2B4–CD48 complexes can be predicted. The dimensions of the CD2–CD48 complex are predicted to be similar to the dimensions of the TCR–MHC complex (14 nm from membrane to membrane [7]). This has led to the suggestion that the maintenance of this intercellular distance is important for initiation of TCR–peptide MHC ligation (7). Interestingly, structure determination of an NK killer inhibitory receptor (KIR [35]) predicts that when bound to MHC class I, the complex will also span a similar distance. Together with functional studies (for a review, see reference 36), this suggests that CD2 on NK cells may contribute to activation through NK receptors. This hypothesis is supported by studies with mAbs and CD58-transfected cells that suggest that CD2 ligation can enhance activation of NK cells (36, 37). By analogy with CD2 (7) and substantiated by experiments with 2B4 mAb (10, 12), the 2B4–CD48 interaction may enhance NK and TCR ligand interaction as well as contributing to signals initiated through NK receptors and the TCR.
The expression on the same lymphocytes of two proteins (2B4 and CD2) that bind the same ligand (CD48) raises the question of whether CD2 and 2B4 have distinct functions. An analogy is exemplified by the well-characterized interactions of the T cell molecules CD28 and CD152, which bind the same ligands, CD80 and CD86 (38). There are several interesting parallels that can be drawn between these two pairs of molecules (CD28/CD152 and 2B4/CD2) that share ligands. First, they have different affinities for the same ligand, i.e., 2B4 and CD2 have different affinities for CD48, and CD152 has a 10-fold higher affinity than CD28 for CD80 (39). Second, their patterns of expression differ. The expression of both 2B4 and CD152 is much enhanced on activated cells (10, 11, 38), whereas both CD2 and CD28 are present on resting and activated cells. Third, they have different cytoplasmic regions. In the case of CD28 and CD152, this results in completely different functions, with CD28 stimulating and CD152 inhibiting (38). Thus, it seems likely that the cytoplasmic portions of 2B4 and CD2 will confer different functions. The more restricted expression of 2B4 on cells associated with non-MHC–restricted killing compared with CD2, which is present on virtually all T cells, suggests the primary role of 2B4 may be in generation of lytic activity. This is supported by the absence of effects of CD48 mAbs on target recognition (29).
Experiments have been done in vitro and in vivo with CD48 and CD2 mAbs that can now be reinterpreted in the light of there being two ligands for mCD48. For example, a CD48 mAb (HM48-1) has been shown to inhibit development of non-MHC–restricted lytic activity, whereas a blocking CD2 mAb had no effect (29). One explanation for this is that it is the 2B4–CD48 and not the CD2–CD48 interaction that has a major role in the development of the lytic population. In vivo, the synergistic effects of blocking CD2 and CD48 mAbs on allograft survival may be explained by disruption of the 2B4–CD48 and the CD2– CD48 interactions (30). Clearly, further experiments are needed to clarify this point. The activating properties (10) and partial inhibition of the 2B4–CD48 interaction make the currently available 2B4 mAb unsuitable for such experiments. Finally, recent observations that there are differences in phenotype between CD48- (40) and CD2-deficient mice (41, 42) can now be investigated with reference to the existence of an alternative ligand for CD48.
Acknowledgments
We thank Martin Wild and Liz Davies for helpful discussions and assistance with BIAcore™ analysis and reagents; Rita Mitnacht-Kraus for assistance with vector construction; Ruth Goddard for assistance with transfections and, with Mike Puklavec, for general cell culture; Amarjit Bhomra for DNA sequencing; Gillian Griffiths for helpful discussions; Dr. Yagita for HM48-1 mAb; and Simon Davis, Nasim Mavaddat, and Don Mason for critical reading of the manuscript.
Abbreviations used in this paper
CD4d3+4
domains 3 and 4 of rat CD4
CD4L
rat CD4 leader
h
human
IgSF
immunoglobulin superfamily
m
mouse
r
rat
RU
response unit(s)
Footnotes
The research was supported by the Medical Research Council and the European Union Biotechnology Programme.
References
- 1.Barclay, A.N., M.H. Brown, S.K.A. Law, A.J. McKnight, M.G. Tomlinson, and P.A. van der Merwe. 1997. The Leucocyte Antigen Factsbook. 2nd ed. Academic Press Ltd., London. 613 pp.
- 2.Preston S, Wright GJ, Starr K, Barclay AN, Brown MH. The leukocyte/neuron cell surface antigen OX2 binds to a ligand on macrophages. Eur J Immunol. 1997;27:1911–1918. doi: 10.1002/eji.1830270814. [DOI] [PubMed] [Google Scholar]
- 3.Hunig T. The cell surface molecule recognized by the erythrocyte receptor of T lymphocytes. Identification and partial characterization using a monoclonal antibody. J Exp Med. 1985;162:890–901. doi: 10.1084/jem.162.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selvaraj P, Plunkett ML, Dustin M, Sanders ME, Shaw S, Springer TA. The T lymphocyte glycoprotein CD2 binds the cell surface ligand LFA-3. Nature. 1987;326:400–403. doi: 10.1038/326400a0. [DOI] [PubMed] [Google Scholar]
- 5.Kato K, Koyanaga M, Okada H, Takanashi T, Wong YW, Williams AF, Okumura K, Yagita H. CD48 is a counter-receptor for mouse CD2 and involved in T cell activation. J Exp Med. 1992;176:1241–1249. doi: 10.1084/jem.176.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Merwe PA, McPherson DC, Brown MH, Barclay AN, Cyster JG, Williams AF, Davis SJ. The NH2-terminal domain of rat CD2 binds rat CD48 with a low affinity and binding does not require glycosylation of CD2. Eur J Immunol. 1993;23:1373–1377. doi: 10.1002/eji.1830230628. [DOI] [PubMed] [Google Scholar]
- 7.Davis SJ, van der Merwe PA. The structure and ligand interactions of CD2: implications for T cell function. Immunol Today. 1996;17:177–187. doi: 10.1016/0167-5699(96)80617-7. [DOI] [PubMed] [Google Scholar]
- 8.de la Fuente MA, Pizcueta P, Nadal M, Bosch J, Engel P. CD84 leukocyte antigen is a new member of the Ig superfamily. Blood. 1997;90:2398–2405. [PubMed] [Google Scholar]
- 9.Wong YW, Williams AF, Kingsmore SF, Seldin MF. Structure, expression, and genetic linkage of the mouse BCM1 (OX45 or Blast-1) antigen. Evidence for genetic duplication giving rise to the BCM1 region on mouse chromosome 1 and the CD2/LFA3 region on mouse chromosome 3. J Exp Med. 1990;171:2115–2130. doi: 10.1084/jem.171.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garni-Wagner BA, Purohit A, Mathew PA, Bennett M, Kumar V. A novel function-associated molecule related to non-MHC-restricted cytotoxicity mediated by activated natural killer cells and T cells. J Immunol. 1993;151:60–70. [PubMed] [Google Scholar]
- 11.Schuhmachers G, Ariizumi K, Mathew PA, Bennett M, Kumar V, Takashima A. 2B4, a new member of the immunoglobulin gene superfamily, is expressed on murine dendritic epidermal T cells and plays a functional role in their killing of skin tumors. J Investig Dermatol. 1995;105:592–596. doi: 10.1111/1523-1747.ep12323533. [DOI] [PubMed] [Google Scholar]
- 12.Schuhmachers G, Ariizumi K, Mathew PA, Bennett M, Kumar V, Takashima A. Activation of murine epidermal gamma delta T cells through surface 2B4. Eur J Immunol. 1995;25:1117–1120. doi: 10.1002/eji.1830250440. [DOI] [PubMed] [Google Scholar]
- 13.Mathew PA, Garni-Wagner BA, Land K, Takashima A, Stoneman E, Bennett M, Kumar V. Cloning and characterization of the 2B4 gene encoding a molecule associated with non-MHC-restricted killing mediated by activated natural killer cells and T cells. J Immunol. 1993;151:5328–5337. [PubMed] [Google Scholar]
- 14.Sewell WA, Brown MH, Owen MJ, Fink PJ, Kozak CA, Crumpton MJ. The murine homologue of the T lymphocyte CD2 antigen: molecular cloning, chromosome assignment and cell surface expression. Eur J Immunol. 1987;17:1015–1020. doi: 10.1002/eji.1830170718. [DOI] [PubMed] [Google Scholar]
- 15.van der Merwe PA, Barclay AN. Transient intercellular adhesion: the importance of weak protein-protein interactions. Trends Biochem Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 16.Brown MH, Preston S, Barclay AN. A sensitive assay for detecting low-affinity interactions at the cell surface reveals no additional ligands for the adhesion pair rat CD2 and CD48. Eur J Immunol. 1995;25:3222–3228. doi: 10.1002/eji.1830251204. [DOI] [PubMed] [Google Scholar]
- 17.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 18.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. . Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 19.Yagita H, Nakamura T, Karasuyama H, Okumura K. Monoclonal antibodies specific for murine CD2 reveal its presence on B as well as T cells. Proc Natl Acad Sci USA. 1989;86:645–649. doi: 10.1073/pnas.86.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korinek V, Stefanova I, Angelisova P, Hilgert I, Horejsi V. The human leucocyte antigen CD48 (MEM-102) is closely related to the activation marker Blast-1. Immunogenetics. 1991;33:108–112. doi: 10.1007/BF00210823. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA. Expression of the Blast-1 activation/adhesion molecule and its identification as CD48. J Immunol. 1991;146:2192–2200. [PubMed] [Google Scholar]
- 22.Brown MH, Barclay AN. Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Protein Eng. 1994;7:515–521. doi: 10.1093/protein/7.4.515. [DOI] [PubMed] [Google Scholar]
- 23.Al-Shamkhani A, Mallett S, Brown MH, James W, Barclay AN. Affinity and kinetics of the interaction between soluble trimeric OX40 ligand, a member of the tumor necrosis factor superfamily, and its receptor OX40 on activated T cells. J Biol Chem. 1997;272:5275–5282. doi: 10.1074/jbc.272.8.5275. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAlister MSB, Brown MH, Barclay AN, Willis AC, Campbell ID, Driscoll PC. Structural studies of the CD5 antigen. Eur J Biochem. 1998;257:131–141. doi: 10.1046/j.1432-1327.1998.2570131.x. [DOI] [PubMed] [Google Scholar]
- 26.van der Merwe PA, Brown MH, Davis SJ, Barclay AN. Affinity and kinetic analysis of the interaction of the cell adhesion molecules rat CD2 and CD48. EMBO J. 1993;12:4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis SJ, Ikemizu S, Wild MW, van der Merwe PA. CD2 and the nature of protein interactions mediating cell-cell recognition. Immunol Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 28.van der Merwe PA, Barclay AN, Mason DW, Davies EA, Morgan BP, Tone M, Krishnam AKC, Ianelli C, Davis SJ. Human cell adhesion molecule CD2 binds CD58 (LFA-3) with a very low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- 29.Chavin KD, Qin L, Lin J, Woodward J, Baliga P, Kato K, Yagita H, Bromberg JS. Anti-CD48 (murine CD2 ligand) mAbs suppress cell-mediated immunity in vivo. Int Immunol. 1994;6:701–709. doi: 10.1093/intimm/6.5.701. [DOI] [PubMed] [Google Scholar]
- 30.Qin L, Chavin KD, Lin J, Yagita H, Bromberg JS. Anti-CD2 receptor and anti-CD2 ligand (CD48) antibodies synergize to prolong allograft survival. J Exp Med. 1994;179:341–346. doi: 10.1084/jem.179.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ianelli CJ, Edson CM, Thorley-Lawson DA. A ligand for human CD48 on epithelial cells. J Immunol. 1997;159:3910–3920. [PubMed] [Google Scholar]
- 32.Sandrin MS, Mouhtouris E, Vaughan HA, Warren HS, Parish CR. CD48 is a low affinity ligand for human CD2. J Immunol. 1993;151:4606–4613. [PubMed] [Google Scholar]
- 33.Arulanandam AR, Moingeon P, Concino MF, Recny MA, Kato K, Yagita H, Koyasu S, Reinherz EL. A soluble multimeric recombinant CD2 protein identifies CD48 as a low affinity ligand for human CD2: divergence of CD2 ligands during the evolution of humans and mice. J Exp Med. 1993;177:1439–1450. doi: 10.1084/jem.177.5.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Merwe PA, McNamee PN, Davies EA, Barclay AN, Davis SJ. Topology of the CD2-CD48 cell-adhesion molecule complex: implications for antigen recognition by T cells. Curr Biol. 1995;5:74–84. doi: 10.1016/s0960-9822(95)00019-4. [DOI] [PubMed] [Google Scholar]
- 35.Fan QR, Mosyak L, Winter CC, Wagtmann N, Long EO, Wiley DC. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors [published erratum 390:315] Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 36.Lanier LL, Corliss B, Phillips JH. Arousal and inhibition of human NK cells. Immunol Rev. 1997;155:145–154. doi: 10.1111/j.1600-065x.1997.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 37.Spruyt LL, Glennie MJ, Beyers AD, Williams AF. Signal transduction by the CD2 antigen in T cells and natural killer cells: requirement for expression of a functional T cell receptor or binding of antibody Fc to the Fc receptor, FcγRIIIA (CD16) J Exp Med. 1991;174:1407–1415. doi: 10.1084/jem.174.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson CB, Allison JP. The emerging role of CTLA-4 as an immune attenuator. Immunity. 1997;7:445–450. doi: 10.1016/s1074-7613(00)80366-0. [DOI] [PubMed] [Google Scholar]
- 39.van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiser, H. 1997. Role of CD48 molecule in lymphocyte activation. Immunology. 92:IS46.
- 41.Killeen N, Stuart SG, Littman DR. Development and function of T cells in mice with a disrupted CD2 gene. EMBO J. 1992;11:4329–4336. doi: 10.1002/j.1460-2075.1992.tb05532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teh SJ, Killeen N, Tarakhovsky A, Littman DR, Teh HS. CD2 regulates the positive selection and function of antigen-specific CD4− CD8+ T cells. Blood. 1997;89:1308–1318. [PubMed] [Google Scholar]