Interleukin (IL)-4–independent Induction of Immunoglobulin (Ig)E, and Perturbation of T Cell Development in Transgenic Mice Expressing IL-13 (original) (raw)
Abstract
Recent studies using interleukin (IL)-4–deficient animals have highlighted the existence of IL-4–independent immunoglobulin (Ig)E induction. We have established transgenic mice expressing IL-13 from a transgene comprising a genomic fragment containing the IL-13 gene and the human CD2 locus control region. The transgenes were expressed in lymphoid tissues and induced by T cell activators, suggesting regulation by elements of the IL-13 promoter. IL-13 transgenic lines expressed 10–100-fold higher levels of serum IgE than their littermate controls, but had normal levels of other serum Ig isotypes. Elevated IgE levels were also detected in sera from IL-4–deficient mice carrying IL-13 transgenes, indicating that IL-4 is not required for IL-13–induced IgE expression in the mouse. Expression of IL-13 also perturbed the development of thymocytes. Although thymocyte development was normal up to 4 wk of age, thymocyte number decreased dramatically thereafter, reaching 10% of normal by 10 wk, and despite normal size and appearance, histological examination demonstrated that transgenic thymi contained only small foci of thymocytes. The reduction in thymocyte number was due mainly to a depletion of CD4+CD8+ thymocytes, and did not affect significantly the composition of peripheral T cell populations. These data indicate that expression of IL-13 transgenes in vivo can regulate IgE production in the mouse, and that IL-13 may also influence thymocyte development.
Keywords: interleukin 13, IgE, interleukin 4, T cells, thymus
Cytokines, it is clear, can modulate B cell function by differentially stimulating specific Ig isotype production. Consequently, if antigenic challenge elicits a Th1 cell response characterized by high levels of IFN-γ, there is an associated increase in the level of IgG2a production. Similarly, immune responses developing a Th2 cell phenotype exhibit high levels of IL-4 and develop elevated serum IgE and IgG1 (for a review, see reference 1). The intrinsic role played by IgE in the development of asthma and allergic responses highlights the importance of understanding the mechanisms controlling its production. IL-4 was believed to be solely responsible for the expression of IgE in the mouse (2–6); however, recent studies using IL-4–deficient mice have clearly shown that IgE can be generated in the complete absence of IL-4 (7, 8), indicating that other mechanisms can mediate IgE production in mice. The discovery of IL-13 and the demonstration that this cytokine could also induce IgE production by human B cells established that a further layer of complexity existed in the regulation of IgE (9–11). IL-13 is a cytokine produced predominantly by Th2 and mast cells, and it shares structural and functional similarities with IL-4 (12, 13). Significantly, addition of IL-13 to in vitro assays has so far failed to induce IgE production by mouse B cells, leading to reports that IL-13 cannot regulate IgE in the mouse (13).
In this paper, we describe the production of IL-13 transgenic mouse lines and present data confirming a role for IL-13 in the development of an IL-4–independent IgE response.
Materials and Methods
Generation of IL-13 Transgenic Mice.
A 6.0-kb BamHI-digested genomic DNA fragment containing the complete mouse IL-13 gene (14) was cloned into a BamHI site upstream of the human CD2 locus control region (LCR; reference 15) in the vector GSE1515 (supplied by Dimitris Kioussis, National Institute for Medical Research, London, UK). The IL-13 and CD2 LCR gene fusion was isolated as an 11.5-kb NotI fragment and injected into eggs. Transgenic mice (strain CBA × C57BL/6) were generated using standard protocols (16). Transgenic mice were identified using Southern blot analysis of genomic DNA hybridized with the CD2 LCR or the mouse IL-13 cDNA (12). In the experiments presented, transgenic lines IL13Tg431 and IL13Tg478 had been backcrossed four times onto strain C57BL/6. IL13Tg431 mice were crossed with IL-4–deficient mice (A.N.J. McKenzie, unpublished data) or IL-4T (5) to derive IL-13Tg+/IL-4−/− offspring. In all experiments presented, transgenic mice were compared with wild-type littermate controls. Mice were maintained in a specific pathogen–free animal facility.
RNA Analysis.
Total RNA was isolated from muscle, kidney, spleen, thymus, bone marrow, heart, and liver using RNAzolB (Biotecx, Biogenesis Laboratories, Poole, UK). Reverse transcription (RT)-PCR primers for IL-13 were 5′-GGGTGACTGCAGTCCTGGCT-3′ and 5′-GCTGGAGACCGTAGTGGG-3′. Hypoxanthine phosphoribosyltransferase (HPRT) primers and conditions were as described previously (17).
Cytokine and Ig ELISAs.
Serum Igs were assayed using sandwich ELISA. 96-well plates were coated with anti-Ig isotype capture mAbs, and bound Ig of diluted serum samples was detected using biotinylated anti-Ig isotype detection mAbs (PharMingen, San Diego, CA). Concentrations were calculated using purified Igs as standards (PharMingen). The IL-13 ELISA was purchased from R&D Systems (Abingdon, UK).
Analysis of IL-13 Production.
LN cells were resuspended at 2 × 106 cells/ml in RPMI 1640 glutamax medium (GIBCO BRL, Paisley, UK), supplemented with 0.05 mM 2-ME, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% FCS, or this medium supplemented with Con A (2 μg/ml). After incubation, supernatants were harvested and assayed for the presence of IL-13 using ELISA.
Immunofluorescent Flow Cytometry.
Single cell suspensions were prepared from thymus, spleen, and LN. Cells were stained with FITC–anti-CD4 Ab (clone H129.19) and PE–anti-CD8 Ab (clone 53-6.7) and analyzed by flow cytometry (FACSCalibur®; Becton Dickinson, San Jose, CA). LN cells were stained with FITC–anti-CD45R/B220 (clone RA3-6B2) and PE–anti-CD23 Ab (clone B3B4). For three-color staining, thymocytes were stained with FITC–anti-CD4, FITC–anti-CD8, FITC–anti-CD3ε (clone 2C11), PE–anti-CD44 (clone IM7), and biotin-conjugated anti-CD25 (clone H129.19, detected with streptavidin–Cy-Chrome). All Abs were purchased from PharMingen.
Results and Discussion
Inducible Tissue-specific IL-13 Expression in IL-13 Transgenic Mouse Lines.
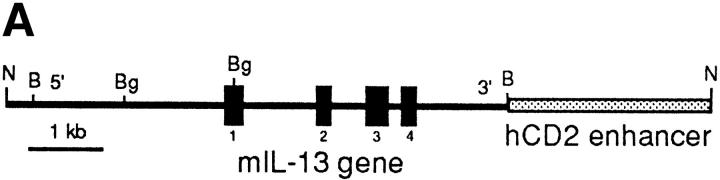
Since IL-13 is primarily produced by T cells, we chose to facilitate transgene expression in the T lymphocyte compartment. To achieve this, we cloned the native mouse IL-13 gene containing ∼3 kb of upstream sequence into the 5′ region of the human CD2 LCR (Fig. 1 A). Using egg injection, several transgenic founders were generated that varied in the number of integrated copies of the IL-13 gene. Three founders were used to establish independent transgenic lines denoted IL13Tg431 (∼6 copies of the transgene), IL13Tg478 (∼10 copies), and IL13Tg520 (∼15 copies), which were analyzed in more detail. Although the data from IL13Tg520 were consistent with those obtained with the other two transgenic lines, they are not presented here because we have determined that the transgenes in this line are on the Y chromosome, thus preventing the use of sex-matched littermate controls.
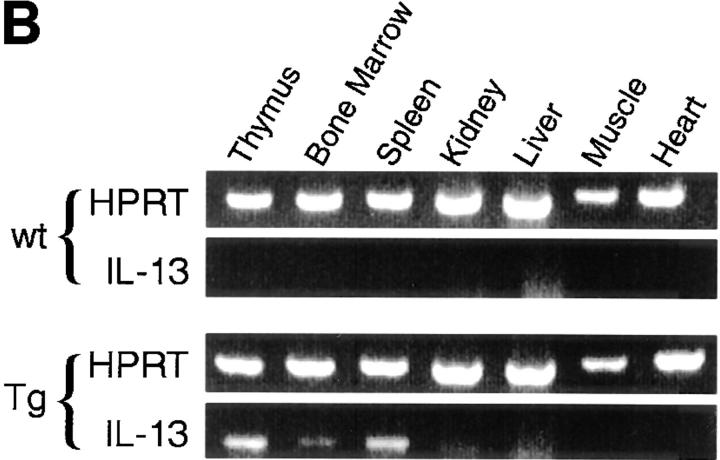
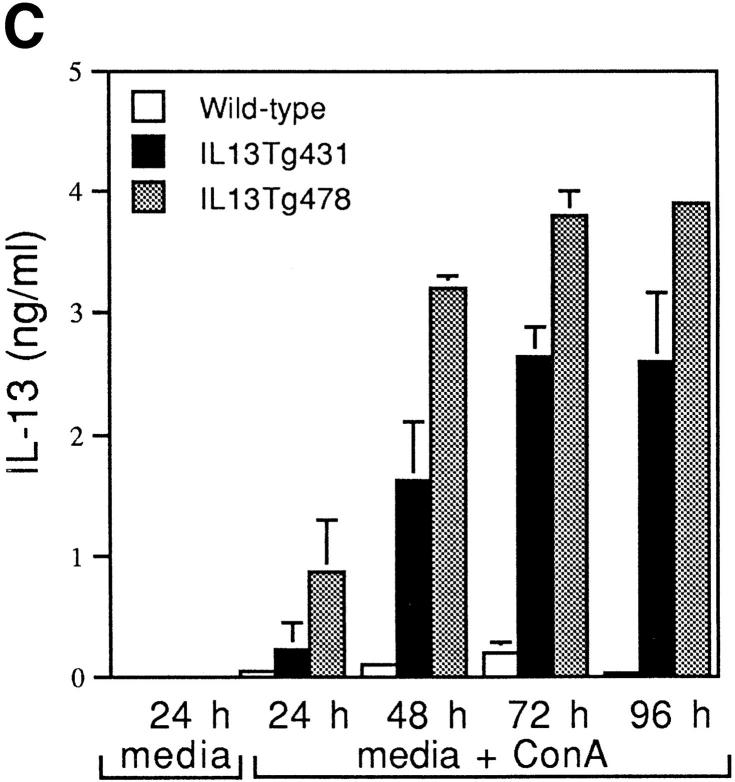
Figure 1.
Construction and expression pattern of IL-13 transgene. (A) Map of the mouse IL-13/human CD2 transgene construct. Black boxes, IL-13 exons. Shaded box, Human CD2 (hCD2) LCR. N, NotI; B, BamHI; Bg, BglII. (B) Total RNA from tissues of IL13Tg431 mice (Tg) and wild-type littermates (wt) analyzed by RT-PCR using IL-13–specific and HPRT primers. (C) IL-13 secretion by activated mesenteric LN cells derived from wild-type mice and IL-13 transgenic littermates. Mesenteric LN cells were cultured in the presence of media, or media containing Con A for the times indicated, and culture supernatants were assayed using IL-13 ELISA. Data were obtained from four wild-type or four transgenic mice in each group.
RT-PCR analysis on cDNA from several tissues clearly demonstrated that expression of IL-13 mRNA was limited to lymphoid tissues, and that this expression was more prevalent in the IL-13 transgenic animals (Fig. 1 B). In addition to the tissue specificity of IL-13 transgene expression, the IL-13 transgene also retained a significant level of activation control. When wild-type and IL-13 transgenic LN cells were cultured in the absence of stimulation, the concentrations of IL-13 protein were below levels of detection (<5 pg/ml). However, upon stimulation with the polyclonal activator Con A, IL-13 expression is induced, with the levels of IL-13 protein produced correlating with the number of copies of the IL-13 genes present, such that IL13Tg478 > IL13Tg431 > wild-type mice (Fig. 1 C). These data demonstrate that the genomic region containing the IL-13 gene retains a capacity for inducible expression, and that IL-13 production is transgene copy number dependent and tissue specific.
Serum Ig Analysis.
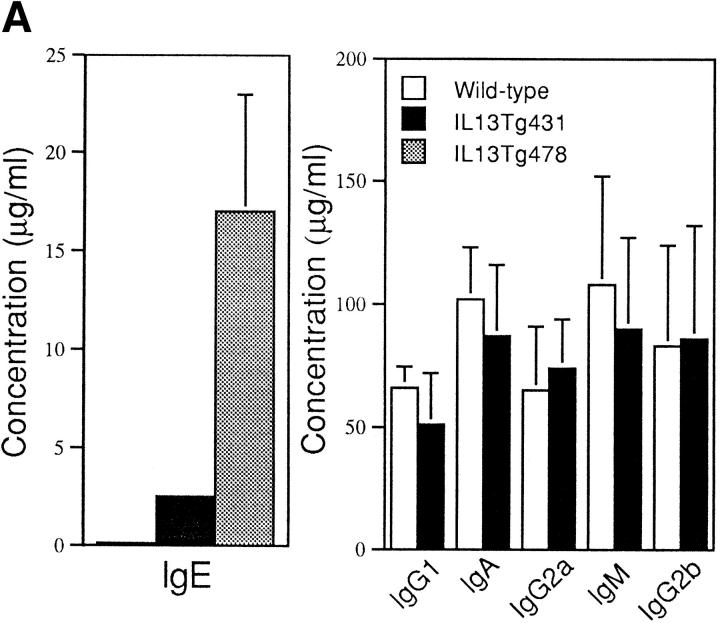
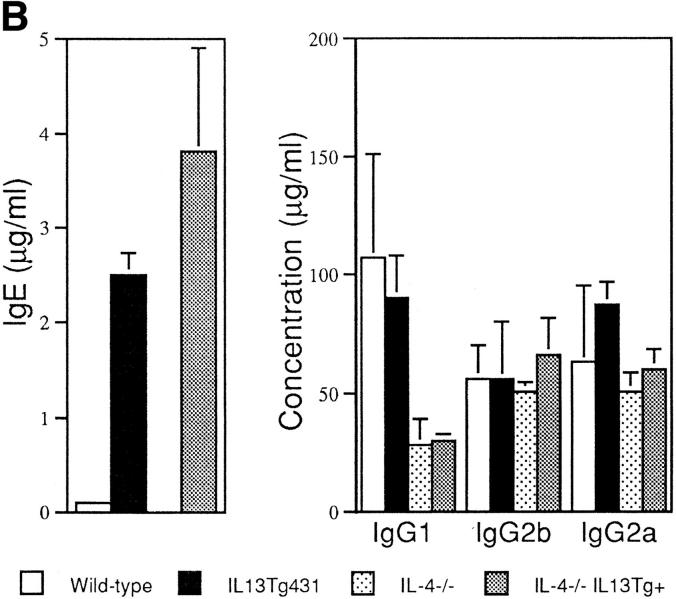
Total serum IgE from naive animals was highly elevated in both of the IL-13 transgenic lines. Indeed, IgE levels were 10-fold greater than normal in IL13Tg431 mice and up to 100-fold higher than normal in the IL13Tg478 animals (Fig. 2 A), indicating that IgE levels increased with the number of integrated copies of the IL-13 gene. In contrast, total serum Ig titers of IgG1, IgG2a, IgG2b, IgA, and IgM were similar in the wild-type, IL13Tg431 (Fig. 2 A), and IL-13Tg478 mice (data not shown). The elevated levels of IgE in the IL-13 transgenics were readily detected in mice at 3, 6, and >10 wk of age (data not shown). Thus, IL-13 expression induces a highly specific upregulation of IgE.
Figure 2.
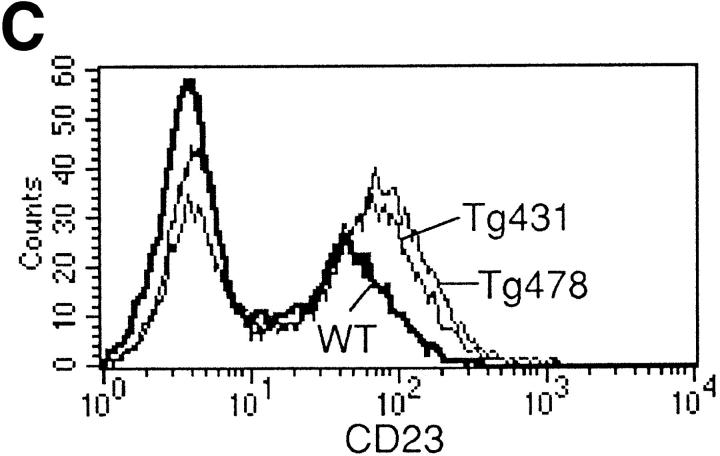
B cell responses to IL-13 transgene expression. (A) Total serum Ig isotype production by wild-type mice and IL-13 transgenic littermates. Represented data were obtained from six wild-type, six IL13Tg431, and four IL-13Tg478 mice. (B) Total serum Ig isotype production by wild-type, IL13Tg431, IL-4−/−, and IL-4−/−IL13Tg+ littermates (five animals per group). (C) Cytofluorometric analysis of CD23 expression on B220+ LN cells. Mesenteric LN cells from IL-13 transgenic mice and nontransgenic littermates were stained for CD23 expression and gated on B220+ cells.
To date, IL-13 has not been demonstrated to regulate IgE switching by mouse B cells in vitro. However, in vivo it is unclear whether IL-13 may mediate its effect directly on B cells or indirectly by modulating other cell types that may influence IgE expression. Although we did not detect a significant increase in total B cell numbers (data not shown), it is possible that IL-13 may be directing the specific outgrowth of IgE-expressing cells in the IL-13 transgenics. Alternatively, and in common with its role on human B cells (11), IL-13 may be acting as an alternative switch factor for IgE transcription. Although IL-4 is a potent IgE differentiation signal for mouse B cells both in vitro (2–6) and in transgenic mice (18, 19), it is also apparent that other factors are able to regulate IgE switching in the mouse. Recent studies have demonstrated that although IL-4–deficient mice generally exhibit undetectable levels of IgE, these animals can still produce IgE after certain antigenic challenge or infection (7, 8). We were unable to detect any changes in the transcription of the IL-4 gene or increased expression of the IL-4 protein from LN cells or splenocytes prepared from the IL-13 transgenic animals (data not shown). To investigate whether IL-13 could still induce IgE expression in the absence of IL-4, we crossed IL-13 transgenics with IL-4–deficient mice. In contrast to naive IL-4−/− animals, in which IgE levels were below the levels of detection, all naive IL-4−/−IL13Tg+ animals expressed high levels of IgE (Fig. 2 B). These data demonstrate that IL-13 induces IgE expression in an IL-4–independent manner. IL-13 transgene expression did not reverse the IgG1 deficiency reported in the IL-4–deficient mice (Fig. 2 B), indicating that IL-13 does not appear to regulate IgG1 production, and correlating with unchanged levels of IgG1 detected in IL-13–deficient mice (20). It is noteworthy that we have detected reduced levels of IgE in IL-13–deficient mice (20), indicating that both upregulation and downregulation of IL-13 can modulate IgE levels in mice. These findings lead us to conclude that IL-13 can modulate IgE levels in mice as well as in humans, but for unknown reasons, this effect cannot be detected using current in vitro mouse B cell differentiation assays (13). Indeed, the contrast between our in vivo results and previous in vitro findings may indicate that IL-13 mediates its role in IgE induction by the regulation of additional cell types.
Coincident with the increase in IgE production, we also observed an elevation in the expression levels of CD23 on B220+ cells from mesenteric LNs (Fig. 2 C) and spleen (data not shown) of IL-13 transgenic mice. These data contrast with a report in which IL-13 failed to modulate CD23 on mouse B cells in vitro (13), but are in agreement with reports that IL-13 can upregulate CD23 on human B cells (11). We have also found that CD23 expression is lower on B cells from IL-13–deficient mice (our unpublished observations). Therefore, our data indicate that in vivo IL-13 can also regulate other facets of the IgE response by B cells.
Perturbation of Thymocyte Development.
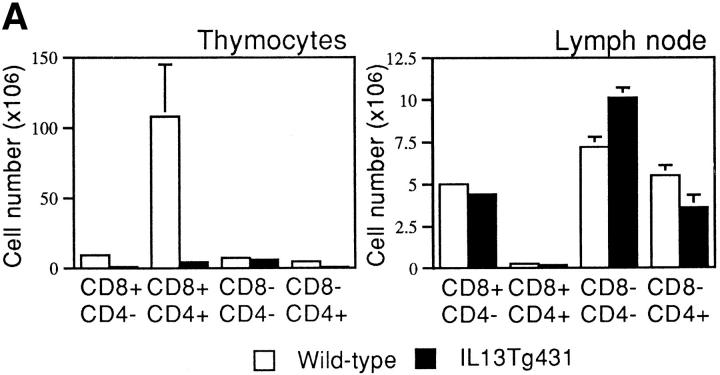
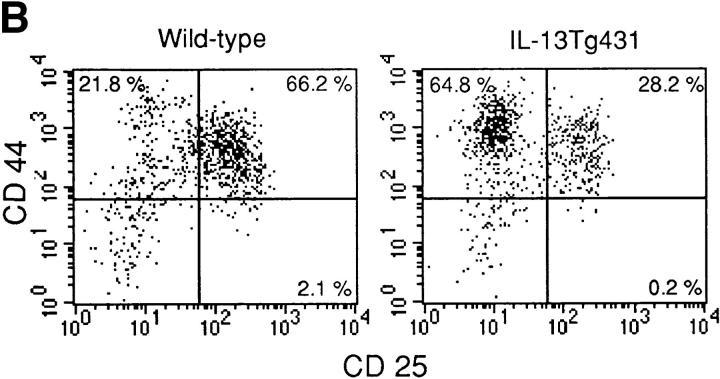
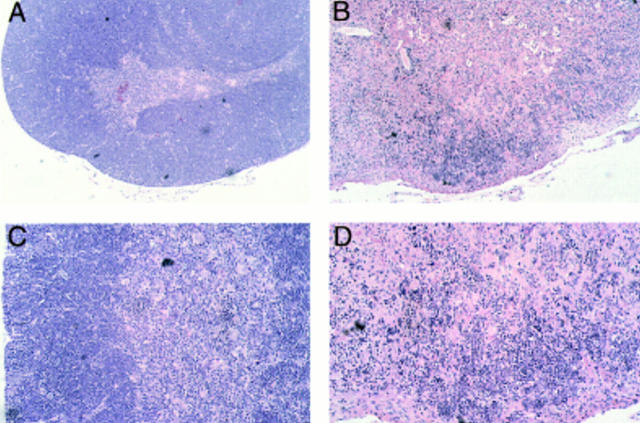
Thymocyte development in IL-13 transgenic mice was severely affected as the mice aged. Thymocyte populations from 1- and 4-wk-old transgenic animals developed normally (data not shown); however, by 6 wk of age, thymocyte numbers in the thymi of IL-13 transgenic animals began to fall to ∼50– 75% of normal. By 10 wk of age, the number of thymocytes was reduced to ∼10% of normal thymocyte number, although thymus size appeared normal. Analysis using the T cell markers CD4 and CD8 indicated that the most significant thymocyte loss was from the CD4+CD8+ subset in the thymi of IL-13 transgenic mice, although CD4+CD8− and CD4−CD8+ cell numbers were also reduced (Fig. 3 A). The numbers of CD4−CD8− cells in the thymus remained equivalent in the IL-13 transgenic mice and their wild-type littermates. However, when CD4−CD8−CD3− cells were analyzed for their expression of the T cell developmental markers CD44 and CD25, the wild-type cells comprised a significantly greater proportion of more mature CD44+CD25+ cells compared with the IL-13 transgenics, which displayed far more cells in the immature CD44+CD25− population (Fig. 3 B). Histologic sectioning of 10-wk-old thymi from IL-13 transgenics and their wild-type littermates revealed a severe breakdown in normal thymus architecture in the IL-13 transgenic animals. Whereas the littermate controls exhibited normal thymic structure consisting of cortex and medulla (Fig. 4, A and C), thymi from IL-13 transgenic mice contained diffuse foci of thymocyte development surrounded by substantial areas of epithelial structure devoid of thymocytes (Fig. 4, B and D).
Figure 3.
Thymocyte number and surface phenotype. (A) Single cell suspensions prepared from thymus and LN were analyzed for surface expression of CD4 and CD8. (B) Thymus samples were also analyzed for surface expression of CD44 and CD25. In this case, CD4−CD8− CD3− thymocytes were analyzed for expression of CD44 and CD25.
Figure 4.
Histologic sections from thymi of wild-type and IL-13 transgenic mice at 10 wk of age after formalin fixation, paraffin embedding, and staining with hematoxylin and eosin. Low power magnification (original magnification ×10) demonstrates that compared with their wild-type littermates (A), significant areas of transgenic thymus lack thymocyte populations (B). Higher power magnification (original magnification ×20) indicates that whereas wild-type thymus contains distinct cortical and medulla structure (C), this is lost in transgenic thymus (D).
Unlike the distorted thymocyte populations observed in the thymus, there was only a small decrease in CD4+CD8− T cells observed in the peripheral mesenteric LNs of the IL-13 transgenics compared with their wild-type littermates (Fig. 3 A) and no discernible differences in the CD4/ CD8 numbers or ratios in the spleen (data not shown). Furthermore, we failed to find any differences in the expression of a number of other cell surface markers (i.e., NK1.1, CD11b, CD117, CD54, or Ly-6G) in spleen, bone marrow, or thymus (data not shown), and we were unable to demonstrate any differences in the growth responses of T or B cells from transgenic mice or wild-type littermates in response to a range of mitogens (data not shown). Therefore, although it is unclear how IL-13 mediates the late onset of thymic depopulation, it is apparent that the number of mature peripheral T cells is not affected significantly, and that macrophage and granulocyte numbers are unchanged (data not shown).
The apparently normal thymocyte profiles generated from the IL-13 transgenic animals up to 4 wk of age demonstrate that normal thymocyte development occurs unhindered during the early development of the thymus; however, as evidenced by the severe depletion of thymocytes from 6 through to 10 wk, IL-13 is able to affect the typical differentiation of immature thymocytes through the CD4+CD8+ stage. Furthermore, we have found that inclusion of IL-13 into in vitro fetal thymic organ cultures also results in an inhibition of thymocyte development (data not shown). However, it remains to be determined whether IL-13 acts directly on the T cell populations or if it mediates its effects by regulating other cell types such as thymic epithelial cells.
Several other transgenic mouse lines expressing a range of factors, including soluble cytokines (21–23), transcription factors (24), or inflammatory molecules (25), have been reported to develop thymus phenotypes similar to that we have observed in the IL-13 transgenics. Hormones such as estrogen can also produce a profound reduction in the numbers of double positive thymocytes (23), as can the administration of glucocorticosteroids (26). Although the final outcome on thymocyte populations may appear similar in these disparate models, it seems likely that they arise by different mechanisms, and that the CD4+CD8+ thymocyte subset is uniquely sensitive to modulatory stimuli for reasons that have yet to be elucidated. It is noteworthy that IL-4 transgenic mice also develop an arrest in the normal development of CD4+CD8+ thymocytes in the thymus, with a coincident breakdown in the typical structure of thymic medulla and cortex (18, 19, 21). However, despite the reduction in numbers of CD4+ CD8+ thymocytes in IL-4 transgenic mice, these animals, unlike the IL-13 transgenics described here, are not reported to have large areas of the thymus devoid of thymocytes. Furthermore, in contrast to the IL-13 transgenics, IL-4 transgenic animals are reported to have losses in peripheral CD4+ T cells (19) or peripheral CD8+ T cells (18, 21) depending on the promoter used to control transgene expression.
Our data demonstrate that expression of IL-13 in vivo can modulate IgE production even in the absence of IL-4, and that the apparent discrepancies between the roles of IL-13 in the murine and human systems appear to be due to features inherent in the in vitro assay systems used for their analysis. Thus, IL-13 transgenic animals represent an important tool for defining the in vivo regulation of IgE by IL-4 and IL-13 in disease.
Acknowledgments
The authors wish to thank Grahame McKenzie for helpful discussions and advice on the manuscript, D. Kioussis for the gift of the human CD2 LCR vector, and Jill Davies for histology.
References
- 1.Coffman RL, Lebman DA, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 2.Finkelman FD, Katona IM, Urban JF, Jr, Snapper CM, Ohara J, Paul WE. Suppression of in vivo polyclonal IgE responses by monoclonal antibody to the lymphokine B-cell stimulatory factor 1. Proc Natl Acad Sci USA. 1986;83:9675–9678. doi: 10.1073/pnas.83.24.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkelman FD, Katona IM, Urban JF, Jr, Holmes J, Ohara J, Tung AS, Sample JV, Paul WE. IL-4 is required to generate and sustain in vivo IgE responses. J Immunol. 1988;141:2335–2341. [PubMed] [Google Scholar]
- 4.Finkelman FD, Urban JF, Jr, Beckmann MP, Schooley KA, Holmes JM, Katona IM. Regulation of murine in vivo IgG and IgE responses by monoclonal anti-IL-4 receptor antibody. Int Immunol. 1991;3:599–607. doi: 10.1093/intimm/3.6.599. [DOI] [PubMed] [Google Scholar]
- 5.Kühn R, Rajewsky K, Müller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 6.Kopf M, Le Gros G, Bachmann M, Lamers MC, Bluethmann H, Köhler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 7.Van der Weld T, Kopf M, Köhler G, Langhorne J. The immune response to Plasmodium chabaudimalaria in interleukin-4-deficient mice. Eur J Immunol. 1994;24:2285–2293. doi: 10.1002/eji.1830241004. [DOI] [PubMed] [Google Scholar]
- 8.Morawetz RA, Gabriele L, Rizzo LV, Noben-Trauth N, Kühn R, Rajewsky K, Müller W, Doherty TM, Finkelman F, Coffman RL, Morse HC., III Interleukin (IL)-4–independent immunoglobulin class switch to immunoglobulin (Ig)E in the mouse. J Exp Med. 1996;184:1651–1661. doi: 10.1084/jem.184.5.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenzie ANJ, Culpepper JA, de Waal R, Malefyt, Briere F, Punnonen J, Aversa G, Sato A, Dang W, Cocks BG, Menon S, et al. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc Natl Acad Sci USA. 1993;90:3735–3739. doi: 10.1073/pnas.90.8.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minty A, Chalon P, Derocq J-M, Dumont X, Guilemot J-C, Kaghad M, Labit C, Leplatois P, Liauzun P, Miloux B, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 11.Punnonen J, Aversa G, Cocks BG, McKenzie ANJ, Menon S, Zurawski G, de Waal R, Malefyt, de Vries JE. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90:3730–3734. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown KD, Zurawski SM, Mosmann TR, Zurawski G. A family of small inducible proteins secreted by leukocytes are members of a new superfamily that includes leukocyte and fibroblast-derived inflammatory agents, growth factors, and indicators of various activation processes. J Immunol. 1989;142:679–687. [PubMed] [Google Scholar]
- 13.Zurawski G, de Vries JE. Interleukin 13, an interleukin 4-like cytokine that acts on monocytes and B cells but not on T cells. Immunol Today. 1994;15:19–26. doi: 10.1016/0167-5699(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 14.McKenzie ANJ, Li X, Largaespada DA, Sato A, Kaneda A, Zurawski SM, Doyle EL, Milatovich A, Francke U, Copeland NG, et al. Structural comparison and chromosomal localization of the human and mouse IL-13 genes. J Immunol. 1993;150:5436–5444. [PubMed] [Google Scholar]
- 15.Greaves DR, Wilson FD, Lang G, Kioussis D. Human CD2 3′-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., R. Beddington, F. Costantini, and E. Lacey. 1994. Manipulation of the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 17.Murphy E, Hieny S, Sher A, O'Garra A. Detection of in vivo expression of interleukin-10 using a semi-quantitative polymerase chain reaction method in Schistosoma mansoniinfected mice. J Immunol Methods. 1993;162:211–223. doi: 10.1016/0022-1759(93)90386-l. [DOI] [PubMed] [Google Scholar]
- 18.Tepper RI, Levinson DA, Stanger BZ, Campos-Torres J, Abbas AK, Leder P. IL-4 induces allergic-like inflammatory disease and alters T cell development in transgenic mice. Cell. 1990;62:457–467. doi: 10.1016/0092-8674(90)90011-3. [DOI] [PubMed] [Google Scholar]
- 19.Erb KJ, Holtschke T, Muth K, Horak I, Schimpl A. T cell subset distribution and B cell hyperreactivity in mice expressing interleukin-4 under the control of major histocompatibility complex class I regulatory sequences. Eur J Immunol. 1994;24:1143–1147. doi: 10.1002/eji.1830240520. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie GJ, Bancroft A, Grencis R, McKenzie ANJ. A distinct role for interleukin-13 in Th2 cell mediated immune responses. Curr Biol. 1998;8:339–342. doi: 10.1016/s0960-9822(98)70134-4. [DOI] [PubMed] [Google Scholar]
- 21.Lewis DB, Yu CC, Forbush KA, Carpenter J, Sato TA, Grossman A, Liggitt DH, Perlmutter RM. Interleukin 4 expressed in situ selectively alters thymocyte development. J Exp Med. 1991;173:89–100. doi: 10.1084/jem.173.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen MM, Skoda RC, Cardiff RD, Campos-Torres J, Leder P, Ornitz DM. Expression of LIF in transgenic mice results in altered thymic epithelium and apparent interconversion of thymic and lymph node morphologies. EMBO (Eur Mol Biol Organ) J. 1994;13:1375–1385. doi: 10.1002/j.1460-2075.1994.tb06391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirahara H, Ogawa M, Kimura M, Iiai T, Tsuchida M, Hanawa H, Watanabe H, Abo T. Glucocorticoid independence of acute thymic involution induced by lymphotoxin and estrogen. Cell Immunol. 1994;153:401–411. doi: 10.1006/cimm.1994.1038. [DOI] [PubMed] [Google Scholar]
- 24.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in δEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabarra B, Casanova M, Paris D, Nicole A, Toyama K, Sinet PM, Ceballos I, London J. Transgenic mice overexpressing the human Cu/Zn-SOD gene: ultrastructural studies of a premature thymic involution model of Down's syndrome (trisomy 21) Lab Invest. 1996;74:617–626. [PubMed] [Google Scholar]
- 26.van Vliet E, Melis M, van Ewijk W. The influence of dexamethasone treatment on the lymphoid and stromal composition of the mouse thymus: a flow cytometric and immunohistological analysis. Cell Immunol. 1986;103:229–240. doi: 10.1016/0008-8749(86)90086-9. [DOI] [PubMed] [Google Scholar]