Density of the Notch ligand Delta1 determines generation of B and T cell precursors from hematopoietic stem cells (original) (raw)
Abstract
Notch signaling regulates multiple cell fate decisions by hematopoietic precursors. To address whether different amounts of Notch ligand influence lineage choices, we cultured murine bone marrow lin−Sca-1+c-kit+ cells with increasing densities of immobilized Delta1ext-IgG consisting of the extracellular domain of Delta1 fused to the Fc domain of human IgG1. We found that relatively lower densities of Delta1ext-IgG enhanced the generation of Sca-1+c-kit+ cells, Thy1+CD25+ early T cell precursors, and B220+CD43−/lo cells that, when cocultured with OP9 stroma cells, differentiated into CD19+ early B cell precursors. Higher densities of Delta1ext-IgG also enhanced the generation of Sca-1+c-kit+ precursor cells and promoted the development of Thy1+CD25+ cells, but inhibited the development of B220+CD43−/lo cells. Analyses of further isolated precursor populations suggested that the enhanced generation of T and B cell precursors resulted from the effects on multipotent rather than lymphoid-committed precursors. The results demonstrate the density-dependent effects of Delta1 on fate decisions of hematopoietic precursors at multiple maturational stages and substantiate the previously unrecognized ability of Delta1 to enhance the development of both early B and T precursor cells.
The detection of Notch receptors in hematopoietic precursors suggested a role for Notch signaling in early hematopoietic development, whereas the potential role of Notch in regulating the self-renewal of multipotent precursors and in determining T versus B cell fates has been demonstrated in both gain- and loss-of-function studies (1–5). Studies using exogenous cell-expressed or soluble Notch ligand forms to activate endogenous Notch receptors have also revealed the effects of Notch signaling on the self-renewal of nonmutant hematopoietic precursors (6, 7). Jagged1, Jagged2, Delta1, and Delta4 have thus all been shown to affect the differentiation of hematopoietic precursors, including repopulating cells. In addition, Delta1 and Delta4 have been shown to promote early T cell differentiation (6–12). The expression of the different Notch ligands and their receptors at different times and in a tissue-specific manner raises the possibility of a unique function for each ligand (13, 14). However, the mechanisms by which specific ligands induce different cellular fates remains unclear.
We investigated whether quantitative differences in the amount of Notch signaling induced by different doses of the Notch ligand Delta1 might account for these selective effects. This is suggested by studies with Drosophila that demonstrate how different functions of Notch can require different threshold levels of signaling. For example, half the wild-type level of Notch gene dosage is insufficient to properly specify the dorsoventral margin of a wing, giving rise to the eponymous “notched wing” phenotype; however, it does suffice for most functions of Notch in the wild type (15, 16).. In vertebrates, a relative reduction in the level of Notch1 in developing T cells can influence their fate to the extent that cells with a single copy of the Notch1 gene are less likely to become αβ T cells than are wild-type cells (17). In developing B cells, a relative reduction in Notch2 results in diminished B1 B cells and a marked reduction in marginal zone B cells (18). The quantitative effects of Notch ligands have also been demonstrated (a) in mice, where the homozygous deletion of Jagged1 led to embryonic lethality though a partial phenotype resulted only in eye defects (19); and (b) in humans, where the haploinsufficiency of Jagged1 appears to be responsible for Alagille syndrome, a pleiotropic disorder involving multiple organ systems (20). We show that the Notch ligand Delta1 can enhance the generation of early T and B cell progenitors from multipotent hematopoietic precursors depending on the density of the ligand. We demonstrate that higher densities of Delta1 mainly promote the adoption of a T cell fate, whereas relatively lower densities enhance the generation of early T- and B-lymphoid precursors.
Results and Discussion
To assess the effect of varying densities of immobilized Delta1ext-IgG on the induction of Notch signaling, we measured the expression of the Notch target gene, Hes1, in lin−Sca-1+c-kit+ (LSK) marrow cells with a Hoechst 33342 dye efflux profile of a “side population” enriched for stem cells (LSK-SP). After culturing for 3 h on plates coated with increasing concentrations of Delta1ext-IgG in the presence of murine stem cell factor (mSCF), human Flt-3 ligand (Flt3L), IL6, and IL11 (4GF), Hes1 mRNA expression was assessed using SYBR green quantitative RT-PCR. Compared with control cells cultured on human IgG1, cells cultured with the ligand plated at 2.5 μg/ml showed a 1.4 ± 0.1-fold increase in Hes1 expression, whereas cells cultured with the ligand plated at 10 μg/ml showed a 2.6 ± 0.2-fold increase (mean ± SEM from four separate experiments). This indicated an increased activation of Notch signaling in a Delta1ext-IgG dose-dependent manner.
To investigate the effect of varying densities of Delta1ext-IgG on hematopoietic cell proliferation and differentiation, we assessed the types and numbers of cells generated after culturing marrow-sorted LSK cells for 14 d with increasing densities of Delta1ext-IgG. We found a 10-fold greater generation of cells in the presence of Delta1ext-IgG plated at concentrations of ≥1.25 μg/ml compared with the number of cells generated with control IgG1 (Fig. 1 A). Additionally, a phenotypic analysis revealed that the number of Sca-1+c-kit+ cells generated was significantly higher in cultures with densities established by plating at ≥2.5 μg/ml of Delta1ext-IgG compared with cultures with control IgG1 or lower densities of Delta1ext-IgG (≤1.25 μg/ml). In contrast, the number of myeloid cells (Gr1+ and/or F4/80+) generated in cultures with lower densities of Delta1ext-IgG (≤2.5 μg/ml) was comparable with that in control IgG1 cultures, but was significantly reduced in cultures with densities of Delta1ext-IgG established by plating at ≥5 μg/ml of Delta1ext-IgG (Fig. 1, B and D).
Figure 1.
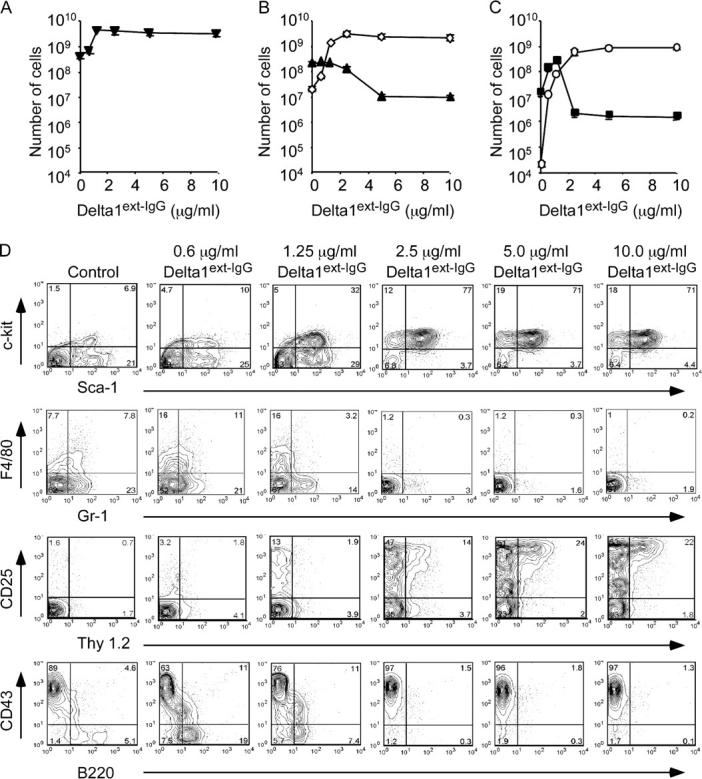
Proliferation and differentiation of LSK cells during culture with various Delta1ext-IgG densities. (A) Total number of cells (▾) generated from LSK cells after 14 d of incubation with Delta1ext-IgG plated at increasing concentrations. (B) Number of cells exhibiting an immature (Sca-1+c-Kit+; ⋄) or myeloid (Gr1+ and/or F4/80+; ▴) phenotype. (C) Cells with an early T cell (Thy1+CD25+; ○) or early lymphoid (B220+ CD43−/lo; ▪) phenotypes were determined based on cell count and FACS analysis. (D) The data shown represent the mean ± SEM of three experiments. The numbers in the corners of the flow cytometry plots represent the percentage of gated events within that quadrant.
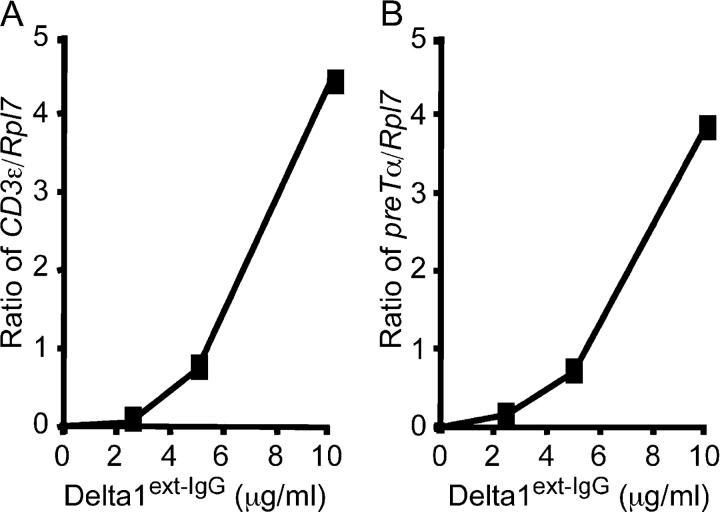
In cultures with an increasing density of Delta1ext-IgG, we found increased numbers of cells expressing Thy1 and CD25, which is suggestive of early T cell differentiation (Fig. 1 C). Compared with control IgG1, there was a 4-log increase in the number of Thy1+CD25+ cells in cultures with Delta1ext-IgG plated at ≥5 μg/ml. Furthermore, the presence of T cell precursors was confirmed by quantitative RT-PCR that revealed the expression of _CD3_ɛ and _pre-T_α mRNA at 28 d of culture in cells cultured on Delta1ext-IgG, but not in control cultures (Fig. 2, A and B).
Figure 2.
Induction of pre-T α and CD3 ɛ expression is dependent on the density of Delta1ext-IgG. (A) _CD3_ɛ and (B) _pre-T_α gene expressions by LSK cells after culture for 28 d with different densities of Delta1ext-IgG determined by quantitative RT-PCR. Data shown represent the ratio of gene expression compared with the housekeeping gene Rpl7.
Studies by us and others have previously reported that B cell differentiation is inhibited in cultures with Delta1 (7, 11, 21). However, our previous studies tested immobilized Delta1ext-IgG plated only at a high concentration (10 μg/ml). To determine whether a culture with lower densities of Delta1ext-IgG allowed for the generation of B lymphoid precursors, cultured LSK cells were also analyzed for the expression of the B cell–associated antigens B220, CD43, CD19, and IgM. Cells expressing CD19 or IgM were not generated in any culture conditions (unpublished data). However, the number of B220+CD43−/lo cells increased substantially in cultures containing lower densities of Delta1ext-IgG (0.6–1.25 μg/ml) compared with control IgG1 or higher density Delta1ext-IgG-containing cultures (≥2.5 μg/ml; Fig. 1, C and D).
To investigate the B cell potential of cells generated with the Notch ligand, LSK cells cultured for 21 d with varying densities of Delta1ext-IgG were transferred onto an OP9 stromal cell culture containing mSCF, Flt3L, and IL7. These cells were then analyzed by FACS analysis at 1 and 2 wk, revealing CD19+ cells only in cultures containing cells previously incubated with low densities of immobilized Delta1ext-IgG (0.6 μg/ml or 1.25 μg/ml; unpublished data). To determine whether the CD19+ cells generated by coculture with OP9 stromal cells were obtained from the B220+CD43−/lo population, we also isolated B220+CD43−/lo and B220− cells from the cultured LSK cells by FACS before transferring them onto OP9 stromal cell cultures. After 7 d, B220+ CD19+ cells were observed only in cultures initiated with B220+CD43−/lo cells obtained from LSK cells that had been cultured in the presence of Delta1ext-IgG plated at 1.25 μg/ml (Fig. 3 A). CD19+ cells were not generated after the culture of the sorted B220− cells on OP9 cells (Fig. 3 B).
Figure 3.
T and B cell potential of Delta1ext-IgG–cultured B220**+**CD43−/lo or B220− populations. After the culture of LSK cells for 21 d with different densities of Delta1ext-IgG, B220+CD43−/lo (A) or B220− (B) cells were isolated by FACS and transferred to cultures containing OP9 stromal cells or 10 μg/ml of Delta1ext-IgG. (C) Early B (B220+CD19+) or early T (Thy1+CD25+) cell phenotypes were determined with FACS analysis. The numbers in the corners of the flow cytometry plots represent the percentage of gated events within that quadrant.
In addition, the T cell potential of the sorted B220+ CD43−/lo and B220− populations generated after the culture of LSK cells with different densities of Delta1ext-IgG was also examined by transferring these sorted cells into wells containing Delta1ext-IgG plated at 10 μg/ml, along with 4GF and IL7. After 7 d of culture, the sorted B220+CD43−/lo cells failed to proliferate (unpublished data). However, the sorted B220− cells from wells established with Delta1ext-IgG plated at ≥2.5 μg/ml did proliferate and generate Thy1+CD25+ cells (Fig. 3 C). These results suggest that only the B220− cells retained the potential to differentiate along the T cell lineage, whereas the B220+CD43−/lo cells were limited to early B cell differentiation. The failure of these B220+CD43−/lo cells to differentiate towards the T cell lineage when cultured with relatively higher Delta1ext-IgG densities further suggests that these cells were committed to B cell differentiation. This finding raises the question of whether the previously demonstrated thymic repopulating ability of B220+ cell results from the B220+ subset that expresses higher levels of CD43 (22).
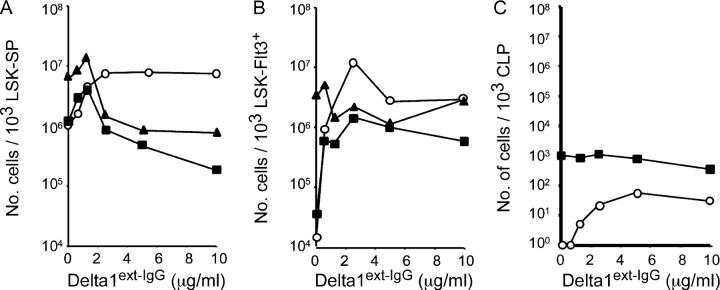
To identify the multipotent or committed precursors that are affected by Notch signaling, we isolated subpopulations of marrow LSK cells, including (a) LSK-SP cells that are enriched for long-term repopulating stem cells (23); (b) LSK cells that express the Flt3 receptor and are enriched for short-term repopulating stem cells (LSK-Flt3+; reference 24); and (c) lin−Sca-1loc-kitloThy1−/loIL7+ cells that are enriched for common lymphoid progenitors (CLPs; reference 25). Similar to LSK cells, LSK-SP cells cultured with lower densities of Delta1ext-IgG generated increased numbers of Thy1+CD25+ early T cell precursors and B220+CD43−/lo early B cell precursors, as well as Gr-1+ and/or F4/80+ myeloid cells. Furthermore, as with LSK cells, LSK-SP cells cultured with densities of ligand generated fewer numbers of B220+CD43−/lo and Gr-1+ and/or F4/80+ cells, but increased numbers of Thy1+CD25+ cells compared with LSK-SP incubated with lower densities of Delta1ext-IgG (Fig. 4 A). Results obtained with LSK-Flt3+ cells were similar to those obtained with LSK-SP cells, except that fewer B cells and myeloid cells were generated at lower ligand densities (Fig. 4 B). This is possibly because the LSK-Flt3+ cells possess less potential for proliferation and self-renewal than the less mature LSK-SP cells. In contrast, CLPs cultured with or without ligand did not generate significantly increased numbers of any type of cells, but primarily differentiated into B220+CD43−/lo cells after 3 d of culture with or without ligand. CLPs cultured with higher densities of Delta1ext-IgG did, however, generate Thy1+ CD25+ cells, although it is not clear whether these cells were derived from uncommitted or committed T-lymphoid precursors (Fig. 4 C).
Figure 4.
Differentiation of LSK-SP, LSK-Flt3**+**, and CLP cells during culture with various Delta1ext-IgG densities. LSK-SP, LSK-Flt3+, and CLP cells isolated by FACS from marrow were incubated with Delta1ext-IgG plated at increasing concentrations. LSK-SP (A) and LSK-Flt3+ (B) cells were incubated for 14 d and CLP (C) cells were incubated for 3 d. Numbers of generated myeloid (Gr1+ and/or F4/80+; ▴), early T cell (Thy1+CD25+; ○), or early lymphoid (B220+ CD43−/lo; ▪) cells were determined based on cell count and FACS analysis.
In this study, we have shown that ligand-induced Notch activation in multipotent hematopoietic precursor cells regulates lineage expression in a density-dependent manner. Our data suggest that the enhanced formation of B and T precursors occurs in the presence of lower densities of Delta1ext-IgG because of the effects on multipotent precursors. In addition, higher densities of ligand lead mainly to T cell differentiation because of the inhibition of early B cell and myeloid differentiation by multipotent precursors and the promotion of T cell differentiation by lymphoid committed precursors. These results represent the first evidence that a single Notch ligand can enhance the development of cells that adopt either a B or a T cell fate.
Previous studies of cell fate outcomes induced by Delta1 have used cell-expressed ligands and were thus unable to address quantitative differences in ligand expression (11, 21, 26). However, our studies suggest that lower densities of Delta1 induce Notch signaling enough to increase precursor cell numbers that then assume the default B cell pathway because the induced Notch signaling was insufficient to further promote development along the T cell pathway and inhibit B cell, as well as myeloid cell, development. Similar differential cell fate choices have been seen in studies comparing Notch signaling induced by Delta versus Jagged (21, 26, 27). The latter differences may result from distinct receptors being induced by the different ligands or they may reflect decreased signal intensity from one or the other ligand. This is supported by studies showing that Notch signaling induced by Jagged may result in decreased signal intensity in the presence of Fringe, where Jagged1 activates Notch2 but not Notch1 (28). Thus, Notch signaling induced by Jagged1 would expectedly be lower than that induced by Delta1 because Delta1 activates both Notch1 and Notch2. The finding that different concentrations of a single Notch ligand leads to differential cell fate outcomes raises the possibility that multiple Notch ligand–receptor combinations simply regulate Notch signaling quantitatively to affect cell fates. However, qualitative differences between Notch1- and Notch2-induced signaling may also play a role.
It remains unclear how quantitative differences in Notch ligand density leads to different cell fate outcomes. It is possible that Notch signaling must exceed specific thresholds in order to instruct particular cell fates. For example, different Hes1 expression levels induced in individual cells might differentially affect downstream genes that have different thresholds for Hes1 regulation. Other Notch target genes, such as Hes5 or Herp2, may also be differentially induced and serve in concert to regulate differentiation programs.
The variable densities of individual Notch ligands in different regions of marrow or thymic stroma, together with the expression of Notch ligands by developing hematopoietic cells that may activate Notch signaling in adjacent cells, suggest the potential importance of ligand density in vivo. In addition to the implications of our findings for growing a variety of stem cell types in vitro for therapeutic purposes, the investigation of physiologic stimuli that might induce variations of Notch ligand expression could provide a novel avenue for manipulating cell development in vivo.
Materials and Methods
Generation of Delta1ext-IgG
Delta1ext-IgG were prepared as previously described (29). Reagent grade purified human IgG1 was purchased from Sigma-Aldrich.
Immobilization of varying Delta1ext-IgG densities
The ligand was immobilized on the plastic surface of the culture vessel as previously described (7). The direct relationship of the plated ligand and the bound ligand concentration was demonstrated by ELISA with an HRP-conjugated Fc specific anti–human-IgG antibody (Sigma-Aldrich) for the detection of Delta1ext-IgG, which demonstrated a linear relationship between the increasing concentration of plated ligand and the increased amounts of bound ligand (R2 = 0.73; P = 0.007).
Cell isolation and immunofluorescence studies
The immunophenotype was analyzed by five-color flow cytometery using an LSR cytometer (Becton Dickinson). Antibodies were purchased from BD Biosciences, unless otherwise noted. Cultured cells were prepared as previously described (6) and stained with the following: (a) FITC-conjugated monoclonal antibodies against Thy1, Gr1, and B220; (b) PE-conjugated monoclonal antibodies against CD25, CD43, CD19, CD127, CD135, and F4/80 (Caltag); and (c) biotinylated antibodies against Sca-1, B220, and IgM secondarily stained with streptavidin-PE-Cy7, and APC monoclonal antibodies against c-kit. LSK cells were cells with a high expression of Sca-1 and c-Kit on bone marrow depleted from the following lineages: CD2, CD3, CD8a, CD5, CD11b, B220, GR-1, and TER-119. LSK cells were obtained using FACS on a Vantage Cell Sorter (Becton Dickinson) as previously described (6). LSK-SP cells were isolated with the addition of Hoechst 33342 dye (Calbiochem) and gating was as described previously (23). LSK-Flt3+ cells were isolated from lineage-depleted bone marrow using FITC-conjugated anti–Sca-1, PE-conjugated anti-CD135 (Flk-2/Flt3), and APC-conjugated anti–c-kit as described previously (24). CLP was sorted using lineage-depleted bone marrow stained with FITC-conjugated anti-Thy1, PE-conjugated anti–CD-127, biotinylated anti–Sca-1, and the APC-conjugated anti–c-kit, and gated as described previously (25). For the isolation of B220+CD43−/lo and B220− cells, cultured cells were stained with FITC-conjugated anti-CD43 antibody and PE-conjugated anti-B220 antibody and isolated by FACS. Sorted cells were ≥95% pure, as determined by a postsort analysis. In all cases, cells were stained with DAPI or propidium iodide (Sigma-Aldrich) to gate for dead cells.
Hematopoietic cell culture
C57BL/6J (Ly5.2) mice (8–10 wk old) obtained from the Jackson Laboratory were maintained and bred at the Fred Hutchinson Cancer Research Center. 103 LSK cells were cultured with immobilized ligand in IMDM supplemented with 20% FBS and 4GF (100 ng/ml each of mSCF, human Flt3L, and human IL6, and 10 ng/ml human IL11; PeproTech) as previously described (7). To test for B cell differentiation, OP9 stromal cell monolayers were prepared 2 d prior by plating 2.5 × 104 OP9 cells/well in a 24-well tissue culture–treated plate (Costar) in αMEM with 20% FBS. Cultured LSK cells were then plated onto an OP9 stromal monolayer in αMEM containing 20% FBS, penicillin, and 10 ng/ml Flt3L, mSCF, and human IL7 (PeproTech). Supplemental media were added every fourth day.
RNA isolation and real-time RT-PCR
The total RNA was extracted with an Absolutely RNA RT-PCR Miniprep Kit (Stratagene) according to the manufacturer's instructions. Single strand cDNA was synthesized with oligo-dT primer using reagents provided in the ThermoScript RT-PCR System (Invitrogen) for 45 min at 50°C. Quantitative PCR was performed using SYBR Green PCR Master Mix on an ABI PRISM 7700 sequence detection system (Applied Biosystems) in the following conditions: an initial denaturation step at 94°C for 10 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s. Transcript quantification was performed in duplicate for every sample, and each gene expression was normalized to the housekeeping gene Ribosomal protein L7 (Rpl7) expression. Primers were designed from sequences of different exons for each gene to prevent the amplification of genomic DNA as follows: Hes1 forward primer, 5′-GGCCTCTGAGCACAGAAAGT-3′; Hes1 reverse primer, 5′-GTGTTAACGCCCTCACACG-3′; _pre-T_α forward primer, 5′-TGGGAGGCAGACTAGCAGAG-3′; _pre-T_α reverse primer, 5′-CCCATAGGTGAAGGCGTCTA-3′; _CD3_ɛ forward primer, 5′-GTCCGCCATCTTGGTAGAGA-3′; _CD3_ɛ reverse primer, 5′-TTGAGGCTGGTGTGTAGCAG-3′; Rpl7 forward primer, 5′-GAAGCTCATCTATGAGAAGGC-3′; and Rpl7 reverse primer, 5′-AAGACGAAGGAGCTGCAGAAC-3′.
Statistical methods
Statistical significances were determined using analysis of variance with Bonferroni posttest.
Acknowledgments
The authors thank Edward Giniger and Roland Walter for their helpful criticism of the manuscript, as well as Ted A. Gooley for performing the statistical analysis.
This work was supported by grants from the National Institutes of Health (P50 HL54881, T32CA09351 and K23 HL077446). I.D. Bernstein was also supported as an American Cancer Society–F.M. Kirby Clinical Research Professor.
The authors have no conflicting financial interests.
References
- 1.Varnum-Finney, B., L. Xu, C. Brashem-Stein, C. Nourigat, D. Flowers, S. Bakkour, W.S. Pear, and I.D. Bernstein. 2000. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 6:1278–1281. [DOI] [PubMed] [Google Scholar]
- 2.Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnell, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, and W.S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, A., H.R. MacDonald, and F. Radtke. 2001. Notch 1–deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- 6.Varnum-Finney, B., L.E. Purton, M. Yu, C. Brashem-Stein, D. Flowers, S. Staats, K.A. Moore, I. Le Roux, R. Mann, G. Gray, et al. 1998. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood. 91:4084–4091. [PubMed] [Google Scholar]
- 7.Varnum-Finney, B., C. Brashem-Stein, and I.D. Bernstein. 2003. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood. 101:1784–1789. [DOI] [PubMed] [Google Scholar]
- 8.Tsai, S., J. Fero, and S. Bartelmez. 2000. Mouse Jagged2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood. 96:950–957. [PubMed] [Google Scholar]
- 9.Lauret, E., C. Catelain, M. Titeux, S. Poirault, J.S. Dando, M. Dorsch, J.L. Villeval, A. Groseil, W. Vainchenker, F. Sainteny, and A. Bennaceur-Griscelli. 2004. Membrane-bound delta-4 notch ligand reduces the proliferative activity of primitive human hematopoietic CD34+ CD38low cells while maintaining their LTC-IC potential. Leukemia. 18:788–797. [DOI] [PubMed] [Google Scholar]
- 10.Jones, P., G. May, L. Healy, J. Brown, G. Hoyne, S. Delassus, and T. Enver. 1998. Stromal expression of Jagged 1 promotes colony formation by fetal hematopoietic progenitor cells. Blood. 92:1505–1511. [PubMed] [Google Scholar]
- 11.Schmitt, T.M., and J.C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 12.Dorsch, M., G. Zheng, D. Yowe, P. Rao, Y. Wang, Q. Shen, C. Murphy, X. Xiong, Q. Shi, J.C. Gutierrez-Ramos, et al. 2002. Ectopic expression of Delta4 impairs hematopoietic development and leads to lymphoproliferative disease. Blood. 100:2046–2055. [PubMed] [Google Scholar]
- 13.Felli, M.P., M. Maroder, T.A. Mitsiadis, A.F. Campese, D. Bellavia, A. Vacca, R.S. Mann, L. Frati, U. Lendahl, A. Gulino, and I. Screpanti. 1999. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 11:1017–1025. [DOI] [PubMed] [Google Scholar]
- 14.Parreira, L., H. Neves, and S. Simoes. 2003. Notch and lymphopoiesis: a view from the microenvironment. Semin. Immunol. 15:81–89. [DOI] [PubMed] [Google Scholar]
- 15.Lindsley, D.L., and G.G. Zimm. 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego. 1133 pp.
- 16.Fehon, R.G., K. Johansen, I. Rebay, and S. Artavanis-Tsakonas. 1991. Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J. Cell Biol. 113:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washburn, T., E. Schweighoffer, T. Gridley, D. Chang, B.J. Fowlkes, D. Cado, and E. Robey. 1997. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 88:833–843. [DOI] [PubMed] [Google Scholar]
- 18.Witt, C.M., W.J. Won, V. Hurez, and C.A. Klug. 2003. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J. Immunol. 171:2783–2788. [DOI] [PubMed] [Google Scholar]
- 19.McCright, B., J. Lozier, and T. Gridley. 2002. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 129:1075–1082. [DOI] [PubMed] [Google Scholar]
- 20.Oda, T., A.G. Elkahloun, B.L. Pike, K. Okajima, I.D. Krantz, A. Genin, D.A. Piccoli, P.S. Meltzer, N.B. Spinner, F.S. Collins, and S.C. Chandrasekharappa. 1997. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16:235–242. [DOI] [PubMed] [Google Scholar]
- 21.Jaleco, A.C., H. Neves, E. Hooijberg, P. Gameiro, N. Clode, M. Haury, D. Henrique, and L. Parreira. 2001. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J. Exp. Med. 194:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, C.H., I. Aifantis, M.L. Scimone, U.H. von Andrian, B. Reizis, H. von Boehmer, and F. Gounari. 2003. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat. Immunol. 4:866–873. [DOI] [PubMed] [Google Scholar]
- 23.Goodell, M.A., K. Brose, G. Paradis, A.S. Conner, and R.C. Mulligan. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183:1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adolfsson, J., O.J. Borge, D. Bryder, K. Theilgaard-Monch, I. Astrand-Grundstrom, E. Sitnicka, Y. Sasaki, and S.E. Jacobsen. 2001. Upregulation of Flt3 expression within the bone marrow Lin(-) Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 15:659–669. [DOI] [PubMed] [Google Scholar]
- 25.Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. [DOI] [PubMed] [Google Scholar]
- 26.Lehar, S.M., J. Dooley, A.G. Farr, and M.J. Bevan. 2005. Notch ligands Delta1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 105:1440–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amsen, D., J.M. Blander, G.R. Lee, K. Tanigaki, T. Honjo, and R.A. Flavell. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 117:515–526. [DOI] [PubMed] [Google Scholar]
- 28.Hicks, C., S.H. Johnston, G. diSibio, A. Collazo, T.F. Vogt, and G. Weinmaster. 2000. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2:515–520. [DOI] [PubMed] [Google Scholar]
- 29.Varnum-Finney, B., L. Wu, M. Yu, C. Brashem-Stein, S. Staats, D. Flowers, J.D. Griffin, and I.D. Bernstein. 2000. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci. 113:4313–4318. [DOI] [PubMed] [Google Scholar]