Hsp40 Facilitates Nuclear Import of the Human Immunodeficiency Virus Type 2 Vpx-Mediated Preintegration Complex (original) (raw)
Abstract
Human immunodeficiency virus type 2 (HIV-2) Vpx is required for nuclear translocation of the viral preintegration complex (PIC) in quiescent cells. In order to decipher the mechanism of action of Vpx, a cDNA library was screened with the yeast two-hybrid assay, resulting in the identification of heat shock protein 40, Hsp40/DnaJB6, as a Vpx-interactive protein. Interaction with Vpx was confirmed by glutathione _S_-transferase (GST) pull-down and coimmunoprecipitation assays. Overexpression of Hsp40/DnaJB6 enhanced Vpx nuclear import, whereas overexpression of a nuclear localization mutant of Hsp40/DnaJB6 (H31Q) or down-regulation of Hsp40/DnaJB6 by small interfering RNA (siRNA) reduced the nuclear import of Vpx. Hsp40/DnaJB6 competed with the Pr55Gag precursor protein for the binding of Vpx and incorporation into virus-like particles. Overexpression of Hsp40/DnaJB6 promoted viral PIC nuclear import, whereas siRNA down-regulation of Hsp40/DnaJB6 inhibited PIC nuclear import. These results demonstrate a role for Hsp40/DnaJB6 in the regulation of HIV-2 PIC nuclear transport.
The human and simian immunodeficiency viruses (HIV and SIV, respectively) have evolved the ability to productively infect nondividing cells, a unique feature that distinguishes lentiviruses from other retroviruses (40). This is mediated by active transport of the viral preintegration complex (PIC) into the nucleus without breakdown of the nuclear envelope during cell division. Components of the PIC that have been implicated in regulating nuclear import include the central DNA flap, as well as viral proteins IN, MA, and Vpr (HIV type 1 [HIV-1]) or Vpx (HIV-2 and SIV) (6, 12, 16, 41). In addition, a mutant CA protein of HIV-1 was reported to have deleterious effects on nuclear targeting and integration, suggesting that HIV-1 PIC nuclear import may depend on proper uncoating of the virus (10). However, it is still poorly understood how the PIC is transported into the nucleus.
Vpx of HIV-2 is a 112-amino-acid protein that is important for efficient infection of nondividing cells (11, 26). Vpx contains a nuclear localization signal (NLS) within amino acids 65 to 72 (3, 20). Moreover, N-terminal (residues 20 to 40) and C-terminal residues (101 to 112) may also affect the nuclear import of Vpx (20, 26, 35). It has been suggested that importins α and β contribute to this activity (35). Although expression of Vpx, in the absence of other viral proteins, results in a predominant nuclear localization, coexpression of the Pr55Gag precursor protein shifts Vpx to the plasma membrane and virus particles (26, 27).
Cellular heat shock proteins (Hsps) are chaperones known to prevent aggregation and participate in the refolding of misfolded proteins during stresses such as heat shock, UV irradiation, and microbial infections (15). In mammalian cells, expression of Hsps is regulated at the transcriptional level (15). Hsps also play essential roles in a variety of other cellular functions, assisting in the folding of newly translated proteins, guiding protein translocation across organelle membranes, assembling and disassembling oligomeric protein complexes, and facilitating the proteolytic degradation of unstable proteins (22). Hsp40s are cochaperones for Hsp70 that regulate its ATPase activity. Members of the Hsp40/DnaJ family of proteins have three distinct domains, (i) a highly conserved J domain of approximately 70 amino acids, often near the N terminus, that is responsible for the interaction with Hsp70, (ii) a glycine and phenylalanine (G/F)-rich region that putatively acts as a flexible linker; and (iii) a cysteine-rich, zinc finger-containing C-terminal domain (25).
Hsp40 affects the replication of diverse viruses. Hsp40 suppresses hepatitis B virus replication through destabilization of viral core and X proteins (36). Hsp40 is also crucial for Nef-mediated enhancement of HIV-1 gene expression and replication (19). Moreover, other Hsps have been examined in HIV-infected cells. Expression of Hsp70 and Hsp27 is enhanced by HIV-1 infection (39). Hsp70 has been suggested to play a role in the nuclear import of the HIV-1 PIC since it promotes the nuclear import, in quiescent cells, of HIV with Vpr deleted (1). Hsp60 and Hsp70 are incorporated into HIV-1 virions and protected from protease digestion (13). Hsp70 is also associated with HIV-2, SIVmac, and SIVagm, but not murine leukemia virus, as a result of a specific interaction with Gag (13). Recent findings suggest that Hsp70 enzymatic activity is required for virion core integrity (14).
In this study, we identified Vpx-interacting factors by the yeast two-hybrid screening method in order to better understand the mechanism of HIV-2 PIC-mediated nuclear import. Our results demonstrated that Vpx interacts with Hsp40/DnaJB6, one member of the Hsp40/DnaJ family. Overexpression of Hsp40/DnaJB6 specifically enhances the nuclear localization of Vpx. Moreover, down-regulation of Hsp40/DnaJB6 by small interfering RNA (siRNA) reduces the nuclear import of viral PICs, and overexpression of Hsp40/DnaJB6 facilitates their nuclear transport. These results suggest that Hsp40/DnaJB6 modulates the targeting of viral PICs to the nuclei of infected cells.
MATERIALS AND METHODS
Plasmids, viruses, cell culture, transfection, and infection.
pTM-Vpx was used to express HIV-2rod Vpx in HeLa and BSC40 cells for immunoprecipitation and immunofluorescence assays (27). The full-length gene for Hsp40/DnaJB6 was amplified from a human lymphocyte cDNA library by PCR, confirmed by DNA sequencing, and cloned into pCNF-myc, a pcDNA3.1 vector with Flag and Myc sequences at the 5′ and 3′ ends of the multiple cloning site, respectively, resulting in pCNF-Hsp40/DnaJB6 (pCNF-B6). pTM-Hsp40/DnaJB6 was made by subcloning the tagged Hsp40/DnaJB6 cDNA into pTM-3. A plasmid containing a Hsp40/DnaJB6 siRNA sequence (pLKO-B6) was constructed by cloning the targeted sequence (5′CCAGTTAAAGTCCTTAACAAT) into pLKO.1, a lentivirus vector (37). Site-directed mutagenesis was used to construct a lentivirus expression clone (pRRL-B6) in pRRL-MagicCpuro for an siRNA-resistant form of Hsp40/DnaJB6 by using the sequence 5′CCAATTGAAATCGTTAACCAT. A negative control plasmid, pLKO-GFP, expressing green fluorescent protein (GFP) siRNA, was a kind gift of S. A. Stewart (Washington University).
For generation of virus stocks, 293T cells were transfected with wild-type or Vpx deletion mutant HIV-2rod10 proviral DNAs, and viral stock concentrations were determined by using an SIV core antigen (p27) enzyme-linked immunosorbent assay (Coulter) (17). Virus stocks used for Alu integration assays were treated with TURBO DNase (30 min at 37°C at 20 U/ml; Ambion). Viral titers were determined by measuring 50% tissue culture infective doses on JC53BL (HeLa-CD4/CCR5) cells by the Reed-Muench accumulation method (8).
Lentiviruses encoding siRNAs for Hsp40/DnaJB6 or GFP were made by transfecting 293T cells with 4 μg pLKO-B6 or pLKO-GFP, 0.5 μg pVSV-G, and 4 μg pGag-Pol (HIV-1) (37). Stable U937 and HeLa siRNA-expressing cell lines were established by infection and selection with G418 (1 mg/ml) for 2 to 3 weeks. A lentivirus expressing Hsp40/DnaJB6 resistant to siRNA was made by transfecting 293T cells with pRRL-B6, pVSVG, and pGag-Pol. Stable U937 cells were obtained by infection and selection of U937 encoding siRNAs for Hsp40/DnaJB6 with both G418 (1 mg/ml) and puromycin (1 μg/ml) for 2 weeks.
293T, HeLa, and BSC40 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 4 mM l-glutamine, 1 mM Na-pyruvate, and 100 μg/ml penicillin-streptomycin. U937 cells were maintained in RPMI medium with the same supplements. Transfections utilized TransIT-LT1 (Mirus Bio).
Yeast two-hybrid screening.
A human lymphocyte MATCHMAKER cDNA library in the pACT2 vector (Clontech) was screened for Vpx-interacting proteins by cotransformation with bait plasmid pGBKT7-vpx into yeast strain AH109. Cotransformants in AH109 were selected for growth on plates with histidine, leucine, and tryptophan dropout. Positive colonies were confirmed by streaking them onto plates also lacking adenine, rescued by transformation into Escherichia coli Top10 competent cells, and transformed back into yeast AH109 containing nonspecific bait, human lamin A, to ensure that the interaction is specific. The positive genes were identified by DNA sequencing.
Immunoprecipitation and Western blot analysis.
BSC40 cells were transfected with pTM-Vpx and pTM-Hsp40/DnaJB6 and then infected with vaccinia virus encoding T7 polymerase (vTF7-3). The cells were harvested at 16 h postinfection and lysed in phosphate-buffered saline (PBS) containing 0.5% NP-40 and protease inhibitors at 4°C. The lysate was cleared by centrifugation at 18,000 × g for 5 min. Lysate proteins were immunoprecipitated overnight at 4°C with mouse anti-Flag antibody (Sigma), mouse anti-Myc antibody (Clontech), or rabbit anti-Vpx antibody, followed by incubation with protein A-agarose for 1 h. The agarose beads were washed, and bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer and resolved by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting. Endogenous Hsp40/DnaJB6 was detected with a mouse polyclonal antibody (Obnova, Taiwan).
Immunofluorescence microscopy.
HeLa cells grown on coverslips were infected with vaccinia virus (vTF7-3) and then transfected with pTM-Vpx and/or pTM-Hsp40/DnaJB6. After 16 h, cells were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and stained with either polyclonal anti-Vpx or monoclonal anti-Flag antibody and then with Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (IgG; Molecular Probes) for Vpx or Alexa Fluor 954-conjugated goat anti-mouse IgG for Hsp40/DnaJB6. Nuclei were visualized by using 0.5 μg/ml Hoechst 33258 pentahydrate (Molecular Probes). Cells were visualized with a fluorescence microscope equipped with an Optronics Magnafire imaging system (Goleta).
Nuclear and cytoplasmic extract preparation.
HeLa cells were infected with vTF7-3 and transfected with pTM-Vpx or pTM-Hsp40/DnaJB6, trypsinized, pelleted, and resuspended in 10 mM HEPES (pH 7.4)-10 mM KCl-1 mM dithiothreitol-0.6% NP-40-protease inhibitor cocktail (Sigma) on ice for 10 min. After centrifugation at 1,000 × g for 5 min, the supernatant was collected as the cytoplasmic fraction and the nuclear fraction was prepared by resuspending the remaining pellet in 100 μl 20 M HEPES (pH 7.4)-150 mM NaCl-1 mM dithiothreitol. Equal amounts of nuclear and cytoplasmic extracts were used for immunoblotting of Vpx, actin, and histone H1.
Alu integration assay.
U937 cells stably expressing either Hsp40/DnaJB6 or GFP siRNA or JC53BL cells containing pCNF-B6 (Hsp40/DnaJB6) or pCNF were infected with HIV-2rod10 for the indicated times and then washed with PBS. DNA was then isolated with the DNeasy tissue kit (Qiagen). Integrated proviral DNA was detected by a nested PCR as described previously (2, 4, 21). The first-round PCR was carried out for 12 cycles with the primer 5′-ATGCCACGTAAGCGAAACTGCGAGGCTGGCAGATTGAGCCCTG-3′(L-Rod) (the 5′ underlined sequence is a linker and is followed by a sequence complementary to the long terminal repeat [LTR] of HIV-2) and two Alu primers (5′-TCCCAGCTACTGGGGAGGCTGAGG and 5′-GCCTCCCAAAGTGCTGGGATTACAG). A second-round real-time PCR was performed with an iQ Supermix reagent (Bio-Rad) and the primers 5′-ATGCCACGTAAGCGAAACTGC (specific to the linker sequence) and 5′-TTACTCAGGTGAACACCGAATGACCAGGC (complementary to the HIV-2 LTR) and the probe 5′-FAM-TCCCATCTCTCCTAGTCGCCGCCT-Iowa Black-3. Genomic DNA from U937 cells chronically infected with HIV-2 served as a standard. Total viral DNA was quantified by amplification of the vpr gene of HIV-2 by real-time PCR with a plasmid containing the HIV-2rod proviral clone as a standard. The sequences of the primers were 5′GGTCTGGTCTAATGGCTGAAGCAC (nucleotides 5671 to 7594) and 5′ACATCCTGCTCTGAAGTGCGTGAA (nucleotides 5924 to 5900).
For the measurement of two-LTR circle products, extrachromosomal DNA was isolated with a QIAprep miniprep kit (Qiagen) by the modified protocol for the isolation of low-copy-number plasmids (33). Two-LTR circles were quantitated by real-time PCR with primers that span the junction generated by ligation of the DNA ends. The sequences of the primers were 5′TGGTCTGTTAGGACCCTTCTTGCT (U5 region 245 to 268) and 5′TCCTCTTGCTTTCAGTCTCGCCTT (U3 region 9314 to 9219). A plasmid containing the two-LTR junction was used as a standard.
RESULTS
HIV-2 Vpx interacts with Hsp40/DnaJB6.
In order to understand the underlying mechanism of Vpx transport into the nucleus, we undertook a genomic screening for interacting proteins with C-terminal Vpx (amino acids 61 to 112) as bait in the yeast two-hybrid system. An initial library screening with full-length Vpx detected only cDNAs encoding invariant chain (not shown) (28). Therefore, we utilized this truncated form of Vpx, which still contains the NLS, to increase the chance of obtaining cDNAs relevant to Vpx nuclear localization (3). Approximately 5 × 106 double-transformant colonies were screened, resulting in the identification of cDNAs encoding Hsp40/DnaJB6. Incomplete Hsp40/DnaJB6 cDNAs of six different lengths were identified 42 times, and all positive clones did not interact with the nonspecific bait, human lamin, suggesting that the interaction was specific (not shown). Hsp40/DnaJB1 and B9 were also identified 20 times and 1 time, respectively, but interaction with Vpx in mammalian cells was not confirmed.
Hsp40/DnaJB6 is a member of the type II group of Hsp40/DnaJ cochaperone proteins. It has two distinct domains, (i) a highly conserved J domain at the amino terminus, which is known to mediate interaction with Hsp70 and regulate its ATPase activity, and (ii) a G/F-rich region which usually follows the J domain and may act as a flexible linker between the J domain and the C-terminal region (25). Two homologs, full-length Hsp40/DnaJB6 and Hsp40/Mrj (a shorter version), have been found in humans as a result of alternative splicing. Both versions were identified in our screenings as Vpx-interactive proteins. Hsp40/Mrj and full-length Hsp40/DnaJB6 are identical in their N-terminal 231 amino acids but differ by 10 and 105 amino acids, respectively, in their C termini.
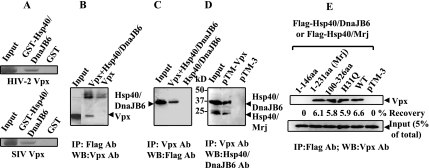
To confirm the interaction between HIV-2 Vpx and Hsp40/DnaJB6, in vitro GST pull-down assays were performed. GST-Hsp40/DnaJB6, but not GST alone, captured radioactively labeled Vpx (Fig. 1A). The rate of HIV-2 Vpx recovery by GST-Hsp40/DnaJB6 was 8%, compared to <0.5% for GST, as measured by densitometry analysis. Since SIV Vpx is highly homologous to HIV-2 Vpx, we also tested Vpx from SIV PBJ for interaction with Hsp40/DnaJB6 by in vitro GST pull-down assay. As expected, SIV Vpx also bound Hsp40/DnaJB6, with a similar capture rate of 8% (Fig. 1A).
FIG. 1.
Vpx binds Hsp40/DnaJB6. (A) GST-DnaJB6, but not GST, binds 35S-labeled HIV-2 and SIV PBJ Vpx synthesized in reticulocyte lysates. Bound complexes were immobilized on glutathione-agarose and resolved by SDS-polyacrylamide gel electrophoresis and autoradiography. The starting amounts of in vitro-translated Vpx for each GST pull-down assay are fivefold greater than that in the input lane, and half of the bound Vpx was loaded on the gel. All samples were run on the same gel and exposed to the same film. (B, C, and D) BSC40 cells (B and C) or 293T cells (D) were transfected with pTM-Vpx, pTM-Hsp40/DnaJB6, and/or pTM-3 and infected with vTF7-3. Cell lysates were immunoprecipitated (IP) with anti-Flag or anti-Vpx antibodies (Ab), and immunoblot assays were performed with anti-Flag, anti-Vpx, or anti-Hsp40/DnaJB6 antibodies. The input lanes represent the Vpx and Hsp40/DnaJB6 proteins in 10% of the lysate, and half of the immunoprecipitation complexes were loaded onto the gel. Proteins in input and immunoprecipitation lanes were detected by the same concentration of antibody, and the blot was exposed for the same amount of time. (E) Vpx was expressed in BSC40 cells with Hsp40/Mrj, various fragments of Hsp40/DnaJB6, or a control plasmid, as indicated. Cell lysates were immunoprecipitated with anti-Flag antibody, and Vpx was detected by anti-Vpx antibody. WB, Western blotting; aa, amino acids.
Coimmunoprecipitation assays were also performed to determine if the interaction of Vpx with Hsp40/DnaJB6 occurs in mammalian cells. Both Vpx and Flag-tagged Hsp40/DnaJB6 were expressed in BSC40 or 293T cells, and then proteins were immunoprecipitated and Western blotted with anti-Flag and anti-Vpx antibodies (Fig. 1B and C). Vpx was detected after immunoprecipitation by the anti-Flag antibody (Fig. 1B). Immunoprecipitation with anti-Vpx was able to recover both overexpressed (Fig. 1C) and endogenous Hsp40/DnaJB6 (Fig. 1D). No cross-reactive bands were detected when expressing either Vpx or Hsp40/DnaJB6 alone, demonstrating a specific interaction. Densitometry analysis of the blots indicated that only a small percentage of coimmunoprecipitated proteins was recovered compared to the input lanes (4% of Vpx, 5% of overexpressed, and 3% of endogenous Hsp40/DnaJB6), possibly due to less-than-saturating concentrations of antibodies and protein A beads and the nonlinear signal amplification of our Western blotting protocol. Nevertheless, the interaction of Vpx and Hsp40/DnaJB6 is specific.
In order to further characterize whether the J domain of Hsp40/DnaJB6 is involved in the binding of Vpx, amino acids 1 to 146 and 100 to 326 of Hsp40/DnaJB6 were expressed and tested for interaction with Vpx by coimmunoprecipitation assay (Fig. 1E). Residues 100 to 326 bound Vpx, while amino acids 1 to 146 did not, indicating that the J domain does not participate in the interaction with Vpx. These results are consistent with the identification of incomplete cDNA clones of Hsp40/DnaJB6 by yeast two-hybrid analysis that lack the J domain (not shown). The interaction region of Hsp40/DnaJB6 (amino acids 146 to 231) falls into the G/F-rich domain which has been reported to facilitate the binding of Hsp40 to Hsp70, but no other structure or function has been identified for this domain (31). Together, these results demonstrate a specific interaction of Hsp40/DnaJB6 with HIV-2 Vpx in a region C terminal of the J domain.
Hsp40/DnaJB6 enhances the nuclear localization of Vpx.
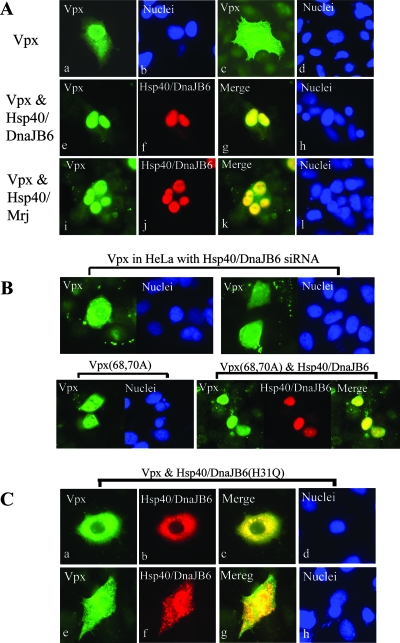
Similar to Vpx, Hsp40/DnaJB6 is a nuclear protein (25). Therefore, we hypothesized that Hsp40/DnaJB6 promoted Vpx nuclear localization. To address this question, we sought to determine whether overexpression of Hsp40/DnaJB6 in HeLa cells would alter Vpx cellular localization. HeLa cells were transfected for 16 h with a Vpx expression plasmid and fixed, and the localization of Vpx was determined by indirect immunofluorescence microscopy. Representative results of the localization of Vpx and Hsp40/DnaJB6 are shown in Fig. 2A. Vpx was found to be primarily localized in the nuclei of 73% (202/277) of the transfected cells when expressed alone (parts a and b). The remaining 27% of the transfected cells showed Vpx in both the cytoplasm and nucleus (part c and d). In contrast, Hsp40/DnaJB6 was exclusively localized in the nuclei of all transfected cells (part f). Interestingly, cotransfection of Vpx and Hsp40/DnaJB6 increased the nuclear localization of Vpx to 92% (314/341) of the cells (Fig. 2A, parts e to h). The difference was statistically significant (P < 0.005) by two-tailed t test. Similarly, coexpression of the Hsp40/Mrj splice variant also increased the nuclear localization of Vpx to 80% (224/280) of the transfected cells (Fig. 2A, parts i to l), which was also a statistically significant difference (P < 0.005), but this enhancement of nuclear localization was not as dramatic as the coexpression of Hsp40/DnaJB6, suggesting that Hsp40/DnaJB6 is much more efficient than Hsp40/Mrj at promoting the nuclear localization of Vpx.
FIG. 2.
Hsp40/DnaJB6 promotes Vpx nuclear import. (A) Confocal microscopy was performed on HeLa cells expressing Vpx (a to d), Vpx and Flag-Hsp40/DnaJB6 (e to h), or Vpx and Flag-Hsp40/Mrj (i to l). Cells were stained with anti-Vpx (a, c, e, and i) or anti-Flag (f and j) antibody, followed by Alexa Fluor 488- or 954-conjugated goat anti-rabbit IgG, respectively. Nuclei were stained with Hoechst 33258 pentahydrate. (B) Confocal microscopic localization of Vpx in HeLa cells expressing Hsp40/DnaJB6 siRNA (top parts) or Vpx mutant K68A/R70A with (bottom right) or without (bottom left) coexpression of Flag-Hsp40/DnaJB6. (C) Confocal micrographs of HeLa cells expressing Vpx and Flag-Hsp40/DnaJB6(H31Q) detected with antibodies to Vpx (a and e) or Flag (b and f) or with Hoechst stain (d and h).
To further assess the role of Hsp40/DnaJB6 in Vpx nuclear localization, we examined the effect of the down-regulation of Hsp40/DnaJB6 on the cellular localization of Vpx. To do this, we constructed a stable HeLa cell line expressing an Hsp40/DnaJB6 siRNA. Characterization of the effects of the expression of the siRNA on endogenous Hsp40/DnaJB6 levels is addressed below (see also Fig. 4A). The localization of Vpx changed dramatically in the Hsp40/DnaJB6 siRNA-expressing cells (Fig. 2B, top parts). Only 10% (27/282) of the transfected cells showed Vpx primarily localized in their nuclei (P < 0.005 compared to cells not expressing siRNA). As a negative control, we assessed Vpx localization in HeLa cells stably expressing an siRNA to GFP. No effect on Vpx subcellular localization was observed (not shown).
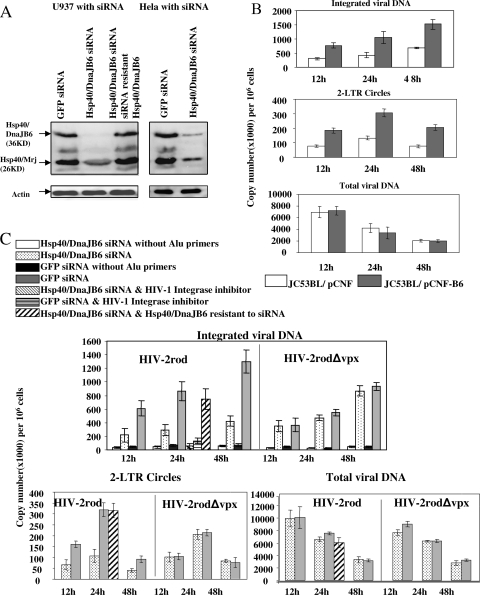
FIG. 4.
Hsp40/DnaJB6 promotes PIC import into the nucleus. (A) Western blot analysis of siRNA knockdown and recovery of Hsp40/DnaJB6 in U937 and HeLa cells. Cells that stably express siRNA to DnaJB6 or GFP and U937 cells with siRNA-resistant Hsp40/DnaJB6 were established as described in Materials and Methods. An anti-actin antibody was used to show that there was similar loading of each sample (bottom part). (B) Analysis of the effect of Hsp40/DnaJB6 overexpression on the infection of JC53BL (HeLa-CD4/CCR5) cells by detection of integration and two-LTR circle accumulation. After transfection with an Hsp40/DnaJB6 expression plasmid (pCNF-B6) or a vector plasmid (pCNF) with Nucleofector (Amaxa Biosystems), JC53BL cells were serum starved for 24 h and then released into complete medium containing 400 μM mimosine. Transfected cells (106) were infected with 100 ng of HIV-2rod for 12, 24, or 48 h, and levels of integrated viral DNA, total viral DNA, and two-LTR circles were measured by real-time PCR analysis. (C) Analysis of the effect of Hsp40/DnaJB6 knockdown on the infection of U937 cells by detection of integration and two-LTR circle accumulation. U937 cells (3 × 106) were infected with 300 ng of HIV-2rod or HIV-2rodΔvpx for 12, 24, or 48 h, and levels of integrated viral DNA, total viral DNA, and two-LTR circles were determined by real-time PCR analysis. Background levels for integration assays were measured by PCR without Alu primers for the first-round reaction. To determine the specificity of the assay, the cells were incubated with an HIV-1 integrase inhibitor (L-870810; Merck) at a concentration of 10 μM and infected for 24 h. For restoration of viral infection levels in Hsp40/DnaJB6 knockdown cells, U937 cells expressing siRNA and Hsp40/DnaJB6 resistant to siRNA were infected for 24 h and levels of integrated viral DNA, two-LTR circles, and total viral DNA were determined as described above. All experiments were repeated three times, and one representative result of each is shown.
Mutation of the NLS of Vpx (K68A, R70A) was reported to inhibit the nuclear translocation of GFP-Vpx (2). Analysis of the localization of this mutant by indirect immunofluorescence showed a diffuse nuclear and cytoplasmic localization (Fig. 2B, lower left part). Surprisingly, overexpression of Hsp40/DnaJB6 facilitated the nuclear localization of the mutant Vpx (Fig. 2B, lower right parts), suggesting that Hsp40/DnaJB6 trans dominantly regulates Vpx nuclear localization.
To further test this hypothesis, we assessed the effect on Vpx cellular localization of overexpression of an Hsp40/DnaJB6 nuclear localization mutant. Mutation of histidine residue 31 to glutamine (H31Q) was previously shown to dramatically reduce Hsp40/DnaJB6 nuclear import (Fig. 2C, parts b and f) (9) but did not disrupt the interaction with Vpx (Fig. 1E). As expected, when Vpx was coexpressed in HeLa cells with Hsp40/DnaJB6 (H31Q), its nuclear localization was significantly reduced (Fig. 2C, parts a and e). In these cells, Vpx was either localized exclusively in the cytoplasm (part a, 70% cells) or evenly distributed in both the nucleus and cytoplasm (part e, 30% cells). This provides further evidence that Hsp40/DnaJB6 trans dominantly regulates Vpx localization.
Since SIV PBJ Vpx also interacts with Hsp40/DnaJB6, we assumed that overexpression of Hsp40/DnaJB6 would similarly enhance the nuclear translocation of SIV Vpx. As expected, the accumulation of SIV Vpx in the nucleus was increased by 20% when it was coexpressed with Hsp40/DnaJB6 plasmid DNA than in its absence (data not shown).
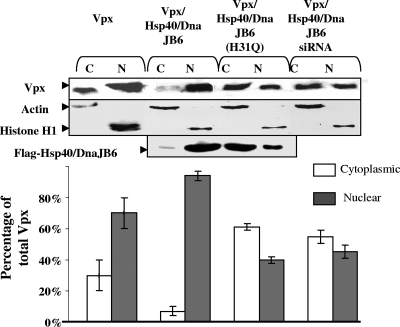
To confirm the effects of Hsp40/DnaJB6 expression on the localization of Vpx, we determined the distribution of Vpx and Hsp40/DnaJB6 by cell fractionation and Western blot analysis (Fig. 3). For this purpose, HeLa cells expressing Vpx alone, Vpx and Hsp40/DnaJB6, Vpx and Hsp40/DnaJB6 (H31Q), or Vpx and Hsp40/DnaJB6 siRNA were subjected to differential centrifugation into nuclear and cytoplasmic fractions. The purity of the fractions was confirmed by immunoblotting for histone H1 (nuclear marker) and actin (cytoplasmic marker; Fig. 3, middle panel). When expressed alone, 70% of the Vpx was found in the nuclear fraction, as determined by densitometry. However, overexpression of Hsp40/DnaJB6 resulted in an increase (to 90%) in Vpx in the nuclear fractions. This confirms our results from the immunofluorescence experiments. In contrast, when the Hsp40/DnaJB6(H31Q) nuclear localization mutant was overexpressed, only 40% of the Vpx was found in the nuclear fraction. This again suggests that Hsp40/DnaJB6 is a _trans_-dominant regulator of Vpx localization. Down-regulation of Hsp40/DnaJB6 by siRNA lowered the amount of Vpx in the nuclear fractions of cells to 45%. Along with the indirect immunofluorescence data, this suggests that endogenous Hsp40/DnaJB6 plays a key role in the nuclear localization of Vpx. In summary, the immunofluorescence and cellular fractionation experiments show that (i) Vpx and Hsp40/DnaJB6 colocalize, (ii) Hsp40/DnaJB6 overexpression increases Vpx nuclear localization and/or retention, and (iii) decreased Hsp40/DnaJB6 expression results in reduced Vpx nuclear localization. Collectively, these data suggest that Hsp40/DnaJB6 is a _trans_-dominant regulator of Vpx nuclear localization.
FIG. 3.
Effect of Hsp40/DnaJB6 on the distribution of Vpx in nuclear and cytoplasmic cellular fractions. HeLa cells were separated into nuclear (N) and cytoplasmic (C) fractions after the expression of Vpx or of Vpx with Hsp40/DnaJB6, DnaJB6(H31Q), or Hsp40/DnaJB6 siRNA and equivalent amounts of each fraction, as determined by Lowry assay, analyzed by immunoblotting with anti-Vpx or anti-FLAG antibody. The purity of the fractions was demonstrated by immunoblotting with antibodies to histone H1 or actin. The amount of Vpx in each band was measured with Alpha Imager software. Only one representative immunoblotting result is presented. Standard deviations are derived from an experiment performed in triplicate.
Hsp40/DnaJB6 promotes nuclear import of the PIC.
It has been shown that HIV-2 lacking Vpx expression fails to infect macrophages and other nondividing cells, suggesting that Vpx plays a critical role in the nuclear translocation of the viral PIC (26). Because Hsp40/DnaJB6 enhances Vpx nuclear localization, we hypothesized that Hsp40/DnaJB6 could modulate the nuclear translocation of HIV-2 PICs. To test this possibility, we established a U937 cell line stably expressing an Hsp40/DnaJB6 siRNA and a GFP siRNA as a negative control. The immunoblot assay shows a specific reduction in the level of Hsp40/DnaJB6 by 94% and that of Hsp40/Mrj by 50%, in comparison to cells expressing the GFP siRNA (Fig. 4A). Stable HeLa cell lines expressing the Hsp40/DnaJB6 siRNA showed a reduction of Hsp40/DnaJB6 expression by 80% and of Hsp40/Mrj expression by 60% compared to cells expressing the GFP siRNA (Fig. 4A, right part).
To assay the effect of Hsp40/DnaJB6 knockdown or overexpression on the nuclear import of the HIV-2 PICs, we examined by real-time PCR the levels of integrated viral genomic DNA and two-LTR circles in HIV-2-infected U937 cells expressing each siRNA (Fig. 4B and C). Omission of Alu primers from the first-round PCR reduced levels of integrated viral DNA by 85 to 97%, whereas incubation of the cells with a HIV-1 integrase inhibitor reduced the level by 81 to 85%, suggesting that the Alu integration assay specifically detects integrated viral genomic DNA (Fig. 4C, black bars). However, we still observed some background even without Alu primers. This is due to fact that the L-Rod oligonucleotide can prime the formation of any single-stranded, LTR-containing viral DNA, leading to inefficient linear amplification. However, since integrated viral DNA increases at later time points, the ratio of the background to the values with Alu primers decreases dramatically. The integrase inhibitor reduced the levels close to the background but did not completely block integration, possibly because the concentration was not optimal or this inhibitor is not as effective against HIV-2 as HIV-1.
As demonstrated by the reduction in the number of integrated copies, and two-LTR circle accumulation, the nuclear import of the viral PIC was impaired over the 48-h time course in U937 cells expressing the Hsp40/DnaJB6 siRNA compared to the negative control cells expressing a GFP siRNA. Figure 4C shows the results at 12, 24, and 48 h after infection (dotted versus shaded bars). A similar depression of integrated viral genomic DNA and two-LTR circle DNA was also seen at 2, 4, and 6 h after infection (not shown). Moreover, the reduction of the nuclear import of the viral PIC was observed regardless of the concentration of input virus (not shown). In contrast, no significant defect was observed in the nuclear transport for Vpx negative virus (Fig. 4C), demonstrating that the reduction of PIC nuclear import was dependent on the presence of Vpx. To verify that the reduction of nuclear import of the PIC is not due to a defect in viral entry or reverse transcription, we quantified the total viral DNA in infected cells. Similar amounts of viral DNA were observed over 48 h postinfection (Fig. 4C), indicating there was no defect in virus entry between the Hsp40/DnaJB6 siRNA-expressing cells and the control GFP siRNA cells.
In order to ensure that the reduced nuclear import of the PIC is caused by loss of Hsp40/DnaJB6, rather than a nonspecific off-target effect of the siRNA, we constructed an expression vector producing Hsp40/DnaJB6 that is resistant to the siRNA and created a stable U937 cell line expressing both Hsp40/DnaJB6 siRNA and the mutant Hsp40/DnaJB6. Immunoblot analysis showed that introduction of the expression vector restored the Hsp40/DnaJB6 protein to a level similar to that of the control cells (Fig. 4A, left part). Infection of these cells was then assayed to determine if the inhibitory effect of the Hsp40/DnaJB6 siRNA could be rescued by expression of the Hsp40/DnaJB6 escape vector. As shown in Fig. 4C, the levels of integrated viral DNA and two-LTR circles in the Hsp40/DnaJB6 rescued cells are similar to those of control cells (cross-hatched versus shaded bars at the 24-h time point). This demonstrates that the reduction in Hsp40/DnaJB6 expression specifically decreased HIV-2 PIC nuclear import.
In order to further examine the effect of Hsp40/DnaJB6 on PIC nuclear import, we examined the infection of arrested JC53BL (HeLa-CD4/CCR5) cells transfected with the Hsp40/DnaJB6 or vector plasmid. JC53BL cells were arrested for 48 h by serum starvation and mimosine treatment and then infected with HIV-2. The nuclear entry of the PIC was measured as described above. The result of this measurement showed that the level of viral DNA integrated into the host genome was increased when Hsp40/DnaJB6 was overexpressed from a plasmid (Fig. 4B).
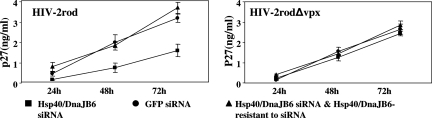
Since inhibition of Hsp40/DnaJB6 caused a reduction of the nuclear import of the PIC, we expected that virus replication would similarly be affected. To verify this possibility, we monitored virus replication in the U937 cell lines expressing Hsp40/DnaJB6 or GFP siRNA, with or without Hsp40/DnaJB6 resistant to siRNA, by measuring the level of capsid protein p27 for a single round of infection (Fig. 5) or reverse transcriptase activities for multiple-round infections (not shown). As expected, HIV-2 replication was delayed in the Hsp40/DnaJB6 siRNA cells compared to the GFP siRNA cells in both cases whereas the introduction of siRNA-resistant Hsp40/DnaJB6 restored the viral replication. In contrast, the virus with vpx deleted replicated at comparable levels in both the Hsp40/DnaJB6- and GFP siRNA-expressing cells. Together, these observations indicate that down-regulation of Hsp40/DnaJB6 expression in cells results in a reduction of PIC nuclear import and virus replication.
FIG. 5.
Hsp40/DnaJB6 is required for efficient HIV-2 replication in U937 cells. U937 cells (106) stably expressing GFP siRNA, Hsp40/DnaJB6 siRNA, and/or Hsp40/DnaJB6 resistant to the siRNA were infected with 2 × 106 50% tissue culture infective doses of HIV-2rod or HIV-2rodΔvpx. Two hours later, the cells were washed with PBS three times and a viral entry inhibitor (T-20, 2.5 μg/ml) was added every 12 h to prevent reinfection by newly synthesized virus. Virus production was examined at the indicated time points by measuring levels of capsid protein (p27) by enzyme-linked immunosorbent assay.
DISCUSSION
Hsps have been shown to participate in the nuclear transport of viral genomes and proteins. For instance, Hsp70 stimulates the nuclear import and replication of Vpr-deficient HIV-1 in macrophages (1) and Hsp90 inhibitors block the nuclear localization of HSV-1 DNA polymerase (7). In this study, we present evidence that Hsp40/DnaJB6 plays a critical role in the nuclear import of the HIV-2 Vpx protein and the viral PIC. Hsp40/DnaJB6 belongs to a family of at least 41 human Hsp40/DnaJ proteins (31), which are found in many cellular compartments and perform diverse functions. Hsp40/DnaJ proteins bind Hsp70 as a cochaperone and facilitate its ATPase activity (38). The interaction of Hsp40 and Hsp70 regulates the activity of Hsp90 on more than 100 client proteins involved in signaling (15). The macromolecular complex, containing Hsp40, -70, and -90 and immunophilin, may then bind to microtubules and/or undergo nuclear import.
Vpx interacts with Hsp40/DnaJB6 and is incorporated into the virus particle through an interaction with the Gag precursor protein (27). To determine if overexpression of Hsp40/DnaJB6 affects the packaging of Vpx into virus-like particles (VLPs) by immunoblot assays, we used BSC40 cells transiently expressing Hsp40/DnaJB6 and Gag-Pol (data not shown). Our results indicated that Vpx incorporation into VLPs was reduced with increasing levels of Hsp40/DnaJB6 expression. No change was observed in the amount of capsid p27 protein in VLPs and the levels of Vpx and Pr55Gag expression in cell lysates with the overexpression of Hsp40/DnaJB6, suggesting that reduced Vpx packaging in VLPs was not due to a reduction of VLP production or Vpx expression. Furthermore, we performed in vitro GST pull-down competition assays (data not shown). In these studies, GST-Gag was used to bind 35S-labeled Vpx in the presence of increasing amounts of His-tagged Hsp40/DnaJB6. The result demonstrated that a reduced amount of Vpx was captured by GST-Gag when His-tagged Hsp40/DnaJB6 was preincubated with Vpx, suggesting that Hsp40/DnaJB6 competes with Gag for binding to Vpx.
Vpx colocalizes with Hsp40/DnaJB6 by indirect immunofluorescence. Interestingly, the nuclear localization of Vpx was modulated by Hsp40/DnaJB6. Overexpression of Hsp40/DnaJB6 increased Vpx nuclear accumulation, whereas the down-regulation of Hsp40/DnaJB6 by siRNA or the expression of a nuclear localization mutant of Hsp40/DnaJB6 reduced the nuclear targeting of Vpx. Since both forms of Hsp40/DnaJB6 bind Vpx, we conclude that the subcellular localization of Vpx is dependent on the localization of Hsp40/DnaJB6. This finding suggests that Vpx may utilize the same pathway as Hsp40/DnaJB6 for its movement to the nucleus. Thus, Vpx may recruit the Hsp90/Hsp70-based chaperone machinery for trafficking through the cytoplasm to the nuclear pore. Because neither Hsp70, Hsp90, nor immunophilins have an NLS, it seems likely that they are carried into the nucleus by multiple NLS-containing client proteins (29, 30). It remains to be determined if Hsp40 is also carried into the nucleus the same way as Hsp90 or if it uses its own NLS to bind importins. Since Vpx has its own NLS and has been shown to interact with importin α/β via the NLS, it is reasonable to assume that Vpx may use importin α/β for its translocation across the nuclear envelope (3, 35). Based on this assumption, enhanced nuclear import of Vpx by Hsp40/DnaJB6 could be a result of an increased local concentration of Vpx at the nuclear pore. It is also possible that Vpx may be carried into the nucleus by clients of Hsp90/Hsp70 complexes through a piggyback mechanism. Alternatively, Hsp70/Hsp40 may travel into the nucleus by an Hsp90-independent mechanism. In agreement with this idea, Izawa and colleagues showed that Hsp40/Mrj interacts with cytokeratins 8 and 18 and regulates their organization into microfilaments (18). Hsp40 was also demonstrated to colocalize with cytoplasmic vimentin filaments (32). These authors hypothesized that there may exist specific Hsp40/DnaJ family proteins that link actin, tubulin, or various intermediate filament proteins to Hsp70.
There is a growing body of evidence indicating that Hsp70 plays a role in the regulation of microtubule formation. Consistent with these observations, Izawa et al. showed that expression of the first 146 amino acids of Hsp40/Mrj disrupted the cytoskeleton network, including actin filaments, microtubules, and vimentin filaments, suggesting that the function of Hsp70 is inhibited by this dominant negative protein (18). Thus, the mislocalization of Vpx caused by overexpression of Hsp40/DnaJB6 (H31Q) might be the consequence of the altered assembly-disassembly dynamics of the cytoskeleton network as a result of Hsp70 inhibition. However, this does not appear to be the cause of reduced nuclear import of PICs in cells expressing siRNA to Hsp40/DnaJB6, because we did not see a significant effect on the nuclear import of PICs from HIV-2Δvpx. It is also noted that the siRNA was designed to target only Hsp40/DnaJB6 and not other Hsp40 homologs; thus, the defect of nuclear transport is not likely due to the inhibition of other Hsp40s. We also observed that Hsp40/Mrj is more abundant than Hsp40/DnaJB6, especially in U937 cells (Fig. 4A, left part), which may explain why these two isoforms were knocked down to such different levels by the same siRNA. In addition, differences in the stability of these two proteins and their mRNAs may also contribute to the discrepancy.
In this study, Hsp40/Mrj was shown to enhance the nuclear localization of Vpx by 7%, which is not as significant as that of Hsp40/DnaJB6 (19%). This difference is possibly caused by the difference in their cellular distribution. Unlike Hsp40/DnaJB6, Hsp40/Mrj is exclusively localized in the nuclei of only 50% of transfected cells, whereas in the remaining cells it is found in both the nucleus and the cytoplasm (data not shown). Therefore, Hsp40/DnaJB6 appears to play a dominant role in enhancing the nuclear localization of Vpx, which may explain the finding that the nuclear import of the PIC was decreased by two- to threefold (Fig. 4C) whereas that of Hsp40/Mrj was reduced only twofold (Fig. 4A, left part).
Vpx is present in viral PICs and regulates their nuclear entry. However, several fundamental questions still remain to be answered. (i) Which pathway does Vpx use for its nuclear import? (ii) Does Vpx have to be a part of the PIC in order to direct the nuclear entry of the PIC? (iii) How does Vpx target the PIC to the nucleus if Vpx works in trans? Since Vpx is a component of the PIC, it may direct the PIC to the nucleus simply by its association with cellular transport machinery. For HIV-1, movement of the viral core initiates within the peripheral regions of the cell cytoplasm by association with the actin cytoskeleton after entry (23). The PIC subsequently translocates toward the nucleus along the microtubule network. The structural basis for interaction between the PIC and microtubules remains to be determined. Although the PIC likely engages a cellular dynein-dependent motor complex, it is unclear what connects the PIC to dynein (5, 23). Interestingly, Mueller et al. demonstrated that Vpx interacts with α-actinin 1, a structural protein that anchors actin filaments to the cell membrane (24). This observation suggests that α-actinin 1 may be an early binding partner of the PIC following uncoating of the virus. Our present findings lead us to speculate that Vpx acts as a specific adaptor protein to link the PIC to microtubules through Hsp90/Hsp70-based chaperone machinery.
Although direct evidence of the interaction of HIV-2 PIC components with actin and microtubules has not been revealed, it is tempting to speculate that HIV-1 may employ a pathway similar to that of HIV-2 for nuclear trafficking. Hsp70 was shown to stimulate HIV-1 ΔVpr nuclear import by enhancing karyophilic properties of weak NLSs present in the matrix protein, since Hsp70 facilitates the interaction between the basic-type NLS and importin α (34). In addition, Vpr does not have to be a part of the PIC to exert its enhancing effect. Surprisingly, Hsp70 and Vpr work in a similar fashion and Hsp70 can even replace Vpr during nuclear import of PIC (1). Because no interaction was detected either between Hsp70 and Vpx or between Hsp40/DnaJB6 and HIV-1 Vpr (data not shown), it would not be surprising if Vpr and Vpx use different mechanisms for enhancing the nuclear import of the PIC. However, it would be interesting to know whether Hsp40/DnaJB6 facilitates the interaction between Vpx and importin α/β.
In conclusion, we present evidence that Hsp40/DnaJB6 interacts with HIV-2 Vpx and promotes the nuclear translocation of both HIV-2 Vpx and the PIC, suggesting an important role for Hsp40/DnaJB6 in the nuclear trafficking of viral proteins. We hypothesize that association of Hsp40/DnaJB6 with Vpx acts as a bridge to connect the viral PIC to cellular transport machinery. These observations will pave the way toward our understanding the diverse function of Hsp40/DnaJ proteins, as well as novel therapeutic approaches.
Acknowledgments
We thank S. A. Stewart and L. Bernal-Mizrachi for assistance with siRNA expression systems and D. Rauch for critical review of the manuscript.
This work was supported by Public Health Service grant AI24745.
Footnotes
▿
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Agostini, I., S. Popov, J. Li, L. Dubrovsky, T. Hao, and M. Bukrinsky. 2000. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp. Cell Res. 259398-403. [DOI] [PubMed] [Google Scholar]
- 2.Belshan, M., L. A. Mahnke, and L. Ratner. 2006. Conserved amino acids of the human immunodeficiency virus type 2 Vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology 346118-126. [DOI] [PubMed] [Google Scholar]
- 3.Belshan, M., and L. Ratner. 2003. Identification of the nuclear localization signal of human immunodeficiency virus type 2 Vpx. Virology 3117-15. [DOI] [PubMed] [Google Scholar]
- 4.Brussel, A., and P. Sonigo. 2003. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J. Virol. 7710119-10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 1882113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., S. Haggerty, M. P. Demsey, N. Sharova, A. G. Adzhubei, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of nondividing cells. Nature 365666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch, A. D., and S. K. Weller. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone Hsp90 for proper localization to the nucleus. J. Virol. 7910740-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coligan, J. E. 2007. Current protocols in immunology. Green Publishing Associates and Wiley-Interscience, New York, NY.
- 9.Dai, Y. S., J. Xu, and J. D. Molkentin. 2005. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol. Cell. Biol. 259936-9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 803712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher, T. M., B. Brichacek, N. Sharova, M. A. Newman, G. Stivathis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1996. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIVsm. EMBO J. 156155-6165. [PMC free article] [PubMed] [Google Scholar]
- 12.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 949825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 764666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurer, C., A. Höglund, S. Höglund, and J. Luban. 2005. ATPγS disrupts human immunodeficiency virus type 1 virion core integrity. J. Virol. 795557-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381571-580. [DOI] [PubMed] [Google Scholar]
- 16.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M.-A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 917311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, W., N. V. Heyden, and L. Ratner. 1989. Analysis of the function of viral protein X (VPX) of HIV-2. Virology 173624-630. [DOI] [PubMed] [Google Scholar]
- 18.Izawa, I., M. Nishizawa, K. Ohtakara, K. Ohtsuka, H. Inada, and M. Inagaki. 2000. Identification of Mrj, a DnaJ/Hsp40 family protein, as a keratin 8/18 filament regulatory protein. J. Biol. Chem. 27534521-34527. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, M., and D. Mitra. 2005. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 28040041-40050. [DOI] [PubMed] [Google Scholar]
- 20.Mahalingam, S., B. VanTine, M. L. Santiago, F. Gao, G. M. Shaw, and B. H. Hahn. 2001. Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J. Virol. 75362-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahnke, L. A., M. Belshan, and L. Ratner. 2006. Analysis of HIV-2 Vpx by modeling and insertional mutagenesis. Virology 348165-174. [DOI] [PubMed] [Google Scholar]
- 22.Matouschek, A., N. Pfanner, and W. Voos. 2000. Protein unfolding by mitochondria. EMBO Rep. 1404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller, S. M., R. Jung, S. Weiler, and S. M. Lang. 2004. Vpx proteins of SIVmac239 and HIV-2ROD interact with the cytoskeletal protein α-actinin 1. J. Gen. Virol. 853291-3303. [DOI] [PubMed] [Google Scholar]
- 25.Ohtsuka, K., and M. Hata. 2000. Mammalian Hsp40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 598-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pancio, H., N. V. Heyden, and L. Ratner. 2000. The C-terminal proline-rich tail of HIV-2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J. Virol. 746162-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancio, H., and L. Ratner. 1998. Human immunodeficiency virus 2 Vpx-Gag interaction. J. Virol. 725271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pancio, H., N. van der Heyden, K. Kosuri, P. Cresswell, and L. Ratner. 2000. Interaction of HIV-2 Vpx with invariant chain. J. Virol. 746168-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratt, W. B., M. D. Galigniana, J. M. Harrell, and D. B. DeFranco. 2004. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell Signal. 16857-872. [DOI] [PubMed] [Google Scholar]
- 30.Pratt, W. B., A. M. Silverstein, and M. D. Galigniana. 1999. A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell Signal. 11839-851. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, X. B., Y. M. Shao, S. Miao, and L. Wang. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 632560-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schietke, R., D. Brohl, T. Wedig, N. Mucke, H. Herrmann, and T. M. Magin. 2006. Mutations in vimentin disrupt the cytoskeleton in fibroblasts and delay execution of apoptosis. Eur. J. Cell Biol. 851-10. [DOI] [PubMed] [Google Scholar]
- 33.Sharkey, M. E., I. Teo, T. Greenough, N. Sarova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Vereyard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 676-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulga, N., P. Roberts, Z. Gu, L. Spitz, M. M. Tabb, M. Nomura, and D. S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singhal, P. K., P. R. Kumar, M. R. K. S. Rao, M. Kyasani, and S. Mahalingam. 2006. Simian immunodeficiency virus Vpx is imported into the nucleus via importin alpha-dependent and -independent pathways. J. Virol. 80526-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohn, S.-Y., J.-H. Kim, K.-W. Baek, W.-S. Ryu, and B.-Y. Ahn. 2006. Turnover of hepatitis B virus X protein is facilitated by Hdj1, a human Hsp40/DnaJ protein. Biochem. Biophys. Res. Commun. 347764-768. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, S. A., D. M. Dykxhoorn, D. Palliser, H. Mizuno, E. Y. Yu, D. S. An, D. M. Sabatini, I. S. Chen, W. C. Hahn, P. A. Sharp, R. A. Weinberg, and C. D. Novina. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takayama, S., Z. Xie, and J. C. Reed. 1999. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperon regulators. J. Biol. Chem. 274781-786. [DOI] [PubMed] [Google Scholar]
- 39.Wainberg, Z., M. Oliveira, S. Lerner, Y. Tao, and B. G. Brenner. 1997. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology 233364-373. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita, M., and M. Emerman. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 34488-93. [DOI] [PubMed] [Google Scholar]
- 41.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101173-185. [DOI] [PubMed] [Google Scholar]