Clinical and Molecular Phenotype of Aicardi-Goutières Syndrome (original) (raw)
Abstract
Aicardi-Goutières syndrome (AGS) is a genetic encephalopathy whose clinical features mimic those of acquired in utero viral infection. AGS exhibits locus heterogeneity, with mutations identified in genes encoding the 3′→5′ exonuclease TREX1 and the three subunits of the RNASEH2 endonuclease complex. To define the molecular spectrum of AGS, we performed mutation screening in patients, from 127 pedigrees, with a clinical diagnosis of the disease. Biallelic mutations in TREX1, RNASEH2A, RNASEH2B, and RNASEH2C were observed in 31, 3, 47, and 18 families, respectively. In five families, we identified an RNASEH2A or RNASEH2B mutation on one allele only. In one child, the disease occurred because of a de novo heterozygous TREX1 mutation. In 22 families, no mutations were found. Null mutations were common in TREX1, although a specific missense mutation was observed frequently in patients from northern Europe. Almost all mutations in RNASEH2A, RNASEH2B, and RNASEH2C were missense. We identified an RNASEH2C founder mutation in 13 Pakistani families. We also collected clinical data from 123 mutation-positive patients. Two clinical presentations could be delineated: an early-onset neonatal form, highly reminiscent of congenital infection seen particularly with TREX1 mutations, and a later-onset presentation, sometimes occurring after several months of normal development and occasionally associated with remarkably preserved neurological function, most frequently due to RNASEH2B mutations. Mortality was correlated with genotype; 34.3% of patients with TREX1, RNASEH2A, and RNASEH2C mutations versus 8.0% RNASEH2B mutation–positive patients were known to have died (_P_=.001). Our analysis defines the phenotypic spectrum of AGS and suggests a coherent mutation-screening strategy in this heterogeneous disorder. Additionally, our data indicate that at least one further AGS-causing gene remains to be identified.
In 1984, Jean Aicardi and Françoise Goutières, two eminent French pediatric neurologists, reported eight children from five families with an early-onset encephalopathy characterized by basal-ganglia calcification, white-matter abnormalities, and a chronic cerebrospinal fluid (CSF) lymphocytosis (MIM 225750).1 The presence of sibling recurrences, affected females, and parental consanguinity suggested that the condition was inherited as an autosomal recessive trait. However, the authors highlighted the risk of misdiagnosis of this disorder as the sequelae of congenital infection, an observation that subsequently led to the finding of raised levels of the antiviral cytokine interferon alpha (IFN-α) in the CSF of affected children.2
Phenotypic variability was recognized in the original description of AGS and has since been confirmed by others.3,4 Patients with apparently static or slowly progressive disease, sometimes presenting after several months of normal development, have been reported,1,5 whereas other children have been reported with a neonatal onset of features and early death.6 Notably, a number of early-presenting cases have exhibited hepatosplenomegaly, thrombocytopenia, and congenital microcephaly, further emphasizing the clinical overlap with acquired in utero infection.3
AGS is a genetically heterogeneous disorder. Elsewhere, we localized the position of a gene (AGS1) causing AGS on chromosome 3p217 and demonstrated that Cree encephalitis, a severe infantile neurodegenerative disorder found among the Canadian Cree Indian tribe of northern Quebec, is allelic to AGS.8 Subsequently, we defined, again by linkage analysis in consanguineous families, a second disease locus (AGS2) on chromosome 13q14-21.9 Recently, we described mutations in genes encoding the 3′→5′ exonuclease TREX1 (or, DNase III [MIM 606609; GenBank transcript AAK07616 and nucleotide sequence NM_033627], which causes AGS1)10 and the three nonallelic subunits of the RNASEH2 protein complex (RNASEH2A [MIM 606034; GenBank transcript AAH11748.1 and nucleotide sequence NM_006397.2], RNASEH2B [MIM 610326; GenBank transcript AAH36744.1 and nucleotide sequence NM_024570.1], and RNASEH2C [MIM 610330; GenBank accession number AAH23588.1 and nucleotide sequence NM_032193.3], which cause AGS4, AGS2, and AGS3, respectively)11 in patients with AGS. The genetic heterogeneity of the disease thus extends to include at least four loci, with strong evidence of a common underlying pathogenic mechanism.
Here, we present clinical and molecular data from a large cohort of AGS cases that define the phenotypic spectrum of AGS and suggest a coherent mutation-screening strategy for this heterogeneous disorder. Our data also indicate that at least one further AGS-causing gene, accounting for the disease in 17% of AGS-affected families in our study, remains to be identified.
Material and Methods
Subjects
Subjects with a clinical diagnosis of AGS from 127 independent pedigrees were recruited internationally through collaborating physicians and the International Aicardi-Goutières Syndrome Association. Twenty-five families were seen by one of us (Y.J.C.). For the remainder, information was provided by the clinician responsible for the care of the patient. Clinical, neuroimaging, and laboratory data were obtained from medical records, magnetic resonance imaging (MRI), and CT scans. Information about every clinical characteristic was not available for all patients. All mutation-negative families and those families in which only one mutation could be identified included at least one child fulfilling the following diagnostic criteria: neurological features of an encephalopathy, intracranial calcification, negative investigations for common prenatal infections, a CSF white-cell count (WCC) ⩾5 white cells/mm3, and/or raised levels of IFN-α (>2 IU/liter or >10 pg/ml) in the CSF. With consent, blood samples were obtained from affected children, their parents, and unaffected siblings. The study was approved by a U.K. Multicentre Research Ethics Committee (reference number 04:MRE00/19).
Mutation Analysis
Genomic DNA was extracted from peripheral-blood leukocytes by standard methods. PCR amplification of all coding exons and conserved splice sites of TREX1 (AGS1), RNASEH2A (AGS4), RNASEH2B (AGS2), and RNASEH2C (AGS3) was performed (primer sequences available on request). Purified PCR-amplification products were sequenced using dye-terminator chemistry and were electrophoresed on ABI 3700 (Applied Biosystems), ABI 3100, or MegaBace500 (Amersham Pharmacia) capillary sequencers. Additionally, high-resolution melting curve analysis (MCA) was employed to screen RNASEH2A and RNASEH2B, with use of a LightScanner (Idaho Technology) operating LightScanner v1.0.364 software. PCR incorporated the fluorescent dye LCGreen+, and, after completion, microtiter plates were transferred to the LightScanner, with fluorescence data collection over the temperature range 70°C–95°C, as samples were melted. Samples were run both “neat” and “spiked” with wild-type PCR product, to ensure detection of homozygous variants. Data were analyzed using the “manual scanning” setting. For DNA fragments with more than one melting domain, as well as examination of the entire melting curve, each separate melting domain was analyzed individually. Samples in which the melting curve deviated from the wild-type control were subjected to DNA sequencing. Mutations were classified as “null” if they were predicted to result in premature protein termination (including frameshifts, nonsense mutations, and mutations altering splice-donor and -acceptor sequences); missense mutations were defined as “those causing the substitution of one amino acid for another.” One hundred European control alleles were sequenced for all four genes, and, in the case of RNASEH2C, 100 South Asian alleles were also analyzed.
Statistical Analysis
χ2 analyses, Kruskal-Wallis tests, and Mann-Whitney U tests for nonparametric variables were performed using the Statistical Package for the Social Sciences for Windows, version 12.01 (SPSS).
Results
Genetic Findings
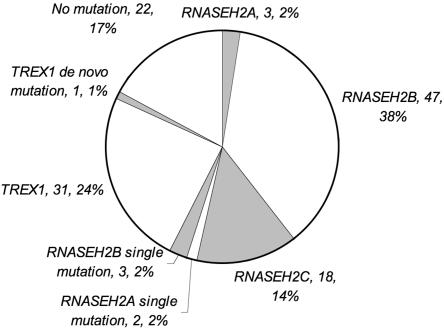
Biallelic mutations in TREX1, RNASEH2A, RNASEH2B, and RNASEH2C were observed in 31, 3, 47, and 18 families, respectively. In three families, we could identify only a single RNASEH2B mutation. In one family, we identified a single mutation in RNASEH2A, and, in one further patient, we saw two putative RNASEH2A mutations on the same allele. In one patient, a single de novo heterozygous TREX1 mutation was observed. Additionally, in 22 families, no mutations were found (fig. 1 and table 1). None of the identified mutations was annotated as a SNP in the available databases, and none was present in alleles from control subjects. Where tested, mutations segregated with the disease in all families, and, except in the case of the de novo heterozygous TREX1 mutation, all available parents were heterozygous for a single mutation.
Figure 1. .
Numbers and percentages of AGS-affected families with biallelic mutations in TREX1, RNASEH2A, RNASEH2B, and RNASEH2C; single RNASEH2A, RNASEH2B, and TREX1 mutations; and those with no identifiable mutation(s).
Table 1. .
Summary Data of Families with No Mutations in TREX1, RNASEH2A, RNASEH2B, or RNASEH2C
| Sample | n |
|---|---|
| Families | 22 |
| Patients | 29 |
| Consanguineous pedigrees | 7 |
| Patients with raised CSF WCC | 26 |
| Patients with raised CSF IFN-α | 17 |
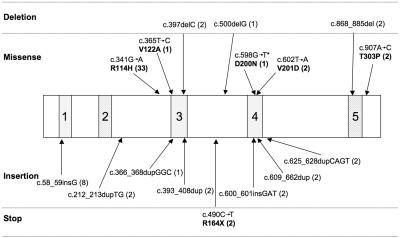
TREX1 is a single-exon gene encoding a 314-aa protein. Mutations were seen throughout the gene (fig. 2). Eleven mutations were null alleles. Eighteen families, 14 of which were of northern European origin, were homozygous (15) or compound heterozygous (3) for a c.341G→A transition (R114H). Notably, all R114H homozygotes were also homozygous for the T allele of a SNP at c.531. This allele exhibits highly significant (χ2 test P<.001) over-representation in patients compared with controls (with a T allele population frequency of 0.4), suggesting that the R114H change might be an ancient founder mutation.
Figure 2. .
Schematic representation of the TREX1 protein, with the corresponding position of the mutations identified in TREX1. Regions 1, 3, and 4 represent the exonuclease regions (Exo1-3), which coordinate the binding of 2 Mg2+ ions required for catalysis. Region 2 represents the polyproline II motif. Region 5 represents the dileucine-repeat region. Numbers in parentheses after mutations represent the number of mutated alleles identified. An asterisk (*) denotes the de novo mutation identified in one family.
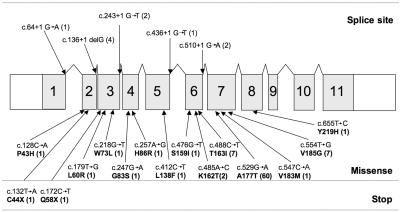
A total of 20 distinct mutations were identified in RNASEH2B (a 308-aa protein) (fig. 3). All were missense changes, except for five splice acceptor/donor mutations and two stop mutations; the 12 affected individuals carrying these were all compound heterozygotes; the second mutation was a missense change. All 50 families with a change identified in RNASEH2B harbored at least one mutation in exon 2, 6, or 7. The recurrent c.529G→A (A177T) mutation was seen in a panethnic cohort of patients, indicating that this is a mutation hotspot. In three families, we could identify only a single RNASEH2B mutation. In one of these families, the mutation had been maternally inherited, whereas, in the other two families, parental samples were unavailable. The two heterozygous mutations observed in these three pedigrees, c.529G→A and c.488C→T, were seen recurrently in other cases.
Figure 3. .
Schematic representation of the RNASEH2B gene, with the position of identified mutations. Shaded areas with large numbers indicate the specified exons. Numbers in parentheses after mutations represent the number of mutated alleles identified. Splice-site variants and stop mutations always occur with a missense mutation.
We identified six distinct RNASEH2C (164-aa) missense mutations in 18 families (fig. 4). Thirteen of these families were of Pakistani origin, all of whom were homozygous for the R69W (c.205C→T) mutation on a common haplotype (data not shown), suggesting an ancient founder effect. The remaining mutations were seen in a panethnic cohort of patients.
Figure 4. .
Schematic representation of the RNASEH2C gene, with the position of identified mutations. Shaded areas with large numbers indicate the specified exons. Numbers in parentheses after mutations represent the number of mutated alleles identified.
Four children from three families were found to have biallelic mutations in RNASEH2A (299 aa) (fig. 5). Four of five mutations, one of which occurred in homozygous form, were missense. In one family, we identified a single mutation, c.704G→A (R236Q), in RNASEH2A that had been inherited. In another patient, we identified two putative RNASEH2A mutations, c.717dup GC and c.719C→T (T241M), on the same allele. Parental samples were unavailable in this case. Finally, a number of polymorphisms were also identified in the RNASEH2A gene.
Figure 5. .
Schematic representation of the RNASEH2A gene, with the position of identified mutations and polymorphisms. Shaded areas with large numbers indicate the specified exons. Numbers in parentheses after mutations represent the number of mutated alleles identified. An asterisk (*) denotes polymorphisms included in the SNP database; a double asterisk (**) denotes synonymous changes not found in controls or annotated as a SNP; ¶ denotes polymorphisms found in controls; † denotes polymorphisms found in combination with RNASEH2B mutations; ‡ denotes putative mutation found in patients with only a single identified RNASEH2A change; a number sign (#) denotes mutations found on one allele in a single patient.
Clinical Findings
Clinical data were collected for 123 individuals from 94 families with mutations in TREX1, RNASEH2A, RNASEH2B, or RNASEH2C. The majority of children were born at term, with a normal birth weight and head circumference, and were discharged to home soon after delivery. However, 23 children (19 with TREX1 mutations, 3 with RNASEH2C mutations, and 1 with a homozygous RNASEH2A mutation) were affected at birth and required immediate support (table 2). These children presented with abnormal neurology, acted jittery, and fed poorly. Eight experienced neonatal seizures, and 15 demonstrated liver involvement, with hepatosplenomegaly and/or raised transaminase levels. Fifteen children had a thrombocytopenia, of whom five required one or more platelet transfusions.
Table 2. .
Features of AGS-Affected Patients with Mutations in TREX1, RNASEH2A, and RNASEH2C Who Presented at Birth[Note]
| Measurementat Birth (percentile) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene andGestation,in wk | Weight | OFC | Neonatal Liver Involvement | Plateletsa | Neonatal Seizures | CSF WCC/mm3 (age) | CSF IFN-α IU/liter (age) | Status (age) |
| TREX1: | ||||||||
| 38 | 9th–25th | 9th–25th | HSM | Low (39) | Yes | 52 (2 wk) | NA | Alive (11 years) |
| 34 | 25th | 50th | HSM and ALFT | Low (40) | Yes | 1 (25 mo); 2 (30 mo) | 50 (25 mo) | Deceased (6 years) |
| 36 | 97th | 97th | No | Low (50) | No | 27 (1 week); 17 (1 mo); 4 (8 mo) | 25–50 (8 mo) | Alive (7 years) |
| NR | NR | NR | No | NR | No | NR | NR | Alive (3 years) |
| NR | NR | NR | No | NR | No | 12 (4 mo) | 200 (4 mo) | Deceased (2 years) |
| 37 | 9th | 2nd | No | Low (38) | Yes | 17 (2 wk) | 400 (2 mo) | Alive (18 mo) |
| NR | NR | 25th | HSM and ALFT | Normal | Yes | NA | NA | Alive (3 years) |
| 40 | 2nd–9th | NR | HSM and ALFT | Normal | No | 0 (11 mo) | 3 (11 mo) | Deceased (6 years) |
| 39 | 9th | 9th | Yes (unspecified) | Normal | No | 18 (14 mo) | 9 (14 mo) | Alive (6 years) |
| 40 | .4th | 2nd | No | Low (115) | No | 3 (2 d); 25 (7 d) | 100 (2 wk); 200 (1 mo) | Deceased (13 years) |
| 38 | 2nd | 2nd | HSM | Low (8)b | Yes | 12 (2 wk) | 200 (2 wk) | Alive (10 mo) |
| NR | NR | NR | No | Normal | No | 14 (4 mo); 25 (11 mo) | 100 (4 mo) | Alive (3 years) |
| 40 | .4th–2nd | NR | ALFT | Low | No | 17 (5 mo); 6 (17 mo) | NA | Alive (9 years) |
| 40 | 2nd–9th | .4th | HSM | Low (53) | No | 57 (1 d); 124 (3 wk); 15 (1 mo); 10 (9 mo) | 50 (9 mo) | Alive (9 years) |
| 40 | <.4th | <.4th | HSM and ALFT | Pancytopeniab | No | 0 (2 mo) | 75 (2 mo) | Alive (6 mo) |
| 38 | 50th | 50th | No | Normal | No | 108 (15 d); 20 (28 d); 6 (37 d) | 36 (1 mo) | Alive (1 years) |
| 31 | .4th | .4th | HSM | Low | No | 0 (1 d); 2 (1 mo) | 20 (1 mo) | Alive (4 mo) |
| 27 | .4th–2nd | 2nd–9th | HSM and ALFT | Low (15)c | No | NA | NA | Deceased (4.5 mo) |
| 36 | 91st | 50th | No | Low (37)c | No | 21 (2 wk) | 200 (2 wk) | Alive (2 mo) |
| RNASEH2C: | ||||||||
| 38 | 9th | 9th–25th | HSM and ALFT | Low (47) | Yes | 25 (11 d); 70 (3 wk); 63 (2 mo) | NA | Deceased (2 years and 11 mo) |
| NR | NR | NR | HSM | Normal | Yes | NA | NA | Deceased (2 years and 8 mo) |
| 40 | 25th | 25th | HSM | Low (30) | No | NR | 150 (3 mo) | Alive (1 years) |
| RNASEH2A: | ||||||||
| 37 | 2nd | <.4th | HSM | Pancytopeniab | Yes | 25 (1 d) | NA | Deceased (7 years) |
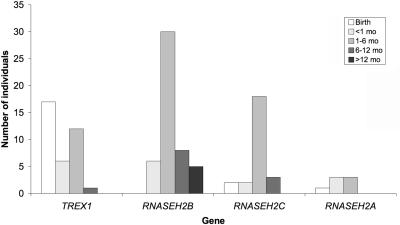
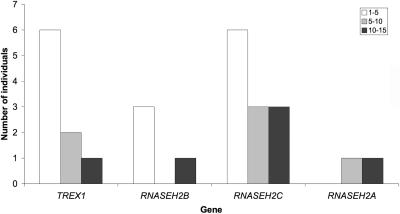
All other children presented at variable periods beyond the first few days of life with, in the majority of cases, the stereotypical subacute onset of a severe encephalopathy characterized by irritability, inconsolable crying, intermittent sterile pyrexias (40%), and a loss of skills. These episodes usually lasted several months, beyond which time the condition stabilized. Thereafter, no further disease progression was generally observed. Children with RNASEH2B mutations presented significantly later than did those with mutations in TREX1, RNASEH2C, or RNASEH2A (Mann-Whitney U test, P<.0005) (fig. 6); five children had onset of symptoms at or beyond age 12 mo, after a completely normal period of development.
Figure 6. .
Age (mo) at presentation by gene for patients with TREX1, RNASEH2A, RNASEH2B, and RNASEH2C mutations. With the Mann-Whitney U test comparing age at presentation for patients with RNASEH2B mutations with that for patients with mutations in TREX1, RNASEH2C, and RNASEH2A, P<.0005. With the Kruskal-Wallis test comparing age at presentation with gene, P<.0005.
The neurological phenotype of all patients was remarkably consistent, although variations were observed in the severity of the associated neurological handicap. Typically, patients were left with peripheral spasticity; dystonic posturing, particularly of the upper limbs; truncal hypotonia; and poor head control. Seizures were reported in 53% of patients. A number of children were noted to demonstrate a marked startle reaction to sudden noise, and, in several cases, the differentiation from epilepsy was uncertain. Almost all patients were severely intellectually and physically impaired. However, six children with RNASEH2B mutations had relatively preserved intellectual function, with good comprehension and at least some retained speech. One of these patients is of normal intelligence at age 19 years. A discrepancy in the severity of the neurological outcome was observed between siblings in several families. Most patients exhibited a severe acquired microcephaly, but, in those children with preserved intellect, the head circumference was normal. Hearing was reported as normal in almost every case. Visual function varied from normal to cortical blindness. Ocular structures were almost invariably normal on examination.
Of all children with TREX1, RNASEH2A, and RNASEH2C mutations, 34.3% were known to have died, the majority (81%) by age 10 years (fig. 7). In contrast, only 4 (8%) of 50 patients with RNASEH2B mutations were deceased (χ2 test, _P_=.001), and 7 individuals with mutations in this gene are known to be aged >18 years (the oldest known patient is aged 25 years).
Figure 7. .
Age at death (in years) of patients with TREX1, RNASEH2A, RNASEH2B, and RNASEH2C mutations; χ2 test comparing number of deaths among children with TREX1, RNASEH2C, and RNASEH2A mutations against number of deaths among children with RNASEH2B mutations (_P_=.001).
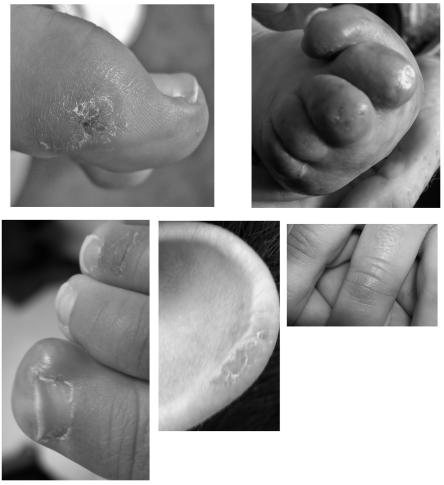
Chilblain lesions were reported in 43% of patients and were associated with mutations in all four genes. The lesions were usually situated on the feet but sometimes also affected the hands and outer rim of the ears (fig. 8). Many parents reported a direct relationship with cold temperatures, so the lesions were considerably worse during winter months.
Figure 8. .
Examples of chilblain lesions seen in patients with AGS
A number of rarer features were observed (table 3), including raised levels of autoantibodies in six children, treated hypothyroidism in an additional two cases, and insulin-dependent diabetes mellitus (IDDM) in two other patients.
Table 3. .
Infrequent Features Seen in AGS-Affected Patients
| No. of Patients with Mutation | ||||
|---|---|---|---|---|
| Phenotype | TREX1 | RNASEH2B | RNASEH2C | RNASEH2A |
| Scoliosis | 0 | 9 | 0 | 0 |
| Cardiomegaly | 4 | 0 | 1 | 1 |
| Abnormal antibody profile | 2 | 3 | 1 | 0 |
| Preserved language | 0 | 6 | 0 | 0 |
| Demyelinating peripheral neuropathy | 1 | 2 | 1 | 0 |
| Congenital glaucoma | 2 | 0 | 1 | 0 |
| Micropenis | 1 | 0 | 1 | 0 |
| Hypothyroidism | 1 | 1 | 0 | 0 |
| IDDM | 1 | 1 | 0 | 0 |
| Transitory deficiency of antidiuretic hormone | 1 | 0 | 0 | 0 |
Radiographic Findings
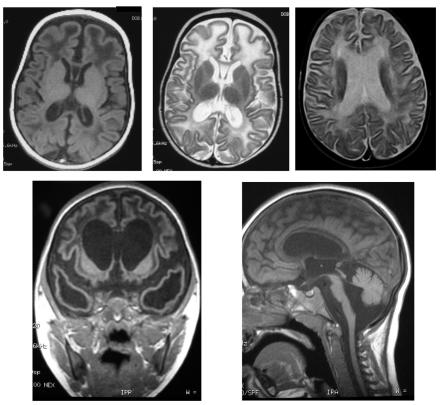
Neuroimaging demonstrated intracranial calcification variably involving the basal ganglia, dentate nuclei of the cerebellum, and deep white matter (fig. 9). Additionally, there were associated abnormalities of the white matter, with, in milder cases, high signal on T2-weighted MRI situated at the anterior and posterior poles of the lateral ventricles and, in more severely affected patients, a striking frontotemporal leukodystrophy with temporal cystic lesions (fig. 10). In those subjects presenting in the perinatal period, calcifications and white-matter disease were evident as early as the 1st d of life. Cortical atrophy was a common feature in later scans, and a number of children demonstrated significant brain-stem and cerebellar atrophy. Thinning and, in one subject, complete absence of the corpus callosum were also observed.
Figure 9. .
Examples of intracranial calcification on CT scan of patients with AGS. Calcification is seen in the basal ganglia (a and b), dentate nuclei of the cerebellum (c), a periventricular distribution (d), and within the deep white matter (e).
Figure 10. .
Spectrum of brain changes seen on MRI of patients with AGS, showing hypointensity on T1-weighted imaging (a), hyperintensity on T2-weighted imaging (b and c) of white matter, extensive bitemporal cystic lesions (d), and significant thinning of the brain stem and cerebellar atrophy (e).
Laboratory Findings
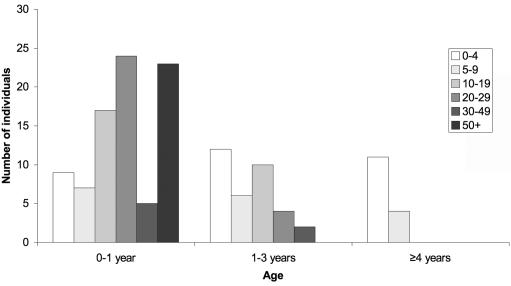
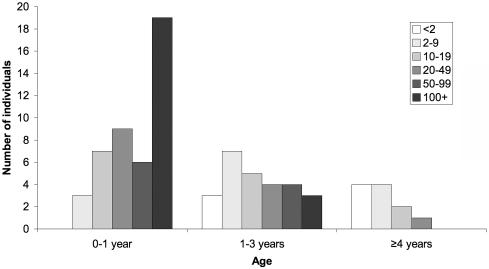
Where assayed, the number of white cells in the CSF was normal (<5 cells/mm3) on 32 of 137 occasions (table 4). The CSF WCC tended to fall with age, and the oldest age at which a level >5 cells/mm3 was documented was 9 years (Kruskal-Wallis test comparing CSF WCC with age group, P<.0005) (fig. 11). CSF IFN-α was recorded as elevated on 74 of 81 occasions tested. Again, levels of IFN-α tended to fall with age (Kruskal-Wallis test comparing CSF IFN-α with age group, P<.0005) (fig. 12). Of note, on 15 of 73 occasions where both were measured, the CSF WCC was normal when the IFN-α level was raised. Conversely, three children demonstrated a normal IFN-α titer with a raised number of CSF white cells (table 5).
Table 4. .
Number of Normal CSF White-Cell and IFN-α Results of Examinations of Mutation-Positive Patients with AGS
| No. (Total Recordings) of Patients | ||
|---|---|---|
| Age(years) | WCC <5/mm3 | IFN-α <2 IU/Liter |
| 0–1 | 9 (87) | 0 (44) |
| 1–3 | 12 (35) | 3 (26) |
| ⩾4 | 11 (15) | 4 (11) |
Figure 11. .
CSF white-cell counts by age in patients with TREX1, RNASEH2A, RNASEH2B, and RNASEH2C mutations. With Kruskal-Wallis test comparing CSF WCC/mm3 with age group, P<.0005.
Figure 12. .
CSF IFN-α titers by age in patients with TREX1, RNASEH2A, RNASEH2B, and RNASEH2C mutations. With Kruskal-Wallis test comparing CSF IFN-α IU/liter with age group, P<.0005.
Table 5. .
Number of CSF Examinations Showing Discordance between Normal/Abnormal Numbers of White Cells and IFN-α Titer
| WCC | n |
|---|---|
| <5/mm3 with IFN-α >2 IU/liter | 15 |
| >5/mm3 with IFN-α <2 IU/liter | 3 |
Pterin levels were recorded in 15 children (table 6). Elevated levels of neopterin correlated well with raised numbers of CSF white cells and CSF IFN-α titers, although, on two occasions, pterin analysis was normal in the presence of a CSF lymphocytosis, whereas, in two cases, there was marked elevation of CSF pterins with no associated increase in CSF white-cell numbers.
Table 6. .
Levels of CSF Pterins, White Cells, and IFN-α in Patients with TREX1, RNASEH2B, and RNASEH2C Mutations[Note]
| Multiple of the Upper Limit of Normal | ||||||
|---|---|---|---|---|---|---|
| Gene andAge atExamination | Neopterin | Biopterin | Tetrahydrobio-Pterin | Dihydrobio-Pterin | WCC/mm3 | IFN-α |
| TREX1: | ||||||
| 11 mo | 16 | 1.5 | NA | NA | 33 | 25 IU/liter |
| 14 mo | 40 | 1.5 | NA | NA | 3 | NA |
| <1 mo | 27 | Normal | NA | NA | 20 | 36 pg/ml |
| 7 mo | 20 | 1.5 | NA | NA | 12 | NA |
| 1 mo | 5 | NA | Normal | 3 | 0 | 20 IU/liter |
| RNASEH2B: | ||||||
| 9 years | Normal | Normal | NA | NA | 8 | 0 IU/liter |
| 18 mo | 25 | 1.5 | NA | NA | 37 | 2 pg/ml |
| 2 mo | 6 | NA | Normal | NA | 51 | NA |
| 2 mo | 20 | 1.5 | NA | NA | 56 | 50 IU/liter |
| 10 mo | 6 | NA | 3 | NA | 5 | NA |
| 17 mo | 7 | NA | 2 | 1.5 | NR | NA |
| 16 years | Normal | NA | NA | NA | 0 | NA |
| 8 years | Normal | NA | NA | NA | 0 | NA |
| 13 mo | 34 | 3.5 | NA | NA | 30 | 150 IU/liter |
| RNASEH2C: | ||||||
| 8 years | Normal | NA | 1.5 | Normal | 6 | NA |
Discussion
AGS is one of a number of disorders whose clinical features mimic the sequelae of in utero viral infection.1,12–16 These conditions are important to recognize because of the associated high risk of recurrence. Our experience indicates that the possibility of a genetic disorder in subjects with AGS is sometimes unrecognized until the birth of a second affected child. In this regard, we note that no mention was made of such genetic disorders in a recent review of congenital infection.17 We suggest that an absence of definitive evidence of an infectious agent in these circumstances should always raise the suspicion of AGS.
A subgroup of AGS-affected patients, typically those with TREX1 mutations, presented at birth with abnormal neurology, hepatosplenomegaly, elevated liver enzymes, and thrombocytopenia, a picture highly reminiscent of congenital infection. All other children presented at variable times beyond the first few days of life, frequently after a period of apparently normal development. The majority of these later-presenting cases exhibited a severe encephalopathy with subacute onset that was characterized by extreme irritability, intermittent sterile pyrexias, a loss of skills, and a slowing of head growth. This encephalopathic phase usually lasted several months. Interestingly, the opinion of most pediatricians involved in the care of these children was that there was no disease progression beyond the encephalopathic period. When death occurred, the death was usually considered not to be due to a regressive process but to be secondary to the neurological damage incurred during the initial disease episode. RNASEH2B mutations were associated with a significantly later age at presentation, at or after age 12 mo in five cases, and a lower mortality, with seven patients known to be alive beyond age 18 years with no signs of disease progression. Also of note, six affected individuals with RNASEH2B mutations demonstrated relatively preserved intellectual function; one patient has a completely normal intelligence quotient and head circumference at age 19 years, and his only feature is a spastic cerebral palsy with associated intracranial calcification.
The observation of changes on brain imaging at birth indicates an in utero onset of the disease process, in keeping with the recording of prenatal raised levels of IFN-α.18 In contrast, our data also highlight the onset of AGS after many months of normal development, raising the possibility that the condition might occur in considerably older individuals too. The stimulus for the disease onset is unknown, and why the disease tends to “burn out” after several months is also not understood.
The cardinal features of AGS on brain imaging were intracranial calcification, a leukodystrophy, and cerebral atrophy. The distribution and extent of the calcification was variable and, in some cases, was observed in a periventricular distribution highly suggestive of congenital infection. Affected sibling pairs have been described as discordant for the presence of intracranial calcification, so this feature should not be considered a prerequisite for the diagnosis of AGS.1 Additionally, in several of our patients, the intracranial calcification became evident only over a period of months. Of importance, intracranial calcification is not always recognized on MRI, the initial imaging modality employed in most medical facilities. Consequently, AGS should be considered in the differential diagnosis of an unexplained leukoencephalopathy, and CT scanning is warranted in cases conforming to the clinical scenarios we have outlined above. Of further note, some patients demonstrated marked frontotemporal white-matter involvement with cyst formation, so that Alexander disease (MIM 203450) and vanishing white-matter disease (MIM 603896) were considered and tested for.
We identified 99 families with biallelic mutations in one of the four genes known to cause AGS. Of these families, 78.8% had mutations in either TREX1 or RNASEH2B. Moreover, all the patients with RNASEH2B mutations harbored at least one mutation in exon 2, 6, or 7. Thus, an initial screen of TREX1 plus these three RNASEH2B exons, together with an analysis for the recurrent c.205C→T (R69W) RNASEH2C mutation in Pakistani patients, is likely to identify 90% of all AGS-affected patients with mutations in one of these four genes. In four families, we were able to identify only a single RNASEH2A (one family) or RNASEH2B (three families) mutation, and one further child carried two putative RNASEH2A mutations on the same allele; these cases are under further study. In addition, we studied 22 families in whom no AGS1–4 mutations could be identified. Given the high detection rate of biallelic mutations in AGS1–4, this finding strongly suggests further genetic heterogeneity. Finally, only a single patient was observed with a de novo heterozygous TREX1 mutation, indicating that such cases are unusual.19
The majority of TREX1 mutations are null alleles that are predicted to produce truncated protein, which might be inactive or disrupt protein complexes in the cell. Our recent description of heterozygous TREX1 mutations as a cause of dominant AGS suggests that some mutations may have a dominant negative effect.19 Additionally, multiple patients are homozygous or compound heterozygous for a recurrent amino acid substitution, R114H, known to be involved in stabilization of the TREX1 dimer20 and that appears to abrogate TREX1 activity.10 In contrast to TREX1, almost all mutations identified in RNASEH2A, RNASEH2B, and RNASEH2C are missense changes, which suggests a hypomorphic effect rather than a complete loss of RNASEH2 enzyme function. Indeed, this mutational spectrum raises the possibility that biallelic null alleles in any of these three genes might be lethal during embryonic development.
Minimum diagnostic criteria for AGS are difficult to define. We have already highlighted above the absence of intracranial calcification in some cases of AGS. A CSF lymphocytosis was originally described as a primary diagnostic feature of the disease and was posited to differentiate AGS from pseudo-TORCH syndrome.14 However, it is well recognized that the level of white cells and IFN-α in the CSF of AGS-affected patients falls to normal over the first few years of life.21 Moreover, in our series, a normal CSF WCC was documented in the presence of elevated CSF IFN-α titers on 15 occasions. Blau et al.22 recently described a possible variant of AGS associated with high levels of CSF pterins. A number of mutation-positive AGS subjects in our series demonstrated a similar pterin profile, which should now be considered a marker of AGS. Whether the cases described by Blau et al.22 have AGS or a separate condition remains to be determined.
The chilblain lesions23 seen in 43% of AGS cases provide a useful clinical marker of the disease and indicate an immune pathology. The observation of a small number of AGS-affected children with autoantibodies, hypothyroidism, and IDDM also suggests immune dysfunction. We recently described heterozygous TREX1 mutations in an autosomal dominant cutaneous form of systemic lupus erythematosus (SLE [MIM 152700]) called “familial chilblain lupus” (MIM 610448).19 The precise functions of TREX1 and the RNASEH2 complex are unknown. We predict that these nucleases are involved in removing nucleic acid species produced during apoptosis and that a failure of this process results in activation of the innate immune system.24,25 This hypothesis would explain the phenotypic overlap of AGS with congenital infection and some aspects of SLE,26–29 where an IFN-α–mediated innate immune response is triggered by viral and host nucleic acids, respectively.30,31
Acknowledgments
We sincerely thank the participating AGS-affected families for the use of genetic samples and clinical information. We thank all clinicians for contributing samples and data not included in this manuscript. We remember Prof. Robin Winter and Dr. Syb van der Meer. We are grateful to Mrs. Fiammetta Boni for her generosity and encouragement. We acknowledge Généthon (Evry, France) for sample storage. This work was supported by the BDF Newlife, the Fondazione Cariplo, the Fondazione Banca del Monte di Lombardia, and the Leeds Teaching Hospitals Charitable Foundation.
Author affiliations are as follows: Leeds Institute of Molecular Medicine (G.R.; R.P.; D.T.B.; I.M.C.; B.E.H.; Y.J.C.) and DNA Laboratory, Department of Clinical Genetics (T.P.; C.F.T; K.F.), St James’s University Hospital, and Cancer Research UK, Mutation Detection Facility (C.F.T.), and Department of Paediatric Neurology, Leeds General Infirmary (C.D.F.; J.H.L.), Leeds; Department of Paediatric Neurology, Erasme Hospital (A.A.) and Children’s Hospital Queen Fabiola (C.D.L.), Brussels; Service de Neuropédiatrie (A.G.), Department of Paediatric Neurology (M.-L.M.), Hôpital Trousseau, Pediatric Neurology Department, Hôpital Bicêtre (P.G.L.), Institut de Myologie, Groupe Hospitalier Pitié-Salpêtrière (T.V.), and Service de Virologie, Hôpital Cochin-St Vincent de Paul (P.L.), Paris (J.A.; F.G.); Departments of Clinical Biochemistry (R.A.) and Barcelona Pediatric Neurology (A.G-C.), Hospital Sant Joan de Déu-Ciberer, Barcelona; Department of Paediatrics, St Luke’s Hospital, Guardamangia, Malta (S.A.M.; D.S.); Department of Molecular and Human Genetics, Baylor College of Medicine, Houston (C.A.B.; Y-H.J.); Serive de Neurologie, Centre Hospitalier, Pau, France (B.B.); Department of Paediatrics, Children's Hospital, Sheffield, United Kingdom (P.B.); Developmental and Metabolic Neurology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda (W.S.B.; R.S.); Department of Human Genetics, Rheinisch-Westfälische Technische Hochschule Aachen University, Aachen, Germany (C.B.); Unit of Molecular Medicine, Bambino Gesù Children’s Research Hospital (E.B.), and Istituto di Ricerca e Cura a Carattere Scientifico (IRCCS) Casa Sollievo della Sofferenza, Mendel Institute (E.M.V.), Rome; Muscular and Neurodegenerative Disease Unit, G. Gaslini Institute, Genova, Italy (R.B.; T.B.); Department of Clinical Genetics, Churchill Hospital, Oxford, United Kingdom (E.M.B.); Division of Clinical Chemistry and Biochemistry, University Children’s Hospital, Zurich (N.B.); Clinical Genetics Unit, Birmingham Women’s Hospital (L.A.B.; H.C.), Department of Paediatrics, Sandwell and West Birmingham NHS Trust (R.J.), and Neurology Department, Birmingham Children’s Hospital (E.W.), Birmingham, United Kingdom; Departments of Human Genetics (H.G.B.; B.C.J.H.) and Pediatric Neurology (M.A.A.W.), Radboud University, Nijmegen, The Netherlands; Department of Paediatric Neurology (C.J.B.) and Genetic Health Queensland (A.Z.), Royal Children’s Hospital, Brisbane, Australia; Serviço de Aconselhamento Genético, Universidade Estadual de São Paulo, Botucatu, Brazil (D.R.C.); Academic Unit of Medical Genetics, St Mary’s Hospital, Manchester, United Kingdom (K.E.C.; H.M.K.); Kinderkrankenhaus auf der Bult, Hannover, Germany (H-J.C.); Department of Paediatrics, Bradford National Health Service (NHS) Trust (P.C.C.; S.S.; Y.J.C.), Bradford, United Kingdom; Developmental Neurology Department, Fondazione Istituto Neurologico “C. Besta,” Milan (S.D.); Grampian Clinical Genetics Centre, Aberdeen (J.D.); Departments of Neonatology (C.D.P.) and Pediatrics (R.N.A.V.C.), University Hospital, Ghent, Belgium; Departments of Clinical Genetics (S.G.M.F.) and Neurology (J.S.H.V.), Maastricht University Hospital, Maastricht, The Netherlands; Department of Paediatrics and Imaging Sciences, Imperial College (F.M.C.), St Mary’s NHS Trust (E.G.H.L.), Department of Ophthalmology (K.K.N.) and North East Thames Regional Genetics Service (E.M.R.), Great Ormond Street Hospital, and Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Trust (M.J.L.), London; Department of Paediatrics, Université Laval Medical School, Québec (C.D.); Clinical Genetics Unit, Hospital de Cruces, Baracaldo, Spain (B.G.); Service de Génétique Médicale (C.G.; D.L.) and Unité de Neurologie de l’Enfant et de l’Adolescent (M.H.), Centre Hospitalier Universitaire Pellegrin Enfants, Bordeaux; National Centre for Medical Genetics, Our Lady’s Hospital (A.J.G.; S.A.L.), and Department of Paediatric Neurology, Children's University Hospital (M.D.K.), Dublin; Departments of Medical Genetics (A.H.) and Paediatrics (M.R.), Rikshospitalet-Radiumhospitalet, Oslo; Department of Pediatrics, Academic Medical Center (R.C.H.), and Department of Child Neurology, Vrije Universiteit Medical Center (M.S.v.d.K.), Amsterdam; Medical Research Council Human Genetics Unit, Western General Hospital, Edinburgh (A.P.J.; A.L.); Department of Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands (S.G.K.); Division of Pediatric Neurology, Oregon Health and Science University, Portland (A.K.); Pediatric Neurology, Klinikum Aschaffenburg, Aschaffenburg, Germany (J.K.); Department of Neurology, Royal Children’s Hospital, Parkville, Australia (A.J.K.); Division of Clinical Genetics, Department for Medical Genetics, Medical University Innsbruck (D.K.), and Department of Pediatrics, Division of Pediatric Neurology and Inborn Errors of Metabolism, Children’s Hospital Innsbruck (K.R.; S.S-B.), Innsbruck, Austria; Klinik für Kinder und Jugendliche, Konstanz, Germany (D.K.; W.K.); Paediatric Neurology, University Hospitals of Gasthuisberg, Leuven, Belgium (L.L.); Department of Child Neurology and Psychiatry, IRCCS Casimiro Mondino Institute of Neurology, Pavia, Italy (G.L.; S.O.; E.F.); Department of Neurogenetics, School of Medicine of Ribeirao Preto, Ribeirao Preto, Brazil (C.M.L.); Greenwood Genetic Center, Greenwood, SC (M.J.L.; R.C.R.); The Raphael Recanati Genetic Institute, Rabin Medical Center, Petach-Tikva, Israel (D.M.); Department of Paediatrics, Crosshouse Hospital, Ayr, United Kingdom (J.P.M.); Fraser of Allander Neurosciences Unit (R.M.; J.B.P.S.; S.M.Z.) and Duncan Guthrie Institute of Medical Genetics (J.L.T.; M.L.W.), Royal Hospital for Sick Children, Glasgow; Division of Medical Genetics, Montreal Children’s Hospital, Montreal (S.B.M.); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, United Kingdom (L.D.M.); University Hospital of Aarhus, Aarhus, Denmark (J.R.Ø.); Division of Pediatric Dermatology, British Columbia’s Children’s Hospital, Vancouver (J.P.); Institut de Pathologie et de Génétique, Gosselies, Belgium (D.R.); Pediatric Neurology Department, Gui de Chauliac Hospital, Montpellier, France (A.R.); Servicio de Pediatría, Hospital Universitario Doctor Peset, Valencia, Spain (A.S.); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa (S.A.S.; R.S.); Neuropediatrics Unit, Complejo Hospitalario de Jean, Jean, Spain (C.S.C.); Department of Paediatrics, Manor Hospital, Walsall, United Kingdom (G.P.S.); Division of Neuropediatrics, University Hospital, Freiburg, Germany (U.T.); Genetic Health Services Victoria, Royal Children’s Hospital, Victoria, Australia (T.T.); Service de Génétique, Hôpital Debrousse, Lyon, France (M.T.); Lancashire Teaching Hospitals Trust, Preston, United Kingdom (P.T.); Neonatal Intensive Care Unit, Arcispedale Santa Maria Nuova, Reggio Emilia, Italy (F.V.); Center for Medical Genetics, Antwerp (N.V.d.A.); Department of Neurology, Children’s National Medical Center, Washington, DC (A.V.); and Department of Neuropediatrics, Humboldt University, Berlin (B.W.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for TREX1 protein [transcript AAK07616 and nucleotide sequence NM_033627, with the A at 2986 as the first base of the initiating ATG codon], RNASEH2A protein [transcript AAH11748.1 and nucleotide sequence NM_006397.2], RNASEH2B protein [transcript AAH36744.1 and nucleotide sequence NM_024570.1], and RNASEH2C protein [accession number AAH23588.1 and nucleotide sequence NM_032193.3])
- International Aicardi-Goutières Syndrome Association, http://www.aicardi-goutieres.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CSF lymphocytosis, TREX1, RNASEH2A, RNASEH2B, RNASEH2C, Alexander disease, vanishing white-matter disease, SLE, and familial chilblain lupus)
References
- 1.Aicardi J, Goutières F (1984) A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol 15:49–54 10.1002/ana.410150109 [DOI] [PubMed] [Google Scholar]
- 2.Lebon P, Badoual J, Ponsot G, Goutieres F, Hemeury-Cukier F, Aicardi J (1988) Intrathecal synthesis of interferon-alpha in infants with progressive familial encephalopathy. J Neurol Sci 84:201–208 10.1016/0022-510X(88)90125-6 [DOI] [PubMed] [Google Scholar]
- 3.Goutieres F, Aicardi J, Barth PG, Lebon P (1998) Aicardi-Goutières syndrome: an update and results of interferon-α studies. Ann Neurol 44:900–907 10.1002/ana.410440608 [DOI] [PubMed] [Google Scholar]
- 4.Lanzi G, Fazzi E, D’Arrigo S, Orcesi S, Maraucci I, Uggetti C, Bertini E, Lebon P (2005) The natural history of Aicardi-Goutieres syndrome: follow-up of 11 Italian patients. Neurology 64:1621–1624 10.1212/01.WNL.0000159864.05826.08 [DOI] [PubMed] [Google Scholar]
- 5.McEntagart M, Kamel H, Lebon P, King MD (1998) Aicardi-Goutieres syndrome: an expanding phenotype. Neuropediatrics 29:163–167 10.1055/s-2007-973555 [DOI] [PubMed] [Google Scholar]
- 6.Sanchis A, Cervero L, Bataller A, Tortajada JL, Huguet J, Crow YJ, Ali M, Higuet LJ, Martinez-Frias ML (2005) Genetic syndrome mimics congenital infection. J Pediatr 146:701–705 10.1016/j.jpeds.2005.01.033 [DOI] [PubMed] [Google Scholar]
- 7.Crow YJ, Jackson AP, Roberts E, van Beusekom E, Barth P, Corry P, Ferrie CD, Hamel BCJ, Jayatunga R, Karbani G, et al (2000) Aicardi-Goutières syndrome displays genetic heterogeneity with one locus (AGS1) on chromosome 3p21. Am J Hum Genet 67:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow YJ, Black DN, Ali M, Bond J, Jackson AP, Lefson M, Michaud J, Roberts E, Stephenson JB, Woods CG, et al (2003) Cree encephalitis is allelic with Aicardi-Goutières syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J Med Genet 40:183–187 10.1136/jmg.40.3.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali M, Highet LJ, Lacombe D, Goizet C, King MD, Tacke U, van der Knaap MS, Lagae L, Rittey C, Brunner HG, et al (2006) A second locus for Aicardi-Goutieres syndrome at chromosome 13q14-21. J Med Genet 43:444–450 10.1136/jmg.2005.031880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van BokHoven H, Brunner HG, Hamel BC, et al (2006) Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38:917–920 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- 11.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38:910–916 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- 12.Hoyeraal HM, Lamvik J, Moe PJ (1970) Congenital hypoplastic thrombocytopenia and cerebral malformations in two brothers. Acta Paediat Scand 59:185–191 [DOI] [PubMed] [Google Scholar]
- 13.Burn J, Wickramasinghe HT, Harding B, Baraitser M (1986) A syndrome with intracranial calcification and microcephaly in two sibs, resembling intrauterine infection. Clin Genet 30:112–116 [DOI] [PubMed] [Google Scholar]
- 14.Reardon W, Hockey A, Silberstein P, Kendall B, Farag TI, Swash M, Stevenson R, Baraitser M (1994) Autosomal recessive congenital intrauterine infection-like syndrome of microcephaly, intracranial calcification, and CNS disease. Am J Med Genet 52:58–65 10.1002/ajmg.1320520112 [DOI] [PubMed] [Google Scholar]
- 15.Knoblauch H, Tennstedt C, Brueck W, Hammer H, Vulliamy T, Dokal I, Lehmann R, Hanefeld F, Tinschert S (2003) Two brothers with findings resembling congenital intrauterine infection-like syndrome (pseudo-TORCH syndrome). Am J Med Genet A 120:261–265 10.1002/ajmg.a.20138 [DOI] [PubMed] [Google Scholar]
- 16.Gardner RJ, Chow CW, Simpson I, Fink AM, Meagher SE, White SM (2005) Severe fetal brain dysgenesis with focal calcification. Prenat Diagn 25:362–364 10.1002/pd.1152 [DOI] [PubMed] [Google Scholar]
- 17.Modlin JF, Grant E, Makar RS, Roberts DJ, Krishnamoorthy KS (2003) Case 25-2003: a newborn boy with petechiae and thrombocytopenia. N Engl J Med 349:691–700 10.1056/NEJMcpc030017 [DOI] [PubMed] [Google Scholar]
- 18.Desanges C, Lebon P, Bauman C, Vuillard E, Garel C, Cordesse A, Oury JF, Crow Y, Luton D (2006) Elevated interferon-alpha in fetal blood in the prenatal diagnosis of Aicardi-Goutieres syndrome. Fetal Diagn Ther 21:153–155 10.1159/000089067 [DOI] [PubMed] [Google Scholar]
- 19.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O’Hara A, et al (2007) Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet 80:811–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T (2007) The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline ii helix for protein partnering. J Biol Chem 282:10537–10543 10.1074/jbc.M700039200 [DOI] [PubMed] [Google Scholar]
- 21.Lebon P, Meritet JF, Krivine A, Rozenberg F (2002) Interferon and Aicardi-Goutieres syndrome. Eur J Paediatr Neurol Suppl A 6:A47–A53 10.1053/ejpn.2002.0574 [DOI] [PubMed] [Google Scholar]
- 22.Blau N, Bonafe L, Krageloh-Mann I, Thony B, Kierat L, Hausler M, Ramaekers V (2003) Cerebrospinal fluid pterins and folates in Aicardi-Goutieres syndrome: a new phenotype. Neurology 61:642–647 [DOI] [PubMed] [Google Scholar]
- 23.Tolmie JL, Shillito P, Hughes-Benzie R, Stephenson JB (1995) The Aicardi-Goutieres syndrome (familial, early onset encephalopathy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis). J Med Genet 32:881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samejima K, Earnshaw WC (2005) Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol 6:677–688 10.1038/nrm1715 [DOI] [PubMed] [Google Scholar]
- 25.Walport MJ (2000) Lupus, DNase and defective dispersal of cellular debris. Nat Genet 25:135–136 10.1038/75963 [DOI] [PubMed] [Google Scholar]
- 26.Dale RC, Tang SP, Heckmatt JZ, Tatnall FM (2000) Familial systemic lupus erythematosus and congenital infection-like syndrome. Neuropediatrics 31:155–158 10.1055/s-2000-7492 [DOI] [PubMed] [Google Scholar]
- 27.Aicardi J, Goutières F (2000) Systemic lupus erythematosus or Aicardi-Goutières syndrome? Neuropediatrics 31:113 10.1055/s-2000-7533 [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen M, Skullerud K, Bakke SJ, Lebon P, Jahnsen FL (2005) Cerebral thrombotic microangiopathy and antiphospholipid antibodies in Aicardi-Goutières syndrome—reports of two sisters. Neuropediatrics 36:40–44 10.1055/s-2004-830532 [DOI] [PubMed] [Google Scholar]
- 29.De Laet C, Goyens P, Christophe C, Ferster A, Mascart F, Dan B (2005) Phenotypic overlap between infantile systemic lupus erythematosus and Aicardi-Goutieres syndrome. Neuropediatrics 36:399–402 10.1055/s-2005-873058 [DOI] [PubMed] [Google Scholar]
- 30.Stetson DB, Medzhitov R (2006) Type I interferons in host defense. Immunity 25:373–381 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 31.Banchereau J, Pascual V (2006) Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25:383–392 10.1016/j.immuni.2006.08.010 [DOI] [PubMed] [Google Scholar]