Caspase-14 reveals its secrets (original) (raw)
Abstract
Caspase-14 is a unique member of the evolutionarily conserved family of cysteinyl aspartate–specific proteinases, which are mainly involved in inflammation and apoptosis. However, recent evidence also implicates these proteases in proliferation and differentiation. Although most caspases are ubiquitously expressed, caspase-14 expression is confined mainly to cornifying epithelia, such as the skin. Moreover, caspase-14 activation correlates with cornification, indicating that it plays a role in terminal keratinocyte differentiation. The determination of in vitro conditions for caspase-14 activity paved the way to identifying its substrates. The recent development of _caspase-14_–deficient mice underscored its importance in the correct degradation of (pro)filaggrin and in the formation of the epidermal barrier that protects against dehydration and UVB radiation. Here, we review the current knowledge on caspase-14 in skin homeostasis and disease.
Introduction
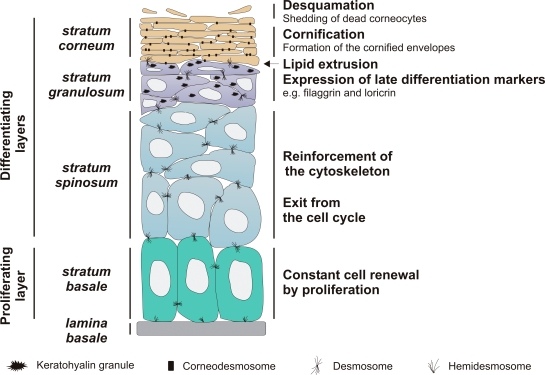
The skin is the largest organ of the body and protects the organism against external physical, chemical, and biological insults such as wounding, UVB radiation, and microorganisms. It also provides a water-impermeable barrier that prevents dehydration. This major barrier resides in the upper layers of the epidermis (for review see Segre, 2006). The epidermis is the upper part of the skin that is continuously renewed. The basal layer, or stratum basale, of the epidermis contains proliferating keratinocytes (Fig. 1). Upon withdrawal from the cell cycle, these basal keratinocytes detach from the basement membrane and undergo a terminal differentiation program to become corneocytes in the outer layers of the epidermis. This process is called cornification. In the intermediate stratum spinosum, the cells reinforce their cytoskeletal keratin filament network, and adjacent cells interact via many desmosomes, a specialized type of cell junction, to resist physical trauma. In the stratum granulosum, the keratinocytes become more flattened and express certain proteins such as profilaggrin and loricrin, which aggregate to form the typical keratohyalin granules of the stratum granulosum. In addition, lipids are produced and stored in lamellar bodies. At the final stage of differentiation, the keratinocytes lose their organelles, including the nucleus, and become the dead, flattened corneocytes of the stratum corneum. During cornification, proteins are cross-linked at the inner side of the cytoplasmic membrane to form a cornified envelope (for review see Candi et al., 2005). In the transitional layer between the stratum granulosum and the stratum corneum, lipids are extruded to form a water-repelling envelope around the cornified envelope, thereby assuring an adequate permeability barrier function of the mammalian epidermis. Improper formation of these envelopes results in an impaired epidermal barrier that cannot protect against dehydration, UVB, and infection. Finally, corneocytes are shed from the skin by a process called desquamation. The signaling cascades involved in epidermal barrier formation are largely unknown, but the many proteases that seem to be involved are currently being intensively studied.
Figure 1.
General structure of the epidermis. See Introduction for details.
Since the cloning of caspase-14 in the late nineties, it has become clear that this protease is a unique member of the caspase family. Unlike apoptotic caspases, which evolved in common ancestors such as hydra, echinodermata, insects, nematodes, and chordates, caspase-14 has so far been found only in terrestrial mammals (Lamkanfi et al., 2002). In contrast to the ubiquitously expressed other members of the caspase family, caspase-14 is expressed and activated mainly in the epidermis and is absent from most other adult tissues (Eckhart et al., 2000b; Lippens et al., 2000). Recently, caspase-14 was found to be involved in epidermal barrier formation (Denecker et al., 2007). In this review, we discuss current knowledge of the expression, regulation, and function of caspase-14.
Caspase-14 expression and regulation
The expression pattern of caspase-14 is unique among the caspases, as it is present mainly in cornifying epithelia, such as the epidermis, the Hassall's bodies of the thymus, and the forestomach of rodents (Lippens et al., 2000, 2005; Denecker et al., 2007). In skin, caspase-14 is expressed only in the differentiating and cornifying layers of the epidermis and the hair follicle (Lippens et al., 2000; Alibardi et al., 2005). This is consistent with the observation that, in vitro_,_ caspase-14 is only expressed in differentiating but not in proliferating keratinocytes (Lippens et al., 2000; Rendl et al., 2002). Remarkably, nail matrix keratinocytes that differentiate into specialized nail corneocytes, the building blocks of the nail plate, do not express caspase-14 (Jager et al., 2007). In addition, caspase-14 is not expressed in the noncornifying keratinocytes of the sweat gland or the mouth epithelium (Lippens et al., 2000; Alibardi et al., 2005). Ultrastructural analysis demonstrated that spatial distribution of caspase-14 in the epidermis and hair follicles is strongly conserved among several mammalian species (Alibardi et al., 2004, 2005). In the granular layer, caspase-14 was found to be associated with the nucleus, the keratohyalin granules, and the desmosomes, whereas in corneocytes, caspase-14 was found in the cytoplasm and was associated with corneodesmosomes (a modified version of desmosomes) and nuclear remnants. These observations suggested a role for caspase-14 in nuclear degradation during cornification, but nuclear degradation was not affected in _caspase-14–_deficient mice (Denecker et al., 2007). The expression of caspase-14 in the Hassall's bodies of the thymus and in the forestomach of rodents is somewhat expected, as they are cornifying structures and express the typical late differentiation markers, such as profilaggrin and loricrin, which are also found in the epidermis (Laster and Haynes, 1986; Favre, 1989; Jarnik et al., 1996). Protein expression of caspase-14 has also been reported in several noncornifying tissues (Lippens et al., 2003; Krajewska et al., 2004, 2005; Kam et al., 2005; Seidelin and Nielsen, 2006; Selicharova et al., 2007). However, these observations should be interpreted carefully, as we have recently shown that the reported expression of caspase-14 in such tissues can be the result of aspecific staining (Denecker et al., 2007).
Remarkably, so far caspase-14 has been found only in terrestrial mammals but not in birds or reptiles. Whereas birds and reptiles have a stiff, dry, scaly epidermis, mammals have a soft stratum corneum because of the larger amounts of histidine-rich late differentiation markers (e.g., profilaggrin; Alibardi, 2003). Interestingly, profilaggrin is a direct substrate of caspase-14 (Denecker et al., 2007). This could indicate that the occurrence of a soft stratum corneum and the caspase-14 gene are associated during evolution.
Although the expression of caspase-14 is very restricted, little is known about the transcriptional regulation of its gene. In vitro, caspase-14 is only expressed when keratinocytes are forced to differentiate by growing them postconfluently or in suspension or by adding vitamin D3 (Eckhart et al., 2000a; Lippens et al., 2000, 2004; Pistritto et al., 2002). In contrast, adding Ca2+ at high concentrations to the medium, a method frequently used to induce differentiation, did not induce caspase-14 expression (Eckhart et al., 2000a; Kuechle et al., 2001; Pistritto et al., 2002). Retinoids, which suppress keratinocyte differentiation, down-regulate caspase-14 expression (Rendl et al., 2002; Lippens et al., 2004).
These results indicate that transcription factors that are specifically active during terminal differentiation are required to regulate caspase-14 expression. Whether caspase-14 expression levels can be regulated at the posttranscriptional level is not known. Down-regulation of several differentiation-associated genes by retinoids has been shown to be mediated by the transrepression of activator protein 1 (AP-1)–mediated gene activation (Fisher and Voorhees, 1996). Indeed, the caspase-14 promoter contains at least two potential AP-1–binding sites (unpublished data). The green tea phenol (−)-epigallocatechin-3-gallate (EGCG) is a potent activator of AP-1 and has been shown to up-regulate caspase-14 in a p38- and JNK-dependent way (Hsu et al., 2005, 2007). AP-1 alone is probably not sufficient to drive caspase-14 expression because TNF and 12-_O_-tetradecanoyl-phorbol 13-acetate, two potent activators of AP-1 in keratinocytes (Arnott et al., 2002), did not induce caspase-14 expression in keratinocytes (Lippens et al., 2004).
Differentiation-dependent expression of caspase-14 in keratinocytes could also result from a strong transcriptional repression in proliferating keratinocytes. This possibility is supported by the observation that mice deficient in nuclear receptor corepressor Hairless (Hr) had 5–10-fold higher levels of caspase-14 and profilaggrin mRNA, starting from postnatal day 6 and progressing during development (Zarach et al., 2004). These alterations in gene expression were detected mainly in the keratinocytes of the utricle, an abnormal pouch-like structure at the upper part of the hair follicle. Increased gene expression occurred before the morphologically distinct utricle could be identified, indicating that the up-regulation was probably a cause rather than a consequence of utricle formation. It would be interesting to analyze both caspase-14 activation and filaggrin processing in these mice. Multiple mutant Hr alleles in mice and in humans show phenotypic variations that include congenital hair loss, skin wrinkling, and papular rash (Cichon et al., 1998; Panteleyev et al., 1998; Sprecher et al., 1998). Whether caspase-14 overexpression is important for the observed phenotypes could be addressed by generating epidermis-specific caspase-14 transgenic mice or by crossing the Hr mice with _caspase-14_–deficient mice.
Activation of caspase-14
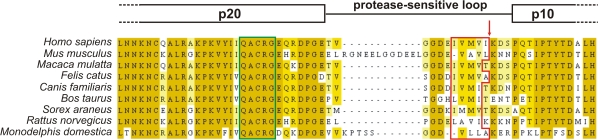
Procaspases consist of a prodomain, a large subunit (p20), and a small subunit (p10). Activation of caspases is induced by dimerization, (auto-)proteolytic cleavage at Asp residues, and/or conformational changes (Lamkanfi et al., 2003). So far, maturation of caspase-14 by proteolytic cleavage into p20 and p10 subunits has been consistently observed only in cornifying epithelia such as the epidermis and the rodent forestomach (Lippens et al., 2000; Denecker et al., 2007). Although some investigators suggested that caspase-8 and -10 can activate caspase-14 in vitro (Ahmad et al., 1998; Van de Craen et al., 1998), this could not be confirmed by others (Lippens et al., 2000; Mikolajczyk et al., 2004). Furthermore, caspase-14 is probably not proteolytically activated by a caspase in vivo, as other caspases are not activated during epidermal differentiation (Eckhart et al., 2000b; Lippens et al., 2000; Raymond et al., 2007), and caspase-14 is not processed at an aspartate residue like other caspases but is processed at Ile152 in man and presumably at Leu167 in the mouse (Chien et al., 2002). Alignment of the protease-sensitive loop between the p20 and p10 subunits of the known mammalian procaspase-14 amino acid sequences (Fig. 2) reveals a conserved hydrophobic patch that is N terminal of the caspase-14 cleavage site. This patch contains P1-preferred amino acids of elastase-like serine proteases, such as Val, Ala, Leu, and Ile (Mallory and Travis, 1975; Vered et al., 1985; Takahashi et al., 1989), suggesting that a serine protease with elastase-like properties could be involved in caspase-14 activation (unpublished data). All together, these data indicate that during skin homeostasis, the caspase-14–activating protease is not a caspase, separating caspase-14 activation from the apoptotic and inflammatory caspase cascades that could be detrimental to epidermal integrity. The precise epidermal layer in which caspase-14 is processed and activated is unknown because antiserum specifically recognizing activated caspase-14 is not yet available. However, the following findings indicate that activation of caspase-14 occurs at the interface between the granular and cornified layers of the epidermis or early during cornification. First, both the proform and activated form of caspase-14 can be found in total epidermal extracts, whereas in the cornified layer only activated caspase-14 is found (Fischer et al., 2004). Second, caspase-14 activation coincides with stratum corneum formation both during embryonic development and in organotypic skin cultures (Eckhart et al., 2000b; Lippens et al., 2004; Fischer et al., 2005). This implies that the main biological function of caspase-14 is exercised in the stratum corneum, as proven by the phenotype of the _caspase-14_–deficient mice (see the next section).
Figure 2.
Alignment of the currently known procaspase-14 amino acid sequences. Sequence analysis indicates that a hydrophobic patch in the protease-sensitive loop is conserved. Only part of the alignment is shown here, including the C-terminal part of the p20 subunit, the protease-sensitive loop, and the N-terminal part of the p10 subunit. The darker the yellow, the more the amino acids are conserved between species. The catalytic QACRG box is delineated with a green box. The conserved hydrophobic patch is delineated with a red box, and the cleavage site in human caspase-14 is indicated with a red arrow. The alignment was performed using ClustalW (Mega version 3.1; Kumar et al., 2004) and was manually optimized in the protease-sensitive loop region. JalView 2.3 (Clamp et al., 2004) was used for visualization.
Identification of caspase-14 substrates has been hampered for a long time by the unavailability of enzymatically active caspase-14. However, it was recently shown that proteolytically processed caspase-14 requires high concentrations of kosmotropic salt to be active in vitro, such as sodium citrate, in addition to proteolytic cleavage between the p20 and p10 subunit (Mikolajczyk et al., 2004). These salts induce both dimerization and ordering of active site loops by partial desolvation of the protein to a more compact, catalytically active protease. The cellular environment of the stratum corneum of the epidermis probably favors caspase-14 activity in a similar way. Indeed, the water content decreases from 45% at the transitional layer to 15–25% at the skin surface (Warner et al., 1988; Caspers et al., 2001).
Surprisingly, human and murine caspase-14 have different substrate preferences. Human caspase-14 preferentially accommodates tryptophan or tyrosine in the S4 subsite, whereas mouse caspase-14 is more tolerant, with almost equal preferences for β-branched and aromatic amino acids (Mikolajczyk et al., 2004). For example, both human and mouse caspase-14 efficiently cleave the fluorescent peptide substrate WEHD-amc, but only mouse caspase-14 cleaves IETD-amc as efficiently (Fischer et al., 2004; Mikolajczyk et al., 2004). These substrate preferences would classify human caspase-14 as an inflammatory caspase and mouse caspase-14 as an inflammatory and apoptotic initiator caspase (Thornberry et al., 1997; Thornberry, 1998). However, human caspase-14 cannot proteolytically activate the inflammatory cytokines pro–interleukin-1β (our unpublished data) or -18 (Mikolajczyk et al., 2004), and there are no data supporting a direct role for caspase-14 in apoptosis (Lippens et al., 2000; Denecker et al., 2007). Whether the substrate preferences of human and murine caspase-14 observed in vitro on peptide substrates also occur in vivo is not clear. Importantly, profilaggrin, a major structural protein in the differentiating epidermis, has been shown to be a physiological substrate of caspase-14 (Denecker et al., 2007). Identification of additional substrates and determination of the cleavage sites will provide more insight into the preferred recognition sequence of caspase-14 in the context of a protein.
Function: caspase-14 is involved in cornification, hydration, and protection against UVB
Keratinocytes can die by two different processes: apoptotic cell death induced by damaging agents such as UVB, chemicals, and cytotoxic cytokines or by a continuous process of differentiation leading to the formation of corneocytes. These processes are clearly distinct pathways executed by different players (for review see Lippens et al., 2005), but the role of caspases in these two cell death programs has long been controversial. Although almost all procaspases are constitutively expressed in the epidermis, only caspase-14 has consistently been shown to be activated during epidermal cornification (Eckhart et al., 2000b; Lippens et al., 2000; Raymond et al., 2007). In addition, knockouts for apoptotic caspases were not reported to have a phenotypic skin anomaly except for _caspase-3_–deficient mice, in which keratinocyte differentiation is delayed in the embryo but normalized at birth (Okuyama et al., 2004). However, others could not confirm the activation of caspase-3 during embryonic epidermal development (Fischer et al., 2005). Although they are not activated during cornification, apoptotic caspases, in contrast to caspase-14, become activated during UVB-, staurosporine-, TNF-, and TNF-related apoptosis-inducing ligand–induced apoptosis of keratinocytes and, thereby, play a role in the apoptotic cell death of keratinocytes (for review see Lippens et al., 2005). Thus, we conclude that apoptotic caspases are not involved in the physiological keratinocyte cell death program leading to cornification. Furthermore, _caspase-14_–deficient epidermal cells can undergo classical apoptosis, which genetically demonstrates that caspase-14 is dispensable for the apoptosis of keratinocytes.
During development, caspase-14 protein expression is detectable from embryonic day (E) 15.5 on, and its processing is observed from E17.5 (Hu et al., 1998; Van de Craen et al., 1998; Fischer et al., 2005), which coincides with stratum corneum formation and establishment of the epidermal barrier. This indicates that caspase-14 might be involved in embryonic barrier formation. However, no differences in outside-in barrier formation of the skin of _caspase-14_–deficient mice during embryogenesis were observed (Denecker et al., 2007). Furthermore, _caspase-14–_deficient mice were born at the expected Mendelian ratios, were fertile, and had long survival rates. Detailed analysis of _caspase-14_–deficient mice indicated that caspase-14 has an important role in cornification, hydration, and UVB protection.
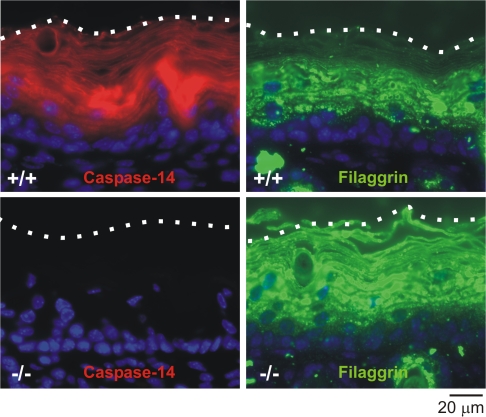
The skin of _caspase-14_–deficient mice was shinier, characterized by deeper skin lines, and had larger scales (Denecker et al., 2007) even though the shape and size of the cornified envelopes themselves were not altered. Biochemical analysis indicated that caspase-14 was responsible for the correct processing and degradation of (pro)filaggrin, as epidermis lacking caspase-14 was characterized by an altered profilaggrin processing and staining pattern (Fig. 3) and by the presence of aberrant keratohyalin granules, the profilaggrin storage granules. Profilaggrin is a large, insoluble protein consisting of a calcium-binding A domain, a B domain, and several tandem repeats of filaggrin units. In the transitional layer, profilaggrin is dephosphorylated and proteolytically processed into its functional filaggrin units (Fig. 4), which aid in the bundling of keratin intermediate filaments and formation of the cornified envelope (for review see Candi et al., 2005). Subsequently, filaggrin is deiminated (conversion of arginine to citrulline by elimination of the imino group of arginine by peptidylarginine deiminases), causing its release from keratin and allowing its degradation into free hygroscopic amino acids that act as natural moisturizing factors of the stratum corneum (Scott and Harding, 1986; Rawlings and Matts, 2005). Therefore, filaggrin plays an important role in skin hydration. Although the filaggrin unit was detected in _caspase-14_–deficient epidermis by Western blot analysis, lower molecular weight filaggrin fragments were also present (Denecker et al., 2007). Immunofluorescence analysis showed that in these mice, filaggrin immunoreactive fragments accumulated in the upper layers of the _s_tratum corneum (Fig. 3). This indicates that the correct degradation of filaggrin into free amino acids was affected in _caspase-14_–deficient skin. Interestingly, caspase-14 was found to be associated with keratohyalin granules in the stratum granulosum and to remain cytoplasmic in the stratum corneum (Alibardi et al., 2004), which could correlate with its possible involvement in the generation of free amino acids.
Figure 3.
Expression of caspase-14 and (pro)filaggrin in wild-type and _caspase-14_–deficient skin. Immunofluorescence staining for caspase-14 (red) and (pro)filaggrin (green) on paraffin sections of 5.5-d-old skin of both wild-type (+/+) and _caspase-14_–deficient (−/−) mice (Denecker et al., 2007). Nuclei are counterstained with DAPI. Fluorescence microscopy was performed on a CellM system (Olympus) with an upright microscope (BX61; Olympus). Observation was performed with a 60× 1.42 NA oil objective. A specific DAPI emission band-pass filter (450–470 nm) and a GFP emission band-pass filter (510–550 nm) were used. Image acquisition and processing were performed with the CellM software using a cooled CCD camera with a 1,344 × 1,024 pixel resolution. Image intensity scaling and color conversion were completed in ImageJ (National Institutes of Health). The dotted lines indicate the outer borders of the stratum corneum. Caspase-14 is expressed mainly in the spinous, granular, and cornified layers of wild-type mice and is absent in _caspase-14_–deficient mice. (Pro)filaggrin is expressed in the granular layer and in the lower cornified layer in wild-type skin. In _caspase-14_–deficient skin, additional filaggrin immunoreactive fragments are detected in the upper layers of the stratum corneum.
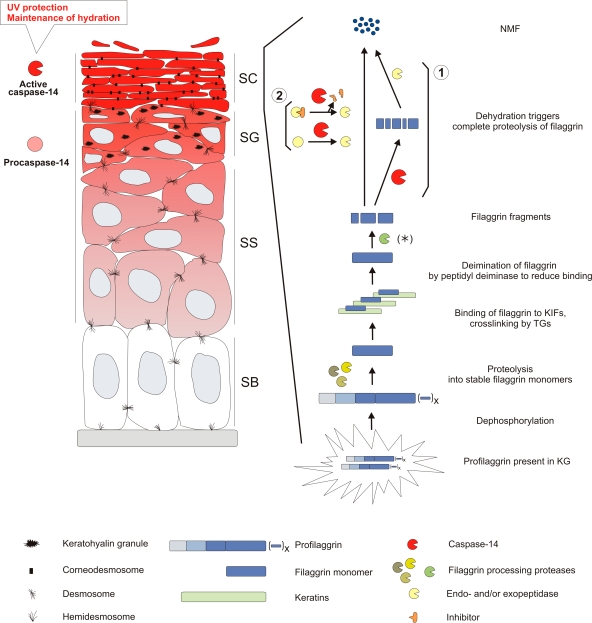
Figure 4.
Caspase-14 protects the skin against UVB photo damage and water loss and is involved in the processing of (pro)filaggrin. Caspase-14 expression starts in the spinous layer (indicated in shades of red), and cleavage into its p20 and p10 subunits occurs at the transition of the granular to the cornified layer. Caspase-14 is active in the dehydrating environment of the cornified layer, where it has an important function in formation of the epidermal barrier leading to protection against UVB and water loss (Denecker et al., 2007). Profilaggrin is a large structural molecule consisting of an N-terminal A domain and a B domain followed by multiple filaggrin repeats and a unique C-terminal sequence (for review see Candi et al., 2005). Profilaggrin undergoes many posttranslational modifications, eventually leading to release from the keratin intermediate filaments (see the section on the function of caspase-14 for details). In the lower stratum corneum, dehydration triggers the degradation of filaggrin monomers into free hygroscopic amino acids. These amino acids compose ∼40% of the natural moisturizing factors present in the stratum corneum and are important for maintaining epidermal hydration (Rawlings and Matts, 2005). In _caspase-14_–deficient skin, accumulating filaggrin fragments are present (Denecker et al., 2007), indicating that an unidentified protease (asterisk) cleaves the filaggrin monomer into these fragments and that caspase-14 is responsible for the further processing and degradation of these fragments into free amino acids. As it is very unlikely that caspase-14 is directly responsible for degradation of the filaggrin fragments into free amino acids, we propose two possible mechanisms: (1) caspase-14 could first cleave these filaggrin fragments, leading to further degradation into free amino acids by another endo- and/or exopeptidase; or (2) caspase-14 could directly or indirectly (by inactivating an inhibitor) activate an endo- and/or exopeptidase that further processes the smaller filaggrin fragments. KG, keratohyalin granule; KIF, keratin intermediate filament; NMF, natural moisturizing factors; SB, stratum basale; SC, stratum corneum; SG, stratum granulosum; SS, stratum spinosum; TG, transglutaminase.
These results, together with the finding that caspase-14 can directly cleave (pro)filaggrin in vitro, demonstrate that caspase-14 has a critical role in the correct processing of (pro)filaggrin during cornification. Whether the degradation of other differentiation-associated proteins is also affected in _caspase-14_–deficient mice remains to be determined. Two possible mechanisms for (pro)filaggrin processing by caspase-14 can be proposed (Fig. 4). First, caspase-14 may cleave the filaggrin fragments and, thereby, expose cleavage sites that can be recognized by other endo- and/or exopeptidases for further degradation. Second, caspase-14 may be the activator of an endo- and/or exopeptidase that cleaves and degrades filaggrin. This could occur directly, or indirectly by inactivating an inhibitor. Direct degradation of filaggrin fragments into free amino acids by caspase-14 can be ruled out, as caspases only cleave after aspartate residues.
The lack of filaggrin processing into free hygroscopic amino acids in _caspase-14_–deficient mice may lead to the reduced epidermal hydration and increased trans-epidermal water loss observed in these mice (Denecker et al., 2007). These results point to an important function of caspase-14 in the maintenance of epidermal hydration. Although a _profilaggrin_-deficient mouse has not been generated, it has been demonstrated that flaky tail (ft/ft) mice, which have an autosomal recessive mutation in the flaky tail gene (probably the profilaggrin gene), lack a functional filaggrin monomer (Presland et al., 2000). These mice have been proposed as a model for the _filaggrin_-deficient skin disease ichthyosis vulgaris because they have dry, flaky skin and irregular scales of variable size. The importance of filaggrin has been underscored recently by human genetic studies demonstrating that loss-of-function mutations in the profilaggrin gene are the primary cause of the skin disease ichthyosis vulgaris (Smith et al., 2006), which is characterized by silvery scales on the abdomen and palmar hyperlinearity. These mutations strongly predispose to atopic dermatitis and asthma (Palmer et al., 2006), possibly as a result of a defect in epidermal barrier function that allows the increased entry of allergens and infectious agents.
Two of the important functions of the skin are prevention of water loss and protection against environmental stress, such as protection against UVB radiation, which are essential for terrestrial life. The development of _caspase-14_–deficient mice revealed that the absence of caspase-14 enhances sensitivity toward UVB-induced photo damage and apoptosis of the skin (Denecker et al., 2007). Importantly, this is not caused by cell-autonomous differences in DNA damage sensitivity and apoptosis between wild-type and _caspase-14_–deficient keratinocytes. Instead, the UVB-filtering capacity of the stratum corneum is severely reduced in _caspase-14_–deficient skin, as higher levels of cyclobutane pyrimidine dimers are detected immediately after UVB irradiation. This indicates that caspase-14 has an indispensable role in the photoprotective function of the stratum corneum. Interestingly, topical application of EGCG, an inducer of caspase-14 expression, has been shown to be photoprotective (Elmets et al., 2001). How caspase-14 alters the structural and biochemical properties of the stratum corneum is currently under investigation.
Caspase-14 and disease
Caspase-14 was shown to be expressed at the protein level in several cancer cell lines (Pistritto et al., 2002; Koenig et al., 2005; Krajewska et al., 2005). In addition, caspase-14 mRNA, together with keratin 1 and profilaggrin mRNA, was decreased in murine UVB-induced squamous cell carcinoma, possibly reflecting reduced differentiation in the tumor (Rundhaug et al., 2005). Furthermore, in some cases, caspase-14 protein expression was associated with highly differentiated cornified areas of lung squamous cell carcinoma and cervix carcinoma (Koenig et al., 2005). However, caspase-14 activation in tumors has not been shown, and so it might not be responsible for the tumor phenotype. Presumably, the ectopic caspase-14 expression is caused by the changed transcriptional activity in these epithelial tumors. Mutations in the caspase-14 gene have not been found in human carcinomas except very rarely in colorectal tumors, which most probably were not the cause of altered caspase-14 expression (Koenig et al., 2005; Yoo et al., 2007).
We as well as other investigators demonstrated that caspase-14 expression is substantially down-regulated in psoriatic lesions but is unaffected in the nonlesional epidermis (Lippens et al., 2000, 2004; Walsh et al., 2005). Psoriasis is an autoimmune disease characterized by the uncontrolled proliferation of keratinocytes and impaired cornification, which results in the aberrant presence of nuclei in the cornified layer, also called parakeratosis. Although caspase-14 is absent in these parakeratotic regions, this is probably not the cause of the development of parakeratotic plaques, as _caspase-14_–deficient mice did not show spontaneous parakeratosis (Denecker et al., 2007). More likely, caspase-14 down-regulation results from the impairment of terminal differentiation or up-regulation of transcriptional repressors. The absence of caspase-14 in psoriatic plaques may lead to the formation of a defective barrier and, therefore, to the aggravation of psoriatic lesions. Treating the parakeratotic plaques of patients with a vitamin D3 analogue results in the up-regulation of caspase-14 and coincides with amelioration of the lesions (Lippens et al., 2004). Likewise, in the flaky skin (fsn/fsn) mouse model of psoriasis, topical EGCG treatment causes the up-regulation of caspase-14 and the amelioration of psoriasis (Hsu et al., 2007). Interestingly, the expression of JunB is strongly down-regulated in psoriatic lesions, and inducible epidermal deletion of both JunB and c-Jun in mice results in a psoriatic phenotype (Zenz et al., 2005). Because caspase-14 might be regulated by these transcription factors, it would be interesting to elucidate whether caspase-14 is down-regulated in _JunB/c-Jun_–deficient mice.
Conclusions
The role of caspase-14 has long been enigmatic. Recent evidence sheds light on the crucial role of caspase-14 in the skin, but several major questions remain. Activation of caspase-14 occurs most probably at the interface between the granular and cornified layer, implicating a role for caspase-14 in the stratum corneum. It is now clear that the caspase-14–activating protease is not a caspase but probably an epidermis-specific serine protease with elastase-like properties. This is not surprising, as it has been known for a long time that serine proteases are of major importance in epidermal homeostasis. Identification of the caspase-14–activating protease remains a major challenge. Determination of the in vitro conditions for caspase-14 activity that mimic stratum corneum conditions, together with the generation of _caspase-14_–deficient mice, led to the identification of (pro)filaggrin as the first known physiological caspase-14 substrate. Importantly, caspase-14 seems to be involved in the correct processing of filaggrin preceding its degradation into free hygroscopic amino acids, which might explain its role in the prevention of water loss from the epidermis. Proteomic approaches could lead to the identification of additional caspase-14 substrates, which would contribute to understanding the role of caspase-14 in the skin. Caspase-14 also protects against UVB-induced damage, which means that it is involved in the establishment of the biochemical or structural properties of the stratum corneum as a UVB filter. How caspase-14 establishes the UVB-filtering capacity of the corneum is not completely understood. An extensive biochemical analysis of _caspase-14_–deficient epidermis could reveal these mechanisms. Whether caspase-14, its activating protease, or its substrates could be used as therapeutic agents or as targets to improve formation of the epidermal barrier is a challenging research goal.
Acknowledgments
We thank A. Bredan for editing the manuscript, S. Lippens, E. Hoste, and L. Eckhart for fruitful discussions, and J. Verspurten for computational sequence analysis.
This work was supported by the Flanders Institute for Biotechnology as well by the European Community (EC) Marie Curie Training and Mobility Program, ApopTrain (grant MRTN-CT-035624), the EC Integrated Project, Epistem (grant LSHB-CT-2005-019067), Interuniversity Attraction Pole 6/18, and the Fonds voor Wetenschappelijke Onderzoek Vlaanderen (grants 3G.0218.06 and G.0133.05). This work was also supported by Ghent University grant BOF-GOA-12.0505.02 and by a Ghent University–cofinanced European Union project (grant I/00001/02). G. Denecker was a postdoctoral fellow at the Fonds voor Wetenschappelijk Onderzoek Vlaanderen; P. Ovaere was supported by an Emmanuel van der Schueren grant and an IWT predoctoral grant.
G. Denecker and P. Ovaere contributed equally to this paper.
Abbreviations used in this paper: AP-1, activator protein 1; EGCG, (−)-epigallocatechin-3-gallate.
References
- Ahmad, M., S.M. Srinivasula, R. Hegde, R. Mukattash, T. Fernandes-Alnemri, and E.S. Alnemri. 1998. Identification and characterization of murine caspase-14, a new member of the caspase family. Cancer Res. 58:5201–5205. [PubMed] [Google Scholar]
- Alibardi, L. 2003. Adaptation to the land: the skin of reptiles in comparison to that of amphibians and endotherm amniotes. J. Exp. Zoolog. B Mol. Dev. Evol. 298:12–41. [DOI] [PubMed] [Google Scholar]
- Alibardi, L., M. Dockal, C. Reinisch, E. Tschachler, and L. Eckhart. 2004. Ultrastructural localization of caspase-14 in human epidermis. J. Histochem. Cytochem. 52:1561–1574. [DOI] [PubMed] [Google Scholar]
- Alibardi, L., E. Tschachler, and L. Eckhart. 2005. Distribution of caspase-14 in epidermis and hair follicles is evolutionarily conserved among mammals. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 286:962–973. [DOI] [PubMed] [Google Scholar]
- Arnott, C.H., K.A. Scott, R.J. Moore, A. Hewer, D.H. Phillips, P. Parker, F.R. Balkwill, and D.M. Owens. 2002. Tumour necrosis factor-alpha mediates tumour promotion via a PKC alpha- and AP-1-dependent pathway. Oncogene. 21:4728–4738. [DOI] [PubMed] [Google Scholar]
- Candi, E., R. Schmidt, and G. Melino. 2005. The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6:328–340. [DOI] [PubMed] [Google Scholar]
- Caspers, P.J., G.W. Lucassen, E.A. Carter, H.A. Bruining, and G.J. Puppels. 2001. In vivo confocal Raman microspectroscopy of the skin: noninvasive determination of molecular concentration profiles. J. Invest. Dermatol. 116:434–442. [DOI] [PubMed] [Google Scholar]
- Chien, A.J., R.B. Presland, and M.K. Kuechle. 2002. Processing of native caspase-14 occurs at an atypical cleavage site in normal epidermal differentiation. Biochem. Biophys. Res. Commun. 296:911–917. [DOI] [PubMed] [Google Scholar]
- Cichon, S., M. Anker, I.R. Vogt, H. Rohleder, M. Putzstuck, A. Hillmer, S.A. Farooq, K.S. Al-Dhafri, M. Ahmad, S. Haque, et al. 1998. Cloning, genomic organization, alternative transcripts and mutational analysis of the gene responsible for autosomal recessive universal congenital alopecia. Hum. Mol. Genet. 7:1671–1679. [DOI] [PubMed] [Google Scholar]
- Clamp, M., J. Cuff, S.M. Searle, and G.J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics. 20:426–427. [DOI] [PubMed] [Google Scholar]
- Denecker, G., E. Hoste, B. Gilbert, T. Hochepied, P. Ovaere, S. Lippens, C. Van den Broecke, P. Van Damme, K. D'Herde, J.P. Hachem, et al. 2007. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell Biol. 9:666–674. [DOI] [PubMed] [Google Scholar]
- Eckhart, L., J. Ban, H. Fischer, and E. Tschachler. 2000. a. Caspase-14: analysis of gene structure and mRNA expression during keratinocyte differentiation. Biochem. Biophys. Res. Commun. 277:655–659. [DOI] [PubMed] [Google Scholar]
- Eckhart, L., W. Declercq, J. Ban, M. Rendl, B. Lengauer, C. Mayer, S. Lippens, P. Vandenabeele, and E. Tschachler. 2000. b. Terminal differentiation of human keratinocytes and stratum corneum formation is associated with caspase-14 activation. J. Invest. Dermatol. 115:1148–1151. [DOI] [PubMed] [Google Scholar]
- Elmets, C.A., D. Singh, K. Tubesing, M. Matsui, S. Katiyar, and H. Mukhtar. 2001. Cutaneous photoprotection from ultraviolet injury by green tea polyphenols. J. Am. Acad. Dermatol. 44:425–432. [DOI] [PubMed] [Google Scholar]
- Favre, A. 1989. Identification of filaggrin in Hassall's corpuscle by histochemical and immunohistochemical methods. Acta Anat. (Basel). 135:71–76. [DOI] [PubMed] [Google Scholar]
- Fischer, H., M. Stichenwirth, M. Dockal, M. Ghannadan, M. Buchberger, J. Bach, A. Kapetanopoulos, W. Declercq, E. Tschachler, and L. Eckhart. 2004. Stratum corneum-derived caspase-14 is catalytically active. FEBS Lett. 577:446–450. [DOI] [PubMed] [Google Scholar]
- Fischer, H., H. Rossiter, M. Ghannadan, K. Jaeger, C. Barresi, W. Declercq, E. Tschachler, and L. Eckhart. 2005. Caspase-14 but not caspase-3 is processed during the development of fetal mouse epidermis. Differentiation. 73:406–413. [DOI] [PubMed] [Google Scholar]
- Fisher, G.J., and J.J. Voorhees. 1996. Molecular mechanisms of retinoid actions in skin. FASEB J. 10:1002–1013. [DOI] [PubMed] [Google Scholar]
- Hsu, S., T. Yamamoto, J. Borke, D.S. Walsh, B. Singh, S. Rao, K. Takaaki, N. Nah-Do, C. Lapp, D. Lapp, et al. 2005. Green tea polyphenol-induced epidermal keratinocyte differentiation is associated with coordinated expression of p57/KIP2 and caspase 14. J. Pharmacol. Exp. Ther. 312:884–890. [DOI] [PubMed] [Google Scholar]
- Hsu, S., D. Dickinson, J. Borke, D.S. Walsh, J. Wood, H. Qin, J. Winger, H. Pearl, G. Schuster, and W.B. Bollag. 2007. Green tea polyphenol induces caspase 14 in epidermal keratinocytes via MAPK pathways and reduces psoriasiform lesions in the flaky skin mouse model. Exp. Dermatol. 16:678–684. [DOI] [PubMed] [Google Scholar]
- Hu, S., S.J. Snipas, C. Vincenz, G. Salvesen, and V.M. Dixit. 1998. Caspase-14 is a novel developmentally regulated protease. J. Biol. Chem. 273:29648–29653. [DOI] [PubMed] [Google Scholar]
- Jager, K., H. Fischer, E. Tschachler, and L. Eckhart. 2007. Terminal differentiation of nail matrix keratinocytes involves up-regulation of DNase1L2 but is independent of caspase-14 expression. Differentiation. 75:939–946. [DOI] [PubMed] [Google Scholar]
- Jarnik, M., T. Kartasova, P.M. Steinert, U. Lichti, and A.C. Steven. 1996. Differential expression and cell envelope incorporation of small proline-rich protein 1 in different cornified epithelia. J. Cell Sci. 109:1381–1391. [DOI] [PubMed] [Google Scholar]
- Kam, D.W., A.K. Charles, and A.M. Dharmarajan. 2005. Caspase-14 expression in the human placenta. Reprod. Biomed. Online. 11:236–243. [DOI] [PubMed] [Google Scholar]
- Koenig, U., W. Sommergruber, and S. Lippens. 2005. Aberrant expression of caspase-14 in epithelial tumors. Biochem. Biophys. Res. Commun. 335:309–313. [DOI] [PubMed] [Google Scholar]
- Krajewska, M., R.E. Rosenthal, J. Mikolajczyk, H.R. Stennicke, T. Wiesenthal, J. Mai, M. Naito, G.S. Salvesen, J.C. Reed, G. Fiskum, and S. Krajewski. 2004. Early processing of Bid and caspase-6, -8, -10, -14 in the canine brain during cardiac arrest and resuscitation. Exp. Neurol. 189:261–279. [DOI] [PubMed] [Google Scholar]
- Krajewska, M., H. Kim, E. Shin, S. Kennedy, M.J. Duffy, Y.F. Wong, D. Marr, J. Mikolajczyk, A. Shabaik, I. Meinhold-Heerlein, et al. 2005. Tumor-associated alterations in caspase-14 expression in epithelial malignancies. Clin. Cancer Res. 11:5462–5471. [DOI] [PubMed] [Google Scholar]
- Kuechle, M.K., H.M. Predd, P. Fleckman, B.A. Dale, and R.B. Presland. 2001. Caspase-14, a keratinocyte specific caspase: mRNA splice variants and expression pattern in embryonic and adult mouse. Cell Death Differ. 8:868–870. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150–163. [DOI] [PubMed] [Google Scholar]
- Lamkanfi, M., W. Declercq, M. Kalai, X. Saelens, and P. Vandenabeele. 2002. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 9:358–361. [DOI] [PubMed] [Google Scholar]
- Lamkanfi, M., W. Declercq, B. Depuydt, M. Kalai, X. Saelens, and P. Vandenabeele. 2003. The caspase family. In Caspases: Their Role in Cell Death and Cell Survival. M. Los and H. Walczak, editors. Landes Bioscience/Kluwer Academic-Plenum Publishers, Georgetown/New York. p 1–40.
- Laster, A.J., and B.F. Haynes. 1986. Characterization of a monoclonal antibody, RTE-21, that binds to keratohyalin granule-associated proteins in epithelial cells of human skin and thymus. Clin. Immunol. Immunopathol. 41:130–144. [DOI] [PubMed] [Google Scholar]
- Lippens, S., M. Kockx, M. Knaapen, L. Mortier, R. Polakowska, A. Verheyen, M. Garmyn, A. Zwijsen, P. Formstecher, D. Huylebroeck, et al. 2000. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 7:1218–1224. [DOI] [PubMed] [Google Scholar]
- Lippens, S., C. VandenBroecke, E. Van Damme, E. Tschachler, P. Vandenabeele, and W. Declercq. 2003. Caspase-14 is expressed in the epidermis, the choroid plexus, the retinal pigment epithelium and thymic Hassall's bodies. Cell Death Differ. 10:257–259. [DOI] [PubMed] [Google Scholar]
- Lippens, S., M. Kockx, G. Denecker, M. Knaapen, A. Verheyen, R. Christiaen, E. Tschachler, P. Vandenabeele, and W. Declercq. 2004. Vitamin D3 induces caspase-14 expression in psoriatic lesions and enhances caspase-14 processing in organotypic skin cultures. Am. J. Pathol. 165:833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens, S., G. Denecker, P. Ovaere, P. Vandenabeele, and W. Declercq. 2005. Death penalty for keratinocytes: apoptosis versus cornification. Cell Death Differ. 12:1497–1508. [DOI] [PubMed] [Google Scholar]
- Mallory, P.A., and J. Travis. 1975. Human pancreatic enzymes: purification and characterization of a nonelastolytic enzyme, protease E. resembling elastase. Biochemistry. 14:722–730. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk, J., F.L. Scott, S. Krajewski, D.P. Sutherlin, and G.S. Salvesen. 2004. Activation and substrate specificity of caspase-14. Biochemistry. 43:10560–10569. [DOI] [PubMed] [Google Scholar]
- Okuyama, R., B.C. Nguyen, C. Talora, E. Ogawa, A. Tommasi di Vignano, M. Lioumi, G. Chiorino, H. Tagami, M. Woo, and G.P. Dotto. 2004. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism. Dev. Cell. 6:551–562. [DOI] [PubMed] [Google Scholar]
- Palmer, C.N., A.D. Irvine, A. Terron-Kwiatkowski, Y. Zhao, H. Liao, S.P. Lee, D.R. Goudie, A. Sandilands, L.E. Campbell, F.J. Smith, et al. 2006. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 38:441–446. [DOI] [PubMed] [Google Scholar]
- Panteleyev, A.A., R. Paus, W. Ahmad, J.P. Sundberg, and A.M. Christiano. 1998. Molecular and functional aspects of the hairless (hr) gene in laboratory rodents and humans. Exp. Dermatol. 7:249–267. [DOI] [PubMed] [Google Scholar]
- Pistritto, G., M. Jost, S.M. Srinivasula, R. Baffa, J.L. Poyet, C. Kari, Y. Lazebnik, U. Rodeck, and E.S. Alnemri. 2002. Expression and transcriptional regulation of caspase-14 in simple and complex epithelia. Cell Death Differ. 9:995–1006. [DOI] [PubMed] [Google Scholar]
- Presland, R.B., D. Boggess, S.P. Lewis, C. Hull, P. Fleckman, and J.P. Sundberg. 2000. Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J. Invest. Dermatol. 115:1072–1081. [DOI] [PubMed] [Google Scholar]
- Rawlings, A.V., and P.J. Matts. 2005. Stratum corneum moisturization at the molecular level: an update in relation to the dry skin cycle. J. Invest. Dermatol. 124:1099–1110. [DOI] [PubMed] [Google Scholar]
- Raymond, A.A., M.C. Mechin, R. Nachat, E. Toulza, R. Tazi-Ahnini, G. Serre, and M. Simon. 2007. Nine procaspases are expressed in normal human epidermis, but only caspase-14 is fully processed. Br. J. Dermatol. 156:420–427. [DOI] [PubMed] [Google Scholar]
- Rendl, M., J. Ban, P. Mrass, C. Mayer, B. Lengauer, L. Eckhart, W. Declerq, and E. Tschachler. 2002. Caspase-14 expression by epidermal keratinocytes is regulated by retinoids in a differentiation-associated manner. J. Invest. Dermatol. 119:1150–1155. [DOI] [PubMed] [Google Scholar]
- Rundhaug, J.E., K.A. Hawkins, A. Pavone, S. Gaddis, H. Kil, R.D. Klein, T.R. Berton, E. McCauley, D.G. Johnson, R.A. Lubet, et al. 2005. SAGE profiling of UV-induced mouse skin squamous cell carcinomas, comparison with acute UV irradiation effects. Mol. Carcinog. 42:40–52. [DOI] [PubMed] [Google Scholar]
- Scott, I.R., and C.R. Harding. 1986. Filaggrin breakdown to water binding compounds during development of the rat stratum corneum is controlled by the water activity of the environment. Dev. Biol. 115:84–92. [DOI] [PubMed] [Google Scholar]
- Segre, J.A. 2006. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116:1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelin, J.B., and O.H. Nielsen. 2006. Expression profiling of apoptosis-related genes in enterocytes isolated from patients with ulcerative colitis. APMIS. 114:508–517. [DOI] [PubMed] [Google Scholar]
- Selicharova, I., K. Smutna, M. Sanda, K. Ubik, E. Matouskova, E. Bursikova, M. Brozova, J. Vydra, and J. Jiracek. 2007. 2-DE analysis of a new human cell line EM-G3 derived from breast cancer progenitor cells and comparison with normal mammary epithelial cells. Proteomics. 7:1549–1559. [DOI] [PubMed] [Google Scholar]
- Smith, F.J., A.D. Irvine, A. Terron-Kwiatkowski, A. Sandilands, L.E. Campbell, Y. Zhao, H. Liao, A.T. Evans, D.R. Goudie, S. Lewis-Jones, et al. 2006. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 38:337–342. [DOI] [PubMed] [Google Scholar]
- Sprecher, E., R. Bergman, R. Szargel, T. Raz, V. Labay, M. Ramon, R. Baruch-Gershoni, R. Friedman-Birnbaum, and N. Cohen. 1998. Atrichia with papular lesions maps to 8p in the region containing the human hairless gene. Am. J. Med. Genet. 80:546–550. [DOI] [PubMed] [Google Scholar]
- Takahashi, L.H., R. Radhakrishnan, R.E. Rosenfield Jr., and E.F. Meyer Jr. 1989. Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate. Biochemistry. 28:7610–7617. [DOI] [PubMed] [Google Scholar]
- Thornberry, N.A. 1998. Caspases: key mediators of apoptosis. Chem. Biol. 5:R97–R103. [DOI] [PubMed] [Google Scholar]
- Thornberry, N.A., T.A. Rano, E.P. Peterson, D.M. Rasper, T. Timkey, M. Garcia-Calvo, V.M. Houtzager, P.A. Nordstrom, S. Roy, J.P. Vaillancourt, et al. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272:17907–17911. [DOI] [PubMed] [Google Scholar]
- Van de Craen, M., G. Van Loo, S. Pype, W. Van Criekinge, I. Van den brande, F. Molemans, W. Fiers, W. Declercq, and P. Vandenabeele. 1998. Identification of a new caspase homologue: caspase-14. Cell Death Differ. 5:838–846. [DOI] [PubMed] [Google Scholar]
- Vered, M., Y. Burstein, and A. Gertler. 1985. Digestion of elastin by porcine pancreatic elastase I and elastase II. Int. J. Pept. Protein Res. 25:76–84. [DOI] [PubMed] [Google Scholar]
- Walsh, D.S., J.L. Borke, B.B. Singh, N.N. Do, S.D. Hsu, M.V. Balagon, and R.M. Abalos. 2005. Psoriasis is characterized by altered epidermal expression of caspase 14, a novel regulator of keratinocyte terminal differentiation and barrier formation. J. Dermatol. Sci. 37:61–63. [DOI] [PubMed] [Google Scholar]
- Warner, R.R., M.C. Myers, and D.A. Taylor. 1988. Electron probe analysis of human skin: determination of the water concentration profile. J. Invest. Dermatol. 90:218–224. [DOI] [PubMed] [Google Scholar]
- Yoo, N.J., Y.H. Soung, S.H. Lee, E.G. Jeong, and S.H. Lee. 2007. Mutational analysis of caspase-14 gene in common carcinomas. Pathology. 39:330–333. [DOI] [PubMed] [Google Scholar]
- Zarach, J.M., G.M. Beaun III, P.A. Coulombe, and C.C. Thompson. 2004. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development. 131:4189–4200. [DOI] [PubMed] [Google Scholar]
- Zenz, R., R. Eferl, L. Kenner, L. Florin, L. Hummerich, D. Mehic, H. Scheuch, P. Angel, E. Tschachler, and E.F. Wagner. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 437:369–375. [DOI] [PubMed] [Google Scholar]