Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells (original) (raw)
Abstract
Characterization of the transcriptional regulatory network of the normal cell cycle is essential for understanding the perturbations that lead to cancer. However, the complete set of cycling genes in primary cells has not yet been identified. Here, we report the results of genome-wide expression profiling experiments on synchronized primary human foreskin fibroblasts across the cell cycle. Using a combined experimental and computational approach to deconvolve measured expression values into “single-cell” expression profiles, we were able to overcome the limitations inherent in synchronizing nontransformed mammalian cells. This allowed us to identify 480 periodically expressed genes in primary human foreskin fibroblasts. Analysis of the reconstructed primary cell profiles and comparison with published expression datasets from synchronized transformed cells reveals a large number of genes that cycle exclusively in primary cells. This conclusion was supported by both bioinformatic analysis and experiments performed on other cell types. We suggest that this approach will help pinpoint genetic elements contributing to normal cell growth and cellular transformation.
Keywords: deconvolution, expression profile
Tight regulation of the cell cycle is necessary for the proper growth and development of all organisms. Dysregulation of cell cycle controls leads to proliferative diseases, most notably cancer. One approach to understanding basic cell cycle processes and their deregulation in cancer has been genome-wide characterization of the cell cycle transcriptional program (1). In these microarray experiments, the RNA levels of every gene is measured in a synchronized cell population at multiple time points. Synchronization is achieved by releasing cells from a cell cycle arrest. This approach was carried out initially to characterize the yeast cell cycle, and, subsequently, it was applied to examine the cell cycle in multiple organisms (reviewed in ref. 2).
Although arrest methods were effective for characterizing cycling genes in a number of species (3–7), they did not lead to complete synchronization, even for yeast cells (8–10). A number of methods were introduced for resynchronizing yeast cells by either matching the profiles for the first and second cycle for each gene (9) or by combining expression and bud count information to reconstruct the expression profile (8). These methods were shown to improve (the already good) yeast cell cycle expression data. However, these methods cannot be directly applied to mammalian cells because of two major differences between yeast and mammalian cells: (i) normal diploid mammalian cells lose their synchronization relatively soon after release of growth arrest (11) and (ii) only 50–70% of wild-type mammalian cells reenter the cell cycle after release from arrest (12). The large percentage of arrested cells and loss of synchronization means that expression values represent a mixed population of cells, which introduces high background noise that confounds differentiation between genuine cell cycle-regulated genes and randomly fluctuating genes. This may have contributed to the problem encountered in a study that used synchronized human fibroblasts for the identification of cycling genes (13, 14).
So far, there have been few attempts to tackle this problem. CheckSum (15), a quality control method for time series expression data, can detect cases in which genes are missed because of synchronization loss. However, CheckSum cannot reconstruct the profiles of the missed genes, and so it cannot recover the cyclic expression patterns in primary cells. A different approach to overcome these difficulties was introduced by Whitfield et al. (16), who used a transformed cell line (HeLa), which is easier to synchronize, to identify cycling genes. However, these cells may not display the normal pattern of gene expression seen in nontransformed human cell types but rather reflect the proliferative nature of transformed cells in culture.
In light of these limitations, we do not have a complete gene expression dataset of the normal human cell cycle. To overcome this problem, we developed a combined experimental and computational approach that addresses the limitations of mammalian cell cycle experiments and leads to the identification of true cell cycle expression profiles from “noisy” data. Our approach uses information about the degree of synchronization of the culture and the percentage of cells reentering the cell cycle (obtained empirically by FACS analysis) to deconvolve the expression profiles of genes. Cycling genes are identified by using these corrected profiles. We reconstructed cell cycle profiles by carrying out new microarray experiments, using (partially) synchronized primary human foreskin fibroblasts. Analysis of the reconstructed profiles and comparison with published expression datasets from transformed cells reveals a large number of genes that cycle in primary cells and not in transformed cells. This list contains both known and new cell cycle genes. We anticipate that further study of the genes cycling specifically in normal cells will advance understanding of both the normal cell cycle and mechanisms leading to cancer-associated deregulation.
Results
Cell Synchronization and FACS Analysis.
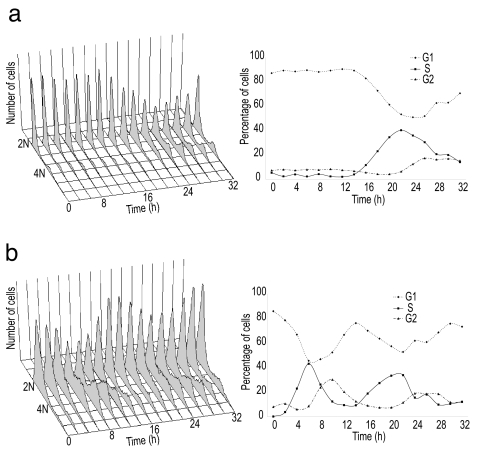
We initiated this study with the knowledge that complete synchronization of primary cells is difficult to achieve. Early passage human foreskin fibroblasts were synchronized by two methods, serum starvation and thymidine block, arresting the cells at G0/G1 and G1/S respectively. Flow cytometry (FACS) analysis shows the limited synchrony that can be achieved with these cells (Fig. 1 a and b); for example, after serum starvation, ≈50% of the cells fail to cycle and remain in the G0/G1 phase of the cell cycle. This information is used when deconvolving the expression profiles.
Fig. 1.
Cell cycle synchronization. The cell cycle distribution of cells synchronized by (a) serum starvation (one cycle) or (b) thymidine block (two cycles) was monitored by FACS. The number of cells (arbitrary units) is plotted against DNA content for time points after release. The percentage of cells in G1, S, and G2 stages of the cell cycle at each time point is shown.
In Silico Synchronization.
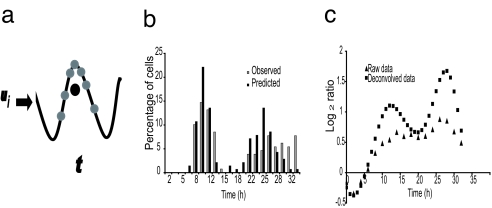
Microarray experiments measure the average RNA level of each gene in a population of cells, and thus are most accurate when using a homogenous population of cells. Partial synchronization causes a severe distortion of microarray results (14). To overcome this problem, we developed a computational approach that takes advantage of the FACS data collected at various time points during the experiment to deconvolve the expression data. The deconvolution algorithm infers gene expression values for the ideal “average single cell”; it does this by using a model learned from the empirically observed distribution of cells and measured expression values recorded at each time point (Fig. 2a). The algorithm is based on the assumption that after release from arrest, each cell proceeds according to its own internal clock. Some of the cells do not emerge from the arrested state, and the remaining cells proceed along the cell cycle at their own rate that, assuming a normal distribution, can be inferred from the FACS data. The inferred “synchronization loss model” can be applied to deconvolve the expression data to generate “single-cell” gene expression profiles. See Materials and Methods and supporting information (SI) Methods for complete details. The synchronization loss model allows estimation of the percentage of cells at each phase at a given moment. The accuracy of this estimation was confirmed by comparing the time until mitotic entry predicted by our model with direct measurements of mitosis made by using time-lapse cinematography. A very strong agreement is observed between the predicted and observed cell division times (Fig. 2b).
Fig. 2.
Data deconvolution. (a) Due to loss of synchronization, cells (gray dots) are distributed around the actual time (t). Using a synchronization loss model, this distribution can be determined. The actual measurement at time t is an average of the expression values of the gene (black dot) in all cells and is thus not an accurate representation of the single-cell expression value for this gene at time t. Using deconvolution on data from multiple time points, we can recover the underlying expression pattern for gene i (u i). (b) A diagram depicting the percentage of cells entering mitosis at each time point after release from the thymidine block as determined by time-lapse cinematography (gray) and as predicted by the synchronization loss model from the FACS data (black). Note the high correlation between the two distributions (R = 0.76, ANOVA P < 10−4). (c) Expression profile of the BIRC5 gene as measured by microarray analysis of the thymidine block experiment. Raw data (gray triangles) and deconvolved data (black squares).
Identifying Cycling Genes.
RNA was isolated from synchronized foreskin fibroblast cells at 2-h intervals after their release from serum starvation or thymidine block arrest. RNA was also isolated from unsynchronized cultures to generate a reference dataset. RNA expression levels were determined by using Affymetrix microarrays U133A 2.0. As mentioned above, measured expression values from the synchronized cultures were corrected to generate deconvolved expression profiles. The resulting profiles represent single-cell expression values for each gene allowing us to identify cycling genes that cannot be identified when relying on uncorrected measured values. For example, a well known cycling gene, BIRC5, shows only a small fluctuation in its RNA level in the raw data, whereas, after the deconvolution process, the cyclical nature of this gene is obvious (Fig. 2c).
Applying a cyclicity score to the data (Materials and Methods) allowed identification of 480 cycling genes. Three lines of evidence support our definition of cycling genes. First, most of the known cycling genes are found among these 480 genes (SI Appendix). Second, applying a gene ontology (GO) annotation analysis to the list revealed a high enrichment for cell cycle related categories such as DNA replication, DNA repair, DNA metabolism, mitosis, cell division, and cell cycle regulation (SI Table 2). Finally, we confirmed the periodic expression of 10 of the identified cell cycle genes, using RT-PCR (SI Appendix).
In addition to genes known to be cycling, the list of 480 cycling genes also includes many genes that were not identified as such in a genome-wide study focused on transformed cells (16), thus vastly expanding the view of cell cycle transcriptional regulation. Assignment of each gene to a cell cycle stage reveals that, as suggested in refs. 4, 7, and 16, the majority of cycling genes are transcribed when they are needed most during the cell cycle (SI Table 3 and SI Appendix).
Categorizing Cycling Genes.
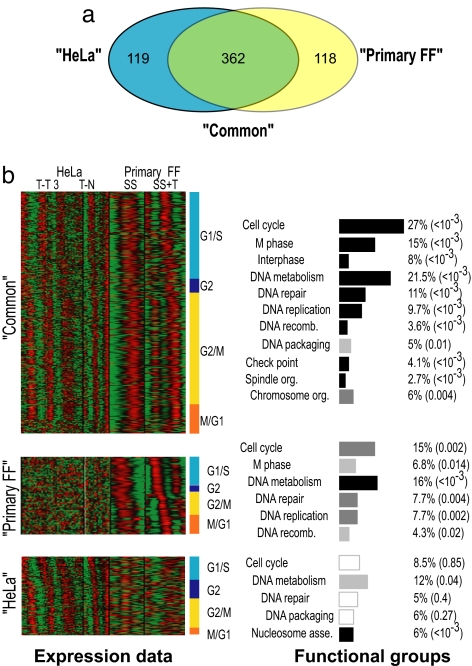
The study in ref. 16, using the cervical carcinoma cell line HeLa, identified >850 genes that show periodic expression across the cell cycle. Among these 850 genes, 550 were measured by our platform, and a significant portion of them (≈40%, P = 0) were identified as cycling in our study as well. To explore the differences between the cell cycles of HeLa and foreskin fibroblast cells as monitored by the two studies, we reanalyzed the Whitfield et al. (16) dataset, using the same criteria used in our data analysis. A gene was defined as cycling if it passed a threshold in at least two of the four datasets analyzed (two from each study; see Materials and Methods). Using randomization analysis, we determined that the false discovery rate (FDR) using this criterion was <7% (SI Appendix). This integrated analysis of both studies resulted in three groups of genes; genes cycling in both primary foreskin fibroblasts and HeLa datasets [“common” (362 genes)], genes cycling only in primary foreskin fibroblasts [“primary FF” (118 genes)], and genes cycling only in the HeLa dataset [“HeLa” (119 genes)] (Fig. 3a).
Fig. 3.
Characterization of three cycling gene groups. (a) Venn diagram depicting the overlap between cycling genes identified in Whitfield's (16) and our datasets. The numbers in each section of the diagram reflect the number of genes in each group (see SI Table 5 for a list of all of the cycling genes). (b) Expression data for each group of cycling genes (log ratio to unsynchronized culture) is represented by color, using a heat map, where red indicates induced expression and green indicates repressed expression. For the HeLa cells, we used data published in ref. 16 of the Thy-Thy 3 (T-T3) and Thy-Noc (T-N) experiments. For the primary FF cells, we used the deconvolved expression data for the serum starvation (SS) and thymidine block (T) experiments. The genes within a group have been ordered (vertically) according to their assigned cell cycle stage, which is indicated on the right of the heat maps. Enrichment analysis of each group's members for functional cell cycle GO categories is represented by rectangles next to that group (for the full analysis, see SI Table 2). The length of a rectangle depicts the percentage of cycling genes that fall into the category (2.7–27%), and the color depicts the significance of the enrichment. The significance levels (in parentheses) were corrected for multiple hypotheses (see Materials and Methods) and are indicated by three levels of gray color for P < 0.05, <0.005 and <0.001. White rectangles indicate no enrichment (_P_ > 0.05).
Characterizing Cycling Gene Groups.
To distinguish potential differences unique to each of the three gene groups, we analyzed their members for GO annotation and transcription factor binding motifs. As expected, the common set was highly enriched for the major cell cycle categories. Similar enrichment, although to a lesser extent, was found in the primary FF set, whereas there was no enrichment in the HeLa set of genes (Fig. 3b). A similar pattern was observed for binding sites of known cell cycle regulators (17). The common and primary FF sets of genes were enriched for the binding sites of E2F transcription factor (E2F) (22%; P < 10−12 and P = 0.01, respectively), nuclear factor Y (NFY) (39%; P < 10−19 and P = 0.02, respectively), and nuclear respiratory factor 1 (NRF1) (36%; P < 0.01 common only). In contrast, the HeLa set of genes were not enriched for motifs of cell cycle regulators. Furthermore, analysis of each group's members, using ChIP on chip data for the transcription factors E2F4, p130, p107 (18), and NFY-B (19), revealed similar findings. Although high percentages of the cycling genes in both common and primary FF datasets are bound by at least one of these factors (27%; P < 10−63 and 24%; P < 10−15, respectively), only a small portion (5%; P = 0.22) of the genes identified in the HeLa set are bound by these factors.
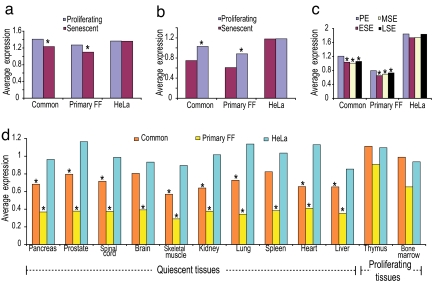
Many cell cycle genes (such as DNA replication genes) are expressed only in proliferating cells, and therefore it is expected that the average expression of cycling genes should be higher in proliferating cells than in arrested cells. To characterize further the cycling gene groups, we used published expression profiles of proliferating and arrested primary fibroblasts (IMR-90) to compare expression levels of the three groups of cycling genes (20). We found that the average expression level of genes in the common and the primary FF groups is significantly higher in primary proliferating cells than in arrested cells (P < 10−59 and P < 10−18, respectively). In contrast, the average expression level of genes in the HeLa group is not significantly higher in proliferating versus arrested cells (P = 0.47; Fig. 4a). Similar results (common, P < 10−20; primary FF, P < 10−7; and HeLa, P = 0.49), were obtained in a reciprocal experiment in which oncogene-induced senescence was bypassed by the transfection of E6/E7 viral proteins (21) (Fig. 4b). The same pattern was also observed in data derived from normal epithelial cells (Fig. 4c and SI Fig. 6). This suggests that the origin of the cells used for the identification of cycling genes (epithelial versus fibroblastic) is not the main cause for the different set of genes identified in each experiment.
Fig. 4.
Expression in normal cells. The average gene expression level for each of the three groups in proliferating and arrested cells is shown. The asterisks designate cases with a significant difference (P < 0.05; paired t test) between proliferating and arrested cells. (a and b) Data of proliferating (blue) and senescent (red) IMR-90 cells from an oncogene-induced senescence experiment (20) (a) and a senescence bypass experiment (21) (b). (c) Data of endometrium tissue (42) in the proliferative (blue) and senescent early, middle, and late secretory (red, yellow, and black, respectively) phases. (d) Average expression levels in a variety of normal tissues (22) for genes in the common (orange), primary FF (yellow) and HeLa (blue) groups. The asterisks designate a significant difference (P < 0.05; t test) between the expression of the genes within a group and the expression of all genes measured in that tissue. Note the different expression pattern of quiescent and proliferating tissues. Similar results were obtained by using an additional dataset (SI Fig. 8).
Further support for these results was obtained from the analysis of the average expression level of genes, using a panel of 12 normal tissues (22). For each tissue, we compared the average expression level of each set of cycling genes to the average expression level of all genes on the array. We found that the expression levels of cycling genes in the common and primary FF groups are significantly low in most tissues and are expressed at considerably higher levels only in the thymus and bone marrow samples, which are the only two tissues in our analysis that contain a high percentage of proliferating cells. In sharp contrast, the genes of the HeLa group do not exhibit this distinctive pattern of tissue expression (Fig. 4d). These results are consistent with the conclusion that our method accurately identifies cycling genes in primary cells.
The Primary FF Group Contains Cycling Genes Unique to Normal Cells.
It is possible that some of the genes identified as cycling in our dataset were not identified as such in the Whitfield study (16), owing to differences in experimental procedures. However, we propose that some of these genes are cycling only in normal cells and not in transformed cells. We explored this supposition as follows. Recent analyses of cancer expression profiles have facilitated classification of genes whose expression correlates with proliferation rates (“tumor proliferation clusters”) (23, 24). These clusters were shown to be highly enriched for genes expressed periodically in HeLa cells (16). Similar analysis of our groups of cycling genes reveals that the common group is significantly more populated with genes from the tumor proliferating clusters than the primary FF group (P = 0.007, Fisher's exact test). In sharp contrast, when comparing our groups of genes to a normal proliferation cluster (25), there is no difference in enrichment between the primary FF and common group (P = 0.5, Fisher's exact test).
To further investigate this, we analyzed published expression data from various normal and transformed cells. We compared the average gene expression levels of each of our gene groups between normal fibroblasts and cancer cells of fibroblast origin (fibrosarcoma) (26). The gene expression profiles of the common and the primary FF groups differ strikingly from one another. Whereas the genes from the common group are expressed at significantly higher levels in cancer cells than in normal cells (P < 0.0007), the expression of genes from the primary FF group is the same in both cell types (P = 0.45; Fig. 5a). The same result was obtained when using a large dataset of normal and transformed tissues (27). Notably, when considering normal cells, the primary FF genes and the common genes have similar expression behavior. In contrast, in cancer cells these gene groups diverge significantly in their expression patterns (P < 0.0002; Fig. 5b). Analysis of additional datasets from normal tissues, cancer tissues, and transformed cell lines reveals similar results (SI Figs. 6–8). These analyses support our premise that the primary FF group contains some genes that are cycling exclusively in normal cells and are dysregulated in transformed cells.
Fig. 5.
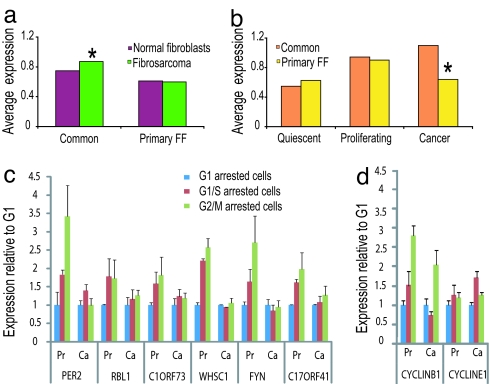
Expression in cancer tissues. (a) The average gene expression level is diagrammed for the common and primary FF groups in normal arrested IMR-90 primary fibroblasts (red, the same data as in Fig. 4b) and fibrosarcoma (26) (green). Significant (P < 0.0007; paired t test) difference between the normal and the cancer samples was observed only for the common group. (b) Expression data from a variety of tissues and cell lines (27) were averaged according to three categories—normal quiescent samples (such as lung, liver, and heart), normal proliferating samples (such as testis, thymus, and bone marrow), and cancer samples. The average expression of genes in the common (orange) and primary FF (yellow) groups is shown. Note that a significant difference (P < 0.0002; t test) between the expression behaviors of the two groups is observed only when considering cancer cells. (c and d) RNA levels of genes from the primary FF (c) and common (d) groups were measured in normal primary endothelial cells, HUVECs (Pr), and a fibrosarcoma cancer cell line, HT1080 (Ca), by semiquantitative RT-PCR at several cell cycle stages (SI Appendix). The level of expression at G1/S and G2/M for each gene is presented relative to its expression at G1 after normalizing for GAPDH. The averages and the standard deviations (error bars) of duplicate measurements are shown. The predicted peak of expression of each gene is G1/S for RBL1, C1ORF73, and CYCLINE1; G2 for FYN; G2/M for PER2, WHSC1, and C17ORF41; and M for CYCLINB1.
To determine whether our conclusion that primary FF genes are only cycling in normal cells is relevant to other cell types, we have measured the RNA level of several such genes in another type of primary cells [human umbilical vein endothelial cells (HUVEC)] and in a fibrosarcoma cell line (HT1080). We observed a sharp difference between the two types of cells, whereas in the primary cells, the RNA levels differ at different cell cycle stages, in the cancer cells, only minor changes were observed (Fig. 5 c and d).
Discussion
To date, technical challenges underlying synchronization of mammalian cells have hampered an accurate description of cell cycle-regulated genes in normal human tissue. We have developed a computational method that overcomes synchrony difficulties in silico and demonstrate its utility by generating a comprehensive list of 480 genes that show periodic expression across the human cell cycle of primary foreskin fibroblasts. Comparison with a previous dataset identified a list of 118 genes that were detected to be cycling only in normal fibroblasts. This list contains several genes that are dysregulated in transformed cells.
Reanalysis of the HeLa cell cycle data (16), using the same criteria applied to our data, allowed us to perform a comparison between the cycling genes identified in each experiment. Cycling genes were categorized into three groups, common, primary FF, and HeLa (Fig. 3), and we used various published datasets to investigate the characteristics of these three cycling gene groups (Figs. 3 and 4). We concluded from these analyses that the common and the primary FF categories contain “genuine” cell cycle genes, whereas the genes identified as cycling only in Whitfield's data (16) (the HeLa group) most likely are not cycling. Thus, it seems that an additional benefit of our analysis is the elimination from the published list of cycling genes many genes whose cyclicity may be due to other cell perturbations and not due to the normal cell cycle.
Further support for our claim that our approach is successful in improving the identification of cycling genes comes from analysis of interspecies conservation. A recent comparison of cell cycle experiments in budding yeast, fission yeast, and human identified a conserved core set of cycling genes (28). Analysis of the conservation in the common set revealed that it is far more enriched for core cycling genes than the original Whitfield list (16) (SI Appendix).
Why were the primary FF genes not identified as cycling in the HeLa dataset? The causes may be differences between the studies, such as different synchronization methods, microarray platforms, and technical variations between laboratories. However, measuring RNA levels of several genes in other normal and cancer cell types (Fig. 5) suggests that this is not the case. Moreover, analysis of complementary high-throughput data suggests that some of these genes are likely to have periodic expression only in normal cells and not in transformed cells, such as HeLa. Explicitly, analysis of a large variety of expression profile datasets revealed significant differences between cancer and normal tissues. In normal tissues, the average expression of genes in the common and the primary FF groups showed a similar pattern (Fig. 4). In contrast, in cancer samples, the two groups differ—the common genes were expressed to a level higher than the primary FF genes (Fig. 5 and SI Figs. 6 and 7). This suggests that some of the primary FF genes are cycling only in a normal cell cycle and are dysregulated in transformed cells. This conclusion is further supported because only a small portion of the primary FF genes are found in the previously defined “cancer proliferating clusters” (23, 24) but are found in the normal proliferation cluster (25).
Cellular transformation is a complex process that causes the perturbation of many genes. It is likely that some of the genes identified in this study as dysregulated in transformed cells exhibit abnormal expression as a consequence of this process. However, some of the primary FF genes may actually play causal roles in the transformation process. Close examination of the biological processes of some of these genes reveals their involvement in processes intimately associated with cancer transformation (see Table 1 for a selected list and SI Appendix for more details).
Table 1.
Cycling genes
| Gene | Phase | Function | Role in cancer, ref(s). |
|---|---|---|---|
| FANCL | G1/S | DNA repair | 29 |
| MRE11A | G2/M | DNA repair | 29 |
| BLM | G1/S | DNA repair | 29 |
| FYN* | G2 | Oncogene | 30 |
| BTG1 | G2 | Antiproliferation | 31 |
| DLEU2 | G2/M | Putative tumor suppressor | 32 |
| ING2 | G1/S | Chromatin | 33 |
| HOXA9 | M/G1 | Transcription | 34, 35 |
| PER2* | G2/M | Circadian clock | 36 |
| WHSC1* | G2/M | Chromatin | 37 |
| RBL1* | G1/S | Cell cycle | 38 |
High-throughput studies have made significant progress in assigning new genes to the cell cycle process. By comparing datasets derived from normal and transformed cells, we can gain new insights into the profound differences between these cellular stages. Using a computational and experimental approach, we were able to obtain high-quality data in primary cells, which led to the identification of genes that may be dysregulated because of cancer transformation. Additional studies of this relatively small set of genes may lead to further characterization of their potential role in the transformation process.
Materials and Methods
Cell Culture and Synchronization.
Early passage human foreskin fibroblasts were grown in DMEM with 10% FCS. For G0/G1 synchronization, cells were arrested with 0.5% FCS (48 h) and then released in 10% FCS. For G1/S synchronization, cells were released from the G0/G1 arrest in the presence of 2 mM thymidine for 24 h, washing the thymidine released the cells. At the time of release and at intervals of 2 h for the next 32 h, RNA was prepared from cells, using RNeasy mini-kit (Qiagen). Synchrony was monitored by FACS of propidium iodide-stained cells and BrdU (50 μM; 1.5 h) incorporation.
Time-Lapse Cinematography.
Cells released from synchronization were photographed every 10 min for ≈36 h. Upon visualization of the data, the timing of 129 mitotic events for the thymidine block release and 74 mitotic events for the serum starvation release were manually recorded.
Microarray.
RNA was reverse transcribed, labeled, and hybridized to Affymetrix microarrays U133A 2.0. The microarray data of each time course was separately analyzed by using the AMARGE suite (39).
Synchronization Loss Model.
The model assumes that cells are not completely synchronized for two primary reasons: (i) a fraction does not reenter cell cycle, and (ii) even for those that do reenter, different cells may progress at different rates resulting in longer or shorter division durations. The model assumes that cell progress rates are distributed as a Gaussian with a mean of 1 (average time). The model has five parameters: percentage of cells reentering cell cycle, three parameters for duration of the three FACS measured phases (G1, S and G2/M), and a parameter for the variance of the progress rate Gaussian. Using FACS, we learned the parameters of the model. These parameters are used for the correction and for the deconvolution discussed below. The parameters learned for our model were further validated by using time-lapse cinematography. See SI Methods for details.
Correcting for Partial Reentry.
After normalization, expression data were corrected for the percentage of cells reentering cell cycle. This percentage was determined in our model, using FACS as discussed above. Let _Y_0 be the measured expression of a gene in time point 0 (before release, G0) and Y t be the measured expression in time point t. If the fraction of cells entering the cell cycle is P, then the measured value at time t is from a mixture of cells, P of which are cycling and the rest are not. This can be formally stated as follows: Y t = PC t + (1 − P)_Y_0.
From this equation, we can derive the expression value for the cycling cells, Ct, which is used in subsequent analysis. We next computed log ratios, using duplicate measurements of unsynchronized populations.
Deconvolving Expression Data.
Using the learned synchronization loss model, the corrected expression data were deconvolved. The goal of a deconvolution algorithm is to obtain the actual expression value for each time point from measurements of cells that are distributed around that time point. The deconvolution method uses continuous representation to determine the underlying expression values. See Fig. 2a and SI Appendix for more details and a discussion of relative peak heights.
Scoring Deconvolved Expression Profiles.
The resulting expression profiles were scored by using Fourier transform (4). To determine a score cutoff, we randomly permuted both datasets and repeated the above steps for each of these (random) datasets. Similarly, we have randomized two datasets from ref. 16 (the T-T3 and the T-N datasets) that showed high levels of synchronization (40). Using scores from the randomized datasets, we have determined a cutoff score. A gene was included in the resulting cycling lists (common, primary FF, and HeLa) if it passed this score for at least two of the datasets. Note that the primary FF group also contains 35 genes that were not measured in the Whitfield experiment (16). Based on the randomization analysis, the false discovery rate was 1% for genes identified as primary FF or HeLa and 6% for genes identified as common. See SI Appendix for a detailed discussion addressing (i) potential synchronization artifacts, (ii) the specificity and sensitivity of the deconvolution method, and (iii) the improvement in data analysis achieved by the deconvolution step.
Phase Assignment.
Genes were assigned to phases by computing their correlation with previously annotated cell cycle genes, as described in ref. 16. Six genes from a list of known cell cycle genes (16) were used (SI Table 4). We computed the correlation of each of the predicted cycling genes and the averages of the known phase genes and assigned the gene to the phase with the highest correlation. This process was repeated for both datasets. For 56% of the genes, the two datasets agreed on the phase assignment. For the majority of the rest, they were assigned to two consecutive phases (such as to G2/M in one and M/G1 in the second). In such cases, we used the assignment from the dataset in which this gene scored higher.
Expression Data Analysis.
Data were downloaded from Gene Expression Omnibus database (accession nos. GSE2487, GSE4888, GDS426, GDS1209, and GDS181) or obtained from the researchers (20). Each experiment was normalized by the average expression in the experiment. Multiple experiments of the same type were combined. Significant differences between groups were assessed by t test. The tumor proliferation clusters of breast (23) and lymphomas (24) were combined and the overlap with the common and primary FF groups was determined. The rank statistics is described in the SI Appendix.
Statistical Analysis of GO and Motif Enrichment.
GO annotation enrichment was calculated by using the STEM program (41), and the reported hypergeometric P values were corrected for multiple hypothesis, using randomization. Motifs enrichment was determined by the PRIMA software (17).
Supplementary Material
Supporting Information
ACKNOWLEDGMENTS.
We thank Shlomit Farkash-Amar for statistical analysis. This work was supported by grants from the Association for International Cancer Research, European Commission Sixth Framework Programme Contract 503576, and the Binational Science Foundation (to I.S.); U.S. National Science Foundation Career Award 0448453 and the Tobacco Settlement Grant from the Pennsylvania Department of Health (to Z.B.-J.); National Institutes of Health Grant CA077245-11 (to B.D.D.); and the German Federal Ministry of Research and Education through the National Genome Research Network Grant 01 GR 0450 (to B.B. and R.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.M. is a guest editor invited by the Editorial Board.
Data deposition: Expression profiles have been deposited in the European Bioinformatics Institute ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. E-\E TABM-263).
References
- 1.Whitfield ML, George LK, Grant GD, Perou CM. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 2.Cooper S, Shedden K. Cell Chromosome. 2003;2:1. doi: 10.1186/1475-9268-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 4.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng X, Karuturi RK, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, et al. Mol Biol Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Joseph Z, Farkash S, Gifford DK, Simon I, Rosenfeld R. Bioinformatics. 2004;20(Suppl 1):i23–i30. doi: 10.1093/bioinformatics/bth915. [DOI] [PubMed] [Google Scholar]
- 9.Lu X, Zhang W, Qin ZS, Kwast KE, Liu JS. Nucleic Acids Res. 2004;32:447–455. doi: 10.1093/nar/gkh205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu P, Jane Wang Z, Ray Liu KJ. Conf Proc IEEE Eng Med Biol Soc. 2005;5:4826–4829. doi: 10.1109/IEMBS.2005.1615552. [DOI] [PubMed] [Google Scholar]
- 11.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 12.Tobey RA, Valdez JG, Crissman HA. Exp Cell Res. 1988;179:400–416. doi: 10.1016/0014-4827(88)90279-0. [DOI] [PubMed] [Google Scholar]
- 13.Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 14.Shedden K, Cooper S. Proc Natl Acad Sci USA. 2002;99:4379–4384. doi: 10.1073/pnas.062569899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon I, Siegfried Z, Ernst J, Bar-Joseph Z. Nat Biotechnol. 2005;23:1503–1508. doi: 10.1038/nbt1164. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 19.Ceribelli M, Alcalay M, Vigano MA, Mantovani R. Cell Cycle. 2006;5:1102–1110. doi: 10.4161/cc.5.10.2777. [DOI] [PubMed] [Google Scholar]
- 20.Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 22.Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 23.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh AA, Staudt LM. Curr Opin Immunol. 2000;12:219–225. doi: 10.1016/s0952-7915(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 25.Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, Mantovani R, Domany E, Rotter V, Pilpel Y. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100030. 0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Detwiller KY, Fernando NT, Segal NH, Ryeom SW, D'Amore PA, Yoon SS. Cancer Res. 2005;65:5881–5889. doi: 10.1158/0008-5472.CAN-04-4078. [DOI] [PubMed] [Google Scholar]
- 27.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Mahony S, Benos PV, Rosenfeld R, Simon I, Breeden LL, Bar-Joseph Z. Genome Biol. 2007;8:R146. doi: 10.1186/gb-2007-8-7-r146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyakhovich A, Surralles J. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 30.Takayama T, Mogi Y, Kogawa K, Yoshizaki N, Muramatsu H, Koike K, Semba K, Yamamoto T, Niitsu Y. Int J Cancer. 1993;54:875–879. doi: 10.1002/ijc.2910540525. [DOI] [PubMed] [Google Scholar]
- 31.Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, Iyengar A, Baranova A, Kashuba V, Merup M, et al. Oncogene. 1997;15:2463–2473. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 33.Sironi E, Cerri A, Tomasini D, Sirchia SM, Porta G, Rossella F, Grati FR, Simoni G. J Cutan Pathol. 2004;31:318–322. doi: 10.1111/j.0303-6987.2004.0187.x. [DOI] [PubMed] [Google Scholar]
- 34.Borrow J, Shearman AM, Stanton VP, Jr, Becher R, Collins T, Williams AJ, Dube I, Katz F, Kwong YL, Morris C, et al. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 35.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 36.Fu L, Pelicano H, Liu J, Huang P, Lee C. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 37.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, Larratt LM, Mant MJ, Reiman T, Belch AR, et al. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewen ME, Xing YG, Lawrence JB, Livingston DM. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 39.Lozano JJ, Kalko SG. Appl Bioinformatics. 2006;5:45–47. doi: 10.2165/00822942-200605010-00006. [DOI] [PubMed] [Google Scholar]
- 40.Wichert S, Fokianos K, Strimmer K. Bioinformatics. 2004;20:5–20. doi: 10.1093/bioinformatics/btg364. [DOI] [PubMed] [Google Scholar]
- 41.Ernst J, Bar-Joseph Z. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA, et al. Endocrinology. 2006;147:1097–1121. doi: 10.1210/en.2005-1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information