Ma-LMM01 Infecting Toxic Microcystis aeruginosa Illuminates Diverse Cyanophage Genome Strategies (original) (raw)
Abstract
Cyanobacteria and their phages are significant microbial components of the freshwater and marine environments. We identified a lytic phage, Ma-LMM01, infecting Microcystis aeruginosa, a cyanobacterium that forms toxic blooms on the surfaces of freshwater lakes. Here, we describe the first sequenced freshwater cyanomyovirus genome of Ma-LMM01. The linear, circularly permuted, and terminally redundant genome has 162,109 bp and contains 184 predicted protein-coding genes and two tRNA genes. The genome exhibits no colinearity with previously sequenced genomes of cyanomyoviruses or other Myoviridae. The majority of the predicted genes have no detectable homologues in the databases. These findings indicate that Ma-LMM01 is a member of a new lineage of the Myoviridae family. The genome lacks homologues for the photosynthetic genes that are prevalent in marine cyanophages. However, it has a homologue of nblA, which is essential for the degradation of the major cyanobacteria light-harvesting complex, the phycobilisomes. The genome codes for a site-specific recombinase and two prophage antirepressors, suggesting that it has the capacity to integrate into the host genome. Ma-LMM01 possesses six genes, including three coding for transposases, that are highly similar to homologues found in cyanobacteria, suggesting that recent gene transfers have occurred between Ma-LMM01 and its host. We propose that the Ma-LMM01 NblA homologue possibly reduces the absorption of excess light energy and confers benefits to the phage living in surface waters. This phage genome study suggests that light is central in the phage-cyanobacterium relationships where the viruses use diverse genetic strategies to control their host's photosynthesis.
The cyanobacterium Microcystis aeruginosa is a toxic, bloom-forming bacteria found in eutrophic freshwaters throughout the world (12). The bacterium produces potent hepatotoxins, cyclic peptides called “microcystins,” which inhibit eukaryotic protein phosphatase types 1 and 2A and can cause hepatocellular carcinoma (42, 53, 82). Blooms of M. aeruginosa can lead to the deaths of livestock and humans and pose serious problems for water management (12, 13).
The mechanisms controlling bloom initiation and termination remain unclear; however, there have been many studies concerning the effects of environmental factors on M. aeruginosa growth (57). Recently, viral mortality of algae was recognized as one of the factors involved in the termination of algal blooms, including M. aeruginosa blooms (10, 52, 71, 74, 75). We previously reported culturing the lytic cyanophage Ma-LMM01 infecting the toxic M. aeruginosa strain NIES298 (81). Currently, M. aeruginosa NIES298 and Ma-LMM01 are the sole culture host/virus system to study interactions between a toxic cyanobacterium and its phage.
Ma-LMM01 is a member of the Myoviridae family with a contractile tail (81), the distinctive morphological feature of this viral family. Myoviridae has at least six subgroups: T4-, P1-, P2-, Mu-, SPO1-, and φH-like phages (23). These subgroups show large genomic diversity, and a recent genome-based phage classification study shows no evidence for grouping of these myovirus subgroups (64). Further, there are many unclassified myoviruses with complete genome sequence data that suggest that myovirus genomes are more diverse despite their shared morphological features (56).
In marine systems, cyanobacteria contribute significantly to global photosynthesis. Recently, the complete genome sequences for seven cyanophages infecting marine cyanobacteria were published (14, 44, 62, 69, 78). These are four cyanomyoviruses (three T4-like [P-SSM2, P-SSM4, and S-PM2] and one unclassified virus [Syn9]) and three cyanopodoviruses (P-SSP7, Syn5, and P60). Interestingly these cyanophage genomes contain various photosynthesis-related genes from cyanobacteria (38, 44, 45, 69, 70). The psbA gene, coding for the photosystem II core reaction center protein D1, was found in all 32 cultured marine cyanomyovirus strains tested by Sullivan et al. (70). The expression of the phage genome-encoded D1 protein appears to maintain photosynthesis during cyanophage infection, thus increasing phage reproduction through the prolongation of the host's energy production (15, 37, 45, 47). Marine cyanobacteria and their cyanophages thus provide excellent models to study coevolutionary processes of viruses and their hosts (15, 16, 37, 38).
Phages are the primary biological entity in both freshwater and marine environments (4, 71). Interestingly, 60 to 80% of the sequences from culture-independent viral metagenomic studies lack significant similarities to known genes. This suggests a large genetic diversity among viruses (71). Therefore, to study this genetic diversity, more phage genomes will need to be sequenced.
Here we describe the first complete genome sequence of a freshwater cyanomyovirus (i.e., Ma-LMM01). Its gene content differs significantly from those of other known phages and provides important clues toward a better understanding of the ecological and evolutionary relationship between the cyanophage and its host.
MATERIALS AND METHODS
Phage propagation and purification.
Phage propagation and purification were performed as previously described (81). Briefly, exponentially growing cultures of M. aeruginosa (1 liter) were inoculated in CB medium (32) at a multiplicity of infection of approximately 0.1 and incubated at 30°C for 4 days using a 12:12 h light:dark cycle of ca. 40 μmol photons m−2 s−1 under cool white fluorescent illumination (FL40SS; Toshiba Co., Ltd.). The virus lysate was mixed with 20 ml chloroform and 40 g NaCl and filtered through a 1-μm PTFE membrane filter (Advantec Co., Ltd). The phage particles were precipitated using 10% (wt/vol) polyethylene glycol 6000 (Nacalai Tesque Co., Ltd.) and resuspended in 5 ml SM buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgSO4·7H2O, and 0.01% gelatin). The particles were then purified using ultracentrifugation with a CsCl step gradient.
Genome isolation and sequencing.
Isolation and sequencing of the Ma-LMM01 genome were performed as previously described (81). Briefly, phage DNA was isolated using the CsCl-purified virions with proteinase K digestion and the phenol-chloroform method. The genomic DNA was digested with HincII or physically sheared using a Hydroshear (Genomic Solutions, Ltd., Cambridgeshire, United Kingdom). The DNA fragments were cloned and sequenced. Sequencing was performed using the dideoxy method with a 3730xl DNA analyzer (Applied Biosystems). Genome fragment sequences were assembled using Phred/Phrap software (20, 21).
Bioinformatics analysis.
The protein-coding genes were predicted using GeneMark (5) and Glimmer (18). Homology searching was performed using BLAST/RPS-BLAST (1) against the UniProt sequence database (76) and the NCBI/CDD database (79). Gene function prediction was performed using the GTOP database (33). The tRNA genes were identified using tRNAscan-SE (41). Predictions of transmembrane proteins and signal peptides were performed using Phobius (30). Multiple-sequence alignments were generated using the GENETYX-WIN program (version 7; Genetyx Co., Tokyo, Japan). A circular genome map was drawn using CGView (68).
Structure of the phage genome.
To determine if the Ma-LMM01 genome was circular, a timed exonuclease Bal31 digest of the phage genome was performed (39). Purified Ma-LMM01 DNA was incubated with Bal31 (Toyobo Co., Ltd.) (0.1 U μl−1) at 30°C for 0, 20, and 40 min according to the manufacturer's recommendations. After ethanol precipitation, the DNA was digested with HindIII (Nippon Gene Co., Ltd.) (0.5 U μl−1) at 37°C for 16 h according to the manufacturer's instructions and electrophoresed in 1% (wt/vol) agarose S (Nippon Gene Co., Ltd.). Following electrophoresis, nucleic acids were visualized after staining for 1 h with SYBR gold (Molecular Probes, Inc., Eugene, OR).
Transcriptional analysis of an intervening DNA region in Ma-LMM01.
To examine for the presence of an intron in the intervening sequence between the DNA polymerase (ORF178) and 3′-5′exonuclease (ORF180) genes (including ORF179), we performed reverse transcription-PCR (RT-PCR) targeting a 671-bp sequence using the following primer pair: 3′ end of ORF180, 5′-GTTAGACCCTCAGGCGATAG, and 5′ end of ORF178, 5′-TAACCTGCAGCAGAAGACAC. Total RNA was isolated from 1 ml of infected Microcystis cells as previously described (80). Briefly, cells were collected by centrifugation and treated with 5% sodium dodecyl sulfate, 1 ml of Sepazol RNAI (Nacalai, Japan) was added, and the RNA was purified using 200 μl of chloroform, followed by two additional phenol and chloroform-isoamylalcohol (49:1) purifications. The RNA was precipitated using 1 volume of isopropanol and was washed with 70% ethanol. The pellet was resuspended in diethyl pyrocarbonate-treated water. After digestion with DNase I, cDNA was synthesized using random primers with the SuperScriptIII first-strand synthesis system (Invitrogen). PCR using the above primer pair was performed on the cDNA and genomic DNA. As a negative control for RT-PCR, PCR was performed with the DNase-treated RNA as the template.
Nucleotide sequence accession number.
The entire genome sequence of Ma-LMM01 has been deposited in the DDBJ database under accession number AB231700.
RESULTS AND DISCUSSION
Genome structure.
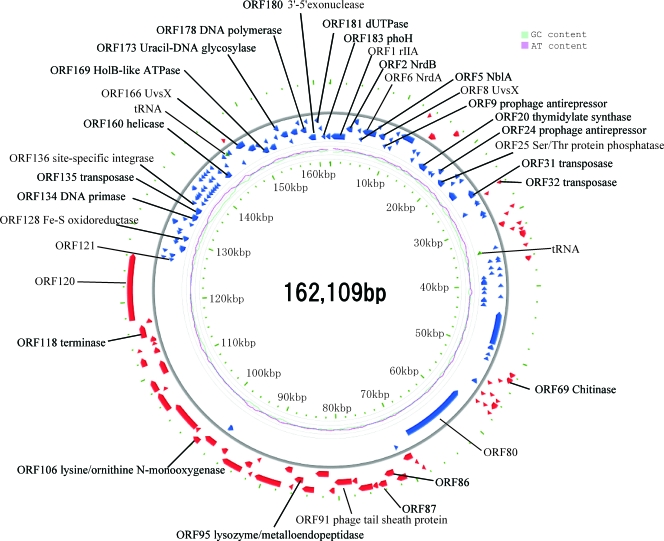
The genomic fragment sequences of Ma-LMM01 were assembled into a circular 162,109-bp sequence in which no physical ends were detected during sequencing (Fig. 1). The Ma-LMM01 phage particles contain a linear double-stranded DNA of about 165 kb (81), indicating that the genome has a terminal redundancy of about 3 kb. When the Ma-LMM01 genomic DNA was Bal31 exonuclease digested followed by a complete digestion using HindIII as described by Loessner et al. (39), simultaneous degradation of all fragments occurred (data not shown), indicating that the packaged genomic DNA is circularly permuted. The average G+C content was 46.0%, which is slightly higher than that of the M. aeruginosa strains (42.1 to 42.8%) (35, 59). A 7.5-kb region (from ORF135 to ORF158) showed a notably lower G+C composition (35.6%) than the rest of the genome (Fig. 1). This region contains a transposase gene (ORF135) and a putative site-specific recombinase gene (ORF136) (described below). We found that part of this genomic region (a 4.7-kb region containing ORF137 to ORF157) contains repeated sequences, which are variable in size (∼10 to 100 bp) and rich in several oligonucleotides (e.g., ATCTTCAT, ATAATAAA, TAATAGGT or their variants).
FIG. 1.
Ma-LMM01 genome organization. Red and blue arrows indicate putative ORFs. Pale blue and pink lines inside the circle show G+C and A+T contents, respectively.
Abundance of novel genes in the Ma-LMM01 genome.
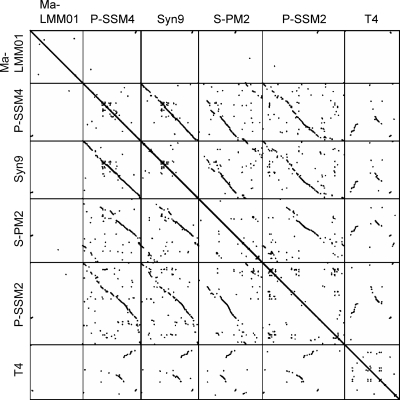
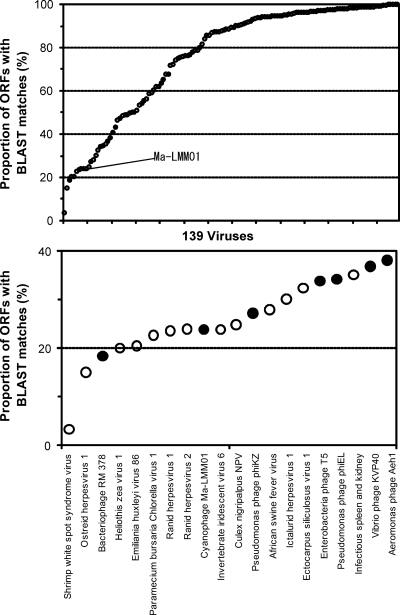
Comparing the Ma-LMM01 genome to previously sequenced genomes of cyanomyoviruses or other Myoviridae, we found no continuous colinearity (Fig. 2). We identified 184 putative protein-coding genes (open reading frames [ORFs]) and two tRNA genes. Eight ORFs were predicted to code for transmembrane proteins. With the exception of ORF9, a prophage antirepressor, all other ORFs coding for predicted transmembrane proteins were of unknown function. ORF120 and ORF57 showed 10 and nine predicted transmembrane helices, respectively. The remaining six ORFs showed one or two predicted membrane-spanning helices. We also found 10 additional ORFs of unknown function having a predicted signal peptide sequence. The proportion of the predicted transmembrane proteins (4.3%) or proteins with signal peptides (5.4%) is comparable to those of other cyanomyoviruses or phage T4 (5 to 11% for predicted transmembrane proteins and 3 to 7% for proteins for signal peptides). A BLAST search (using an E value threshold of 10−5) found database homologues for only 44 (24%) of the 184 ORFs, leaving 140 ORFs with no detectable homologues in the databases. Such a large proportion of the sequence having “novel” ORFs is among the highest for viruses with sequenced genomes larger than 100 kbp (using the same E value threshold) (Fig. 3). Two unclassified myoviruses (Rhodothermus phage RM378 and Pseudomonas phage φKZ) show a similar proportion of novel genes. Only four Ma-LMM01 ORFs (ORF1, ORF41, ORF108, and ORF109) had their best database sequence homologues in other viruses. Based on these features, we propose that Ma-LMM01 represents a member of a new subgroup of the Myoviridae family. We assigned putative functions for 28 ORFs (Table 1).
FIG. 2.
Genomic dot plots for five cyanomyoviruses and T4. Dots correspond to high-scoring segments (>150 bp in length) detected by TBLASTX (i.e., translated query against translated target) with an E value threshold of 10−5.
FIG. 3.
Proportions of ORFs with database homologues for large DNA viruses. (Top) A total of 139 viruses with genomes larger than 100 kbp. Filled and open circles correspond to 28 phages and 111 eukaryotic viruses, respectively. (Bottom) Magnified view for the viruses with the smallest proportions of ORFs with homologues. For the identification of homologues, we used a BLAST E value of 10−5, excluding self matches. Note that for viral genomes with closely related genomes represented in the databases, the proportions of ORFs with database homologues become large.
TABLE 1.
Putative ORFs of the Ma-LMM01 genome with homologues in the databases
| ORF no. | Position | Product size (amino acids) | Predicted function | Significant matchb | E value |
|---|---|---|---|---|---|
| 1a | 9-2417 | 803 | rIIA-like protein | rIIA of Aeromonas phage25 | 4E−22 |
| 2a | 2449-3495 | 349 | Ribonucleoside-diphosphate reductase beta subunit | Slr0591 protein of Synechocystis sp. strain PCC 6803 | 7E−91 |
| 5a | 4899-5138 | 80 | PBS degradation protein NblA | PBS degradation protein of Anabaena sp. strain PCC 7120 | 2E−04 |
| 6a | 5113-7341 | 743 | Ribonucleoside-diphosphate reductase alpha subunit | Ribonucleoside-diphosphate reductase alpha subunit of Synechocystis sp. strain PCC 6803 | 0 |
| 8a | 7777-8841 | 355 | UvsX (RecA-like recombinase) | RecA protein 1 of Myxococcus xanthus | 2E−46 |
| 9a | 8925-9413 | 163 | Prophage antirepressor | Similar to bacteriophage antirepressor protein of Photorhabdus luminescens | 0.001 |
| 14a | 11048-13135 | 696 | Unknown | SAP DNA-binding domain-containing protein of Dictyostelium discoideum | 3E−07 |
| 19 | 14713-15717 | 335 | Unknown | ORF19 of Xestia c-nigrum granulovirus | 4E−21 |
| 20a | 15717-17192 | 492 | Putative thymidylate synthase ThyX | Alternative thymidylate synthase-like of synthase Acidobacteria bacterium (strain Ellin345) | 7E−27 |
| 22a | 17967-18128 | 34 | Unknown | Hypothetical protein XCC2823 of Xanthomonas campestris (pv. campestris) | 1E−04 |
| 24a | 18772-19584 | 271 | Prophage antirepressor | Putative antirepressor of Streptococcus pyogenes (serotype M18) | 2E−20 |
| 25a | 19926-21146 | 407 | Serine/threonine protein phosphatase | Ser/Thr protein phosphatase YjbP of Bacillus licheniformis strain ATCC 14580 | 8E−14 |
| 29a | 22172-22759 | 196 | Unknown | Hypothetical protein CwatDRAFT_4471 of Crocosphaera watsonii WH 8501 | 2E−04 |
| 31a | 24416-25645 | 410 | Transposase | Transposase, IS_605_ OrfB of Crocosphaera watsonii WH 8501 | E−100 |
| 32 | 25710-26129 | 140 | Transposase | Transposase, IS_200_-like of Crocosphaera watsonii WH 8501 | 4E−39 |
| 37a | 28028-28765 | 246 | Unknown | Hypothetical protein MA4278 of Methanosarcina acetivorans | 1E−45 |
| 39 | 29163-29630 | 156 | Unknown | Hypothetical protein SO2944 of Shewanella oneidensis | 2E−04 |
| 40 | 29780-30208 | 143 | Unknown | Hypothetical protein PA2G02937 of Pseudomonas aeruginosa 2192 | 5E−21 |
| 41 | 30742-31506 | 255 | Unknown | Gp7 of cyanophage P-SSM4 | 2E−06 |
| 44 | 32400-32639 | 80 | Unknown | Hypothetical protein BBta_p0029 of Bradyrhizobium sp. strain BTAi1 | 8E−11 |
| 45 | 32748-33686 | 313 | Unknown | COG1637; predicted nuclease of the RecB family | 5E−04 |
| 51 | 36090-36770 | 227 | Unknown | Hypothetical protein slr7057 of Synechocystis sp. strain PCC 6803 | 5E−12 |
| 62a | 41682-42509 | 276 | Unknown | Hypothetical protein DUF323 of Trichodesmium erythraeum IMS101 | 3E−100 |
| 68 | 51615-52256 | 214 | Unknown | Hypothetical protein of Synechococcus sp. strain PCC 6301 | 9E−19 |
| 69 | 52314-53069 | 252 | Lysozyme | Putative lysozyme of Pseudomonas syringae (pv. tomato) | 6E−31 |
| 72 | 54515-55231 | 239 | Unknown | Gp97 of Mycobacterium phage Che12 | 6E−30 |
| 91 | 78218-80542 | 775 | Putative phage tail sheath protein | Phage-related contractile tail sheath protein of Xylella fastidiosa ATCC 700964 | 2E−04 |
| 94 | 83187-84752 | 522 | Unknown | Hypothetical protein precursor of Synechococcus sp. strain WH8102 | 9E−14 |
| 95 | 84752-85945 | 398 | Lysozyme/metalloendopeptidase | Lipoprotein, NlpD of Synechocystis sp. strain PCC 6803 | 8E−36 |
| 101 | 92000-92980 | 327 | Unknown | XkdT, uncharacterized homologue of phage Mu protein gp47 | 1E−06 |
| 104a | 96683-97483 | 267 | Unknown | Slr1033 protein of Synechocystis sp. strain PCC 6803 | 3E−39 |
| 105 | 97622-99355 | 578 | Unknown | Putative cell wall surface anchor family protein precursor of Bdellovibrio bacteriovorus | 3E−13 |
| 106 | 99480-100493 | 338 | Putative lysine/ornithine N-monooxygenase | LucD of _Klebsiella pneumoniae N_-monooxygenase | 4E−04 |
| 108 | 101054-105133 | 1360 | Unknown | Hypothetical protein KgORF65 of Staphylococcus phage K | 6E−17 |
| 109 | 105120-107642 | 841 | Unknown | Hypothetical protein KgORF65 of Staphylococcus phage K | 8E−06 |
| 118 | 115106-116884 | 593 | DNA terminase | Terminase_1; phage terminase | 7E−09 |
| 128a | 129804-130721 | 306 | Putative Fe-S oxidoreductase | Radical SAM superfamily enzyme of Thermus thermophilus phage YS40 | E−09 |
| 134a | 133470-134663 | 398 | Putative DNA primase | DNA primase of Azoarcus sp. strain EbN1 | 3E−11 |
| 135a | 134712-135884 | 391 | Transposase | Transposase of Crocosphaera watsonii WH 8501 | E−135 |
| 136a | 135862-136470 | 203 | Putative site-specific recombinase | MerR (resolvase, N-terminal) of Crocosphaera watsonii WH 8501 | E−106 |
| 160a | 142680-143828 | 383 | Putative helicase | RRM3/PIF1 helicase homologue precursor of Bdellovibrio bacteriovorus | 4E−25 |
| 164 | 146035-146295 | 87 | Unknown | Hypothetical protein PSSM4_160 of cyanophage P-SSM4 | 4E−05 |
| 166a | 146894-148345 | 484 | UvsW | Predicted DNA/RNA repair helicase of Pyrococcus kodakaraensis (Thermococcus kodakaraensis) | 2E−19 |
| 169a | 150291-151394 | 368 | DNA polymerase III γ/τ subunit | DNA polymerase III γ/τsubunits of Synechococcus elongatus | 4E−13 |
| 171a | 151605-152753 | 383 | Unknown | All7027 protein of Anabaena sp. strain PCC 7120 | 4E−94 |
| 173a | 153269-154024 | 252 | Uracil-DNA glycosylase | Uracil-DNA glycosylase of Thermus thermophilus (strain ATCC BAA-163) | 2E−06 |
| 174a | 154027-155163 | 379 | Unknown | All2778 protein of Anabaena sp. strain PCC 7120 | 3E−35 |
| 175a | 155187-155558 | 124 | Unknown | Asl2779 protein of Anabaena sp. strain PCC 7120 | 3E−12 |
| 178a | 157270-158232 | 321 | DNA polymerase | POLAc; DNA polymerase I of Caulobacter crescentus | 3E−14 |
| 180a | 158352-159635 | 428 | 3′-5′ exonuclease | 3′-5′ exonuclease of Rhodopseudomonas palustris | 2E−06 |
| 181a | 159632-160306 | 225 | dUTPase | Deoxyuridine 5′-triphosphate nucleotidohydrolase of Helicobacter hepaticus | 3E−10 |
| 183a | 160545-161216 | 224 | PhoH | PhoH-like protein of Rhodopirellula baltica starvation-inducible protein | 2E−30 |
Gene organization.
The Ma-LMM01 genome can be divided into two parts in ORF orientations. The upper side of the genome map, the “UG region,” contains 144 ORFs (ORF121 to ORF184 and ORF1 to ORF80), where most (82%) are found in a counterclockwise direction (Fig. 1). Conversely, the lower side of the map, the “LG region,” contains 40 ORFs (ORF81 to ORF120), found mostly (95%) in the clockwise direction.
Of the 28 ORFs with putative function assignments (Table 1), 24 were found in the UG region, including 13 ORFs involved in DNA processing and nucleotide metabolism. The LG region contains four ORFs with putative functions. These correspond to a DNA terminase homologue (ORF118) involved in the packaging of double-stranded DNA into viral procapsids (31), a tail sheath protein homologue (ORF91), a putative lysozyme (ORF95), and a putative lysine/ornithine _N_-monooxygenease (ORF106). Except for ORF91 (the tail sheath protein gene), we found no ORFs in the genome that are similar to genes previously described for structural components (e.g., the major capsid proteins) of other phages.
Previously, we showed that Ma-LMM01 contains four major polypeptides of 84, 47, 38, and 26 kDa by using SDS-PAGE (81). Here, we determined the N-terminal sequence of the 47-kDa (SDIPS) and 38-kDa (SIHNV) polypeptides. They correspond to ORF86 and ORF87, respectively (except for the initial methionine). The quantity of the major head protein is usually three to four times more than that of tail sheath or tail tube proteins (25, 40). The concentrations of the 47- and 38-kDa polypeptides appear to be higher than that of the 26-kDa polypeptide and just slightly higher than that of the 84-kDa polypeptide (81). This suggests that ORF86 and ORF87 code for the major head proteins and that the head of the Ma-LMM01 virion consists of two major components, similar to the case for the lambda phage (77). The 84-kDa polypeptide may correspond to the putative tail sheath protein (ORF91) with a predicted molecular mass of 83.6 kDa (although the N-terminal sequence of the 84-kDa polypeptide was not determined). Tail tube protein is often encoded just downstream of the tail sheath genes (44, 56, 69). Downstream of ORF86 to -91, we found three ORFs (ORF88 [25.2 kDa], ORF90 [26.3 kDa], and ORF92 [26.3 kDa]) showing predicted molecular masses close to 26 kDa.
The large number of ORFs for DNA/nucleotide processing in the UG region suggests that the genes in this region constitute early transcription units. In contrast, the LG region putatively codes for several phage structural proteins and may correspond to late transcriptional units; however, a putative consensus sequence for a late promoter as found in cyanophage S-PM2 (NATAAATA) (44) was not found in the Ma-LMM01 genome.
The PBS degradation gene.
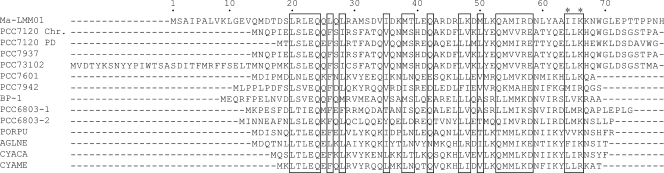
The Ma-LMM01 genome has no homologues for the core photosystem reaction center genes (psbA and psbD), which are prevalent in marine cyanophages (70). However, we found that Ma-LMM01 possesses a homologue (ORF5) of the nblA (_n_on_bl_eaching) genes of cyanobacteria and red algae (Table 1). In cyanobacteria, NblA (a small protein of about 7 kDa) plays an central role in the degradation of the phycobilisomes (PBSs), the major light-harvesting complex of the M. aeruginosa photosynthetic apparatus (3). The PBSs are attached to the cytoplasmic surface of the photosynthetic membrane and constitute up to 50% of the total cellular soluble proteins. During nitrogen starvation, cyanobacterial PBSs are proteolytically degraded, causing “bleaching.” This helps to prevent photodamage caused by the absorption of excess light energy and provides substrates for protein synthesis during acclimation processes (3, 26). NblA directly binds to the main rod structures of PBSs (6), where this is considered to trigger PBS degradation (3, 17, 26). In Synechococcus elongatus PCC 7942, nblA is expressed at low levels in nutrient-replete cells and is highly up-regulated under nitrogen starvation (17). The Ma-LMM01 NblA-like sequence (ORF5, 80 amino acids) is readily aligned with cellular NblA homologues with no additional sequence domains (Fig. 4). The Ma-LMM01 NblA-like sequence shows all the residues conserved in previously identified NblA homologues, including the two amino acid residues (i.e., I64 and K66 in the Ma-LMM01 NblA sequence) used for phycobiliprotein-binding (6). Thus, this evidence suggests that Ma-LMM01 ORF5 codes for a functional polypeptide capable of interacting with phycobiliproteins. Angly et al. found psbA gene sequences in their marine viral metagenomic data (2). In contrast, we found no homologues for nblA genes in viral (2, 7-9, 11) or marine microbial (65) metagenomic sequence data sets. This is the first identification of a homologue of nblA in a viral genome.
FIG. 4.
Sequence alignment of NblA homologues. PCC7120, Nostoc sp. (Chr, chromosome [accession no. BAB76216]; PD, plasmid Delta [BAB77423]); PCC7937, Ananbaena variabilis (ABA22990); PCC73102, Nostoc punctiforme (ZP00345234); BP-1, Thermosynechococcus elongates (NP680824); PCC7601, Tolypothrix sp. strain PCC7601 (CAD28153); PCC6803, Synechocystis sp. (1, BAA17955; 2, BAA17954); PCC7942, Synechococcus sp. (ABB58157); PORPU, Porphyra purpurea (NP053976); AGLNE, Aglaothamnion neglectum (P48446); CYACA, Cyanidium caldarium (NP045072); CYAME, Cyanidoschyzon merolae (BAC76137). Conserved residues among the NblAs are shown in boxes (6). Amino acid residues binding to the PBS are marked with asterisks (6).
Potential lysogeny and insertion sequence (IS) elements.
Ma-LMM01 forms clear plaques on host lawns, with no turbid plaques observed (data not shown), and thus only shows lytic behavior under laboratory conditions (81). However, the Ma-LMM01 genome (ORF136) codes for a homologue of a site-specific recombinase (int) used by temperate phages to integrate the phage genome into the bacterial chromosome (27). ORF136 has a high level of sequence similarity (i.e., 81 to 98% amino acid sequence identity) to site-specific recombinases found in cyanobacteria, e.g., Crocosphaera watsonii, Cyanothece sp., and Nodularia spumigena. Indeed, the genome sequences of Ma-LMM01 and Cyanothece sp. (GenBank accession no. NZ_AAXW01000043) show a colinear region of about 1,800 bp (nucleotide positions 134715 to 136525 for Ma-LMM01 compared to nucleotide positions 12730 to 14550 for Cyanothece genome, at about 82% nucleotide identity). This region contains the int gene and a flanking transposase gene (ORF135 for Ma-LMM01; pfam01385 E value = 10−45). This Ma-LMM01 genomic region shows a high nucleotide sequence similarity (∼67%) to several regions of the genome of the marine cyanobacterium Trichodesmium erythraeum (GenBank accession no. NC_008312). The int gene and the transposase gene are located within a 7.5-kb region with an atypically low G+C content (Fig. 1). Further, Ma-LMM01 possesses two ORFs (ORF9 and ORF24) for the putative prophage antirepressors, which are implicated in prophage induction of temperate phages (67). The presence of these genetic elements (int and prophage antirepressor genes) suggests that Ma-LMM01 may exhibit a prophage state in some hosts, although transfer of those genes by transposable elements is also possible. In a previous study, the lytic cyanopodovirus P-SSP7 was reported to have an int gene (69). A 42-bp sequence identical to part of the host leucine tRNA gene (i.e., Prochlorococcus MED4) was identified downstream of the P-SSP7 int gene, suggesting the use of the tRNA gene as an integration site recognized by the P-SSP7 int product (69). Such a feature was not found in the flanking sequences of the Ma-LMM01 int gene.
In addition to the putative int (ORF136) and the transposase gene (ORF135), we found that Ma-LMM01 has four ORFs showing high similarities (53 to 70% amino acid sequence identities; E value, <10−38) to ORFs from C. watsonii WH 8501 and other cyanobacteria (Table 1). The high levels of sequence similarity suggest that these ORFs originated from recent horizontal transfers. Two of these ORFs (ORF31 [pfam01385 E value = 4 × 10−41] and ORF32 [COG1943 E value = 2 × 10−27]) are similar to known transposase genes, whereas the remaining two (ORF62 and ORF171) are of unknown functions. IS elements are rare in phage genomes, possibly reflecting their disadvantageous effects on phage replication (66). In addition to Ma-LMM01, only the c-st phage infecting Clostridium botulinum is known to have multiple IS elements (12 copies); eight other sequenced phage genomes harbor a single IS element (66).
DNA replication, repair, and recombination.
The T4 replisome contains seven proteins encoded in the viral genome (i.e., DNA polymerase, sliding clamp loader, sliding clamp, DNA helicase, DNA primase, and single-strand DNA binding protein) (48); however, only two homologues were found in the Ma-LMM01 genome: DNA polymerase I (ORF178) and DNA primase (ORF134) (Table 1). Ma-LMM01 possesses an additional ORF (ORF169) showing a significant sequence similarity to the N-terminal region of the DNA polymerase III γ/τ subunit. Genes for DNA polymerase III subunits are rarely found in the genomes of phages (61). The replisome of Ma-LMM01 may contain host proteins. Alternatively, the phage genome may encode DNA replication proteins lacking detectable sequence similarity to the T4 replisome components.
The amino acid sequence determined from ORF178 corresponds to a polymerase domain from DNA polymerase I but lacks the exonuclease domains. We found that ORF180 is similar to 3′-5′exonuclease domain sequences. Exonuclease domains and polymerase domains are usually encoded in a single gene in phages and bacteria. The unusual split gene organization for the putative Ma-LMM01 DNA polymerase prompted us to examine for the presence of introns between the two ORFs. In phage K, DNA polymerase I is encoded by a gene with two group I introns (56). Each of the two introns codes for a homing endonuclease. In contrast, the intervening sequence (including ORF179) between ORF180 and ORF178 showed no detectable similarity to known homing endonuclease or self-splicing intron sequences. We performed RT-PCR and PCR experiments using primers targeting the intervening sequence (i.e., from the 3′ end of ORF180 to the 5′ end of ORF178) and confirmed that the size of the RT-PCR products from the total RNA from the infected host was the same as the size of the PCR products from genomic DNA (Fig. 5). No putative Shine-Dalgarno sequence was found upstream of the putative translation start site of ORF180; however, this is a common feature of cyanobacterial and chloroplast genomes (78). This suggests that the two domains are coded in separate genes and translated as individual polypeptide chains.
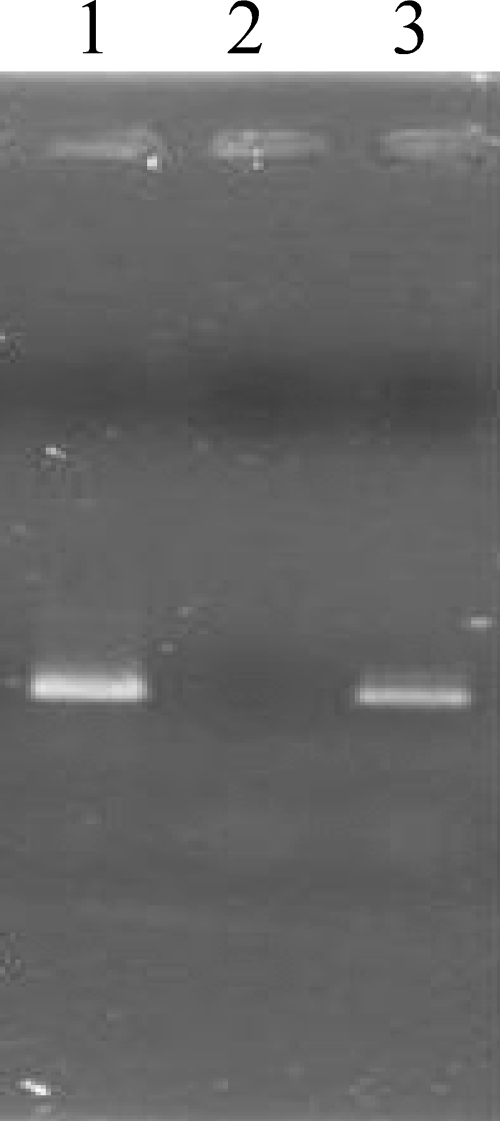
FIG. 5.
Transcription analysis of an intervening region between the DNA polymerase and 3′-5′exonuclease genes of Ma-LMM01. PCR targeting a 671-bp sequence between primer pairs at the 3′ end of ORF180 and 5′ end of ORF178 was performed using the following templates: lane 1, cDNA from host cells infected by Ma-LMM01; lane 2, DNase-treated RNA from host cells infected by Ma-LMM01; lane 3, DNA from Ma-LMM01.
The Ma-LMM01 genome contains four ORFs associated with putative DNA recombination and repair functions. ORF166 is a homologue of the T4 uvsW gene. T4 UvsW protein is an RNA-DNA helicase playing a role in the shift from an origin-dependent replication mode during early infection to a recombination-dependent replication mode during the late infection stages (19, 50). ORF8 may encode a homologue of UvsX (RecA-like recombinase), an essential enzyme for both recombination and recombination repair in T4 (29). ORF160 corresponds to a putative ATP-dependent RecD-like helicase similar to the T4 Dda helicase. In Escherichia coli, RecD is a subunit of an ATP-dependent exonuclease (ExoV) involved in the repair of double-strand breaks (36). UvsX recombinase and Dda helicase are reported to rescue stalled T4 replication forks in vitro (29). Finally, ORF173 encodes a homologue of uracil-DNA glycosylases that removes uracil in DNA that can arise either by misincorporation of dUTP during DNA synthesis or by the spontaneous deamination of cytosine (22).
Nucleotide metabolism.
Ma-LMM01 possesses two ORFs for the α (ORF6) and β (ORF2) subunits of ribonucleotide reductase (RNR), as previously reported (81). RNR is an essential enzyme that generates precursors of DNA by converting ribonucleoside diphosphates into deoxyribonucleoside diphosphates (28). RNR genes are commonly found in previously sequenced lytic myoviruses and are considered essential for the rapid replication found in lytic phages (14). Marine cyanomyoviruses possess RNR subunit genes with higher sequence similarities to other T4-like phages than to cyanobacterial homologues (69). In contrast, the RNR genes of Ma-LMM01 are more closely related to cyanobacteria (81), suggesting lateral exchange of this key enzyme gene between the lineages of Ma-LMM01 and cyanobacteria. Similar to other lytic myoviruses (44, 46, 69), the Ma-LMM01 genome encodes a homologue (ORF20) of flavin-dependent thymidylate synthase ThyX that produces thymidylate (dTMP) de novo from dUMP (51). Finally, Ma-LMM01 possesses an ORF (ORF181) for dUTPase that converts dUTP into dUMP to avoid misincorporation of uracil into genomic DNA.
Phosphate stress gene.
Ma-LMM01 ORF183 shows significant sequence similarity to E. coli phoH, which codes for an ATPase induced under phosphate starvation. phoH homologues are widely distributed among bacteria and archaea (34). All previously sequenced marine cyanomyovirus genomes show phoH homologues (14, 44, 69). The biological functions of phoH products have not been characterized (34). It has been proposed that phoH may be important for phage propagation in phosphorus-poor environments such as marine systems (69). However, this hypothesis does not account for the presence of the phoH homologue in the Ma-LMM01 genome, as the habitat for this phage is in eutrophic phosphate rich waters.
A short sequence similar to the microcystin synthetase gene loci.
Microcystins are synthesized by nonribosomal enzyme complexes encoded on a cluster of 10 bidirectionally transcribed genes (mcyA to -C and mcyD to -J) in M. aeruginosa (54, 55, 72). We found no homologues (or pseudogenes) for the microcystin biosynthetic genes in the Ma-LMM01 genome. Toxic and nontoxic Microcystis strains are randomly distributed on the phylogenetic trees constructed from the sequences of the rRNA-intergenic spacer and the phycocyanin locus intergenic spacer regions (58, 73). The sporadic distribution of toxic/nontoxic strains may be a result of loss of genes in different lineages, as the peptide synthetase genes are likely to have been present in the common ancestor of Microcystis (63). However, Tillett et al. show clear incongruities between the phylogenetic tree based on mcyA sequences and another based on the phycocyanin locus spacers (73), suggesting the possible horizontal transfer of mcyA genes among strains. Such horizontal transfers have significant implications in the spread of toxic blooms. Accumulating evidence suggests that marine cyanophages are important agents of horizontal gene transfer among cyanobacteria (44, 69, 70). We showed that Ma-LMM01 has several “host-like” genes (i.e., nblA, RNR genes, and other ORFs strikingly similar to genes found in cyanobacteria), suggesting that DNA transfers between Ma-LMM01 and its host have occurred. Further, we found in the 3′-region of ORF104 a 41-bp sequence (TCATACTAAATCTGGTTATTAAAAACTGATTATTTATTTCC) that is 90% identical to part of the intergenic sequence between the transposase-like gene (trp2, pfam01609) and mcyJ in Microcystis viridis NIES102 (GenBank accession no. AB254436; E value = 6 × 10−6 using BLASTN comparisons in GenBank). The 41-bp sequence also shows 88% identity (E value = 10−3) to a downstream sequence of a transposase gene (uma4, COG3464) located adjacent to the M. aeruginosa PCC7806 mcyA-C/mcyD-J gene cluster (GenBank accession no. AF183408). These sequences may provide recognition sites for transposases or integrases responsible for the transfer of DNA, possibly including the mcyA-C/D-J locus, among certain Microcystis strains and their viruses.
Potential role for the phage-encoded NblA-like protein.
The Ma-LMM01 _nblA_-like gene represents another example of the capture of “host” genes incorporated into a phage genome and suggests that the lateral transfer of photosynthesis system-associated genes plays a major role in the coevolutionary process of cyanophages and their hosts (15, 37, 38).
The Ma-LMM01 genome carries no homologues for psbA and psbD, which are prevalent in marine cyanophage genomes (37, 38, 44, 45, 69, 70). The psbA and psbD genes encode the photosystem II core reaction center proteins D1 and D2, respectively. Previously, the phage-carried psbA gene was shown to be expressed during phage infections (15, 37, 38). These phage-encoded photosynthetic proteins appear to sustain photosynthesis during infection after the decline of host protein synthesis and thus provide the energy for viral replication (45). Sullivan et al. proposed that cyanophages lacking psbA may have a short latent period of infection (∼1 h), which is not long enough for psbA expression to be beneficial to the phages (70). However, this hypothesis does not account for the lack of psbA in Ma-LMM01, as the latent period of Ma-LMM01 (6 to 12 h) (81) is as long as those of the other cyanophages having psbA.
We propose that the Ma-LMM01 NblA homologue probably functions to reduce the absorption of excess light energy through the degradation of the PBS. In the M. aeruginosa/Ma-LMM01 host/parasite system, the benefit to the phage conferred by the degradation of the PBS (and the decline of photosynthesis) may be greater than that gained by sustaining host photosynthesis (for instance, by having psbA). M. aeruginosa is commonly found in surface waters due to the buoyancy provided by its subcellular structures (i.e., gas vesicles) (49). In this environment, the availability of light is several orders of magnitude greater than in lower parts of the ocean's photic layer (60), which is dominated by diverse marine cyanobacteria (24). After the decline in the host metabolism through Ma-LMM01 infection, decreasing the absorption of light energy may be necessary to prevent photodamage during phage replication. Further, the PBS is a large multiprotein complex that can constitute up to 50% of the soluble protein in the cell (26). Mann (43) proposed that the PBS may be an important source of amino acids for phage protein synthesis. Rapid degradation of the PBS during the early infectious stages may be advantageous for the efficient production of Ma-LMM01.
As an alternative possibility, the Ma-LMM01 NblA-like protein may provide a way to sustain host photosynthesis by interfering with the function of the host NblA that initiates the degradation of PBS in response to the stress induced by phage infection. However, several lines of evidence argue against this hypothesis. The protein sequence derived from the Ma-LMM01 _nblA_-like gene shows all of the highly conserved residues found in the cyanobacterial NblA protein sequence and possesses no extra sequence domains. This suggests a similar function for the Ma-LMM01 _nblA_-like gene product and the cellular NblA. Our preliminary experiments show that Ma-LMM01-encoded nblA is expressed in the infected host and host PBS is degraded during infection (data not shown); however, we have not demonstrated that the Ma-LMM01 NblA is responsible for the degradation of the PBS.
Conclusions.
The genome of Ma-LMM01 is significantly different from previously sequenced genomes of marine cyanophages and possesses several traces of horizontal gene transfers from cyanobacteria. The co-option of the nblA homologue in this genome points to diverse adaptive strategies for cyanophages and further suggests that light is central in the phage-cyanobacterium relationships where the viruses use different genetic strategies to control their host's photosynthesis. Given the large number of ORFs lacking detectable database homologues, much work remains to be performed to characterize the phage-host interactions for this cultured toxic-cyanobacterium/lytic-phage model system.
Acknowledgments
We thank Jean-Michel Claverie (head of Information Génomique et Structurale) for laboratory space and support for the computer analyses. Thanks are also due to Ken Nishikawa (Maebashi Institute of Technology) and Fumio Arisaka for providing the GTOP analysis.
H.O. was supported in part by Marseille-Nice Genopole and the French National Genomic Network (RNG). This study was supported in part by the Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization of Japan (NEDO) and the Research Foundation of Fukui Prefecture for the Promotion of Science.
Footnotes
▿
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baier, K., H. Lehmann, D. P. Stephan, and W. Lockau. 2004. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 1502739-2749. [DOI] [PubMed] [Google Scholar]
- 4.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340467-468. [DOI] [PubMed] [Google Scholar]
- 5.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 292607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienert, R., K. Baier, R. Volkmer, W. Lockau, and U. Heinemann. 2006. Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. J. Biol. Chem. 2815216-5223. [DOI] [PubMed] [Google Scholar]
- 7.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 1856220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 9914250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brussaard, C. P. 2004. Viral control of phytoplankton populations—a review. J. Eukaryot. Microbiol. 51125-138. [DOI] [PubMed] [Google Scholar]
- 11.Cann, A. J., S. E. Fandrich, and S. Heaphy. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30151-156. [DOI] [PubMed] [Google Scholar]
- 12.Carmichael, W. W. 1995. Toxic Microcystis and the environment, p. 1-11. In M. F. Watanabe, K. Harada, W. W. Carmichael, and H. Fujiki (ed.), Toxic Microcystis. CRC Press, Boca Raton, FL.
- 13.Carmichael, W. W., S. M. Azevedo, J. S. An, R. J. Molica, E. M. Jochimsen, S. Lau, K. L. Rinehart, G. R. Shaw, and G. K. Eaglesham. 2001. Human fatalities from cyanobacteria: chemical and biological evidence for cyanotoxins. Environ. Health Perspect. 109663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 682589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clokie, M. R., J. Shan, S. Bailey, Y. Jia, H. M. Krisch, S. West, and N. H. Mann. 2006. Transcription of a ‘photosynthetic’ T4-type phage during infection of a marine cyanobacterium. Environ. Microbiol. 8827-835. [DOI] [PubMed] [Google Scholar]
- 16.Coleman, M. L., M. B. Sullivan, A. C. Martiny, C. Steglich, K. Barry, E. F. Delong, and S. W. Chisholm. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 3111768-1770. [DOI] [PubMed] [Google Scholar]
- 17.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 131039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 274636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudas, K. C., and K. N. Kreuzer. 2001. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol. Cell. Biol. 212706-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 21.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 22.Farez-Vidal, M. E., C. Gallego, L. M. Ruiz-Perez, and D. Gonzalez-Pacanowska. 2001. Characterization of uracil-DNA glycosylase activity from Trypanosoma cruzi and its stimulation by AP endonuclease. Nucleic Acids Res. 291549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 24.Garczarek, L., A. Dufresne, S. Rousvoal, N. J. West, S. Mazard, D. Marie, H. Claustre, P. Raimbault, A. F. Post, D. J. Scanlan, and F. Partensky. 2007. High vertical and low horizontal diversity of Prochlorococcus ecotypes in the Mediterranean Sea in summer. FEMS Microbiol. Ecol. 60189-206. [DOI] [PubMed] [Google Scholar]
- 25.Govind, R., J. A. Fralick, and R. D. Rolfe. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J. Bacteriol. 1882568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman, A. R., M. R. Schaefer, G. G. Chiang, and J. L. Collier. 1993. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57725-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335667-678. [DOI] [PubMed] [Google Scholar]
- 28.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 6771-98. [DOI] [PubMed] [Google Scholar]
- 29.Kadyrov, F. A., and J. W. Drake. 2004. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J. Biol. Chem. 27935735-35740. [DOI] [PubMed] [Google Scholar]
- 30.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 3381027-1036. [DOI] [PubMed] [Google Scholar]
- 31.Kanamaru, S., K. Kondabagil, M. G. Rossmann, and V. B. Rao. 2004. The functional domains of bacteriophage T4 terminase. J. Biol. Chem. 27940795-40801. [DOI] [PubMed] [Google Scholar]
- 32.Kasai, F., M. Kawachi, M. Erata, and M. M. Watanabe. 2004. NIES—collection list of strains, 7th ed. The National Institution for Environmental Studies, Tsukuba, Japan.
- 33.Kawabata, T., S. Fukuchi, K. Homma, M. Ota, J. Araki, T. Ito, N. Ichiyoshi, and K. Nishikawa. 2002. GTOP: a database of protein structures predicted from genome sequences. Nucleic Acids Res. 30294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazakov, A. E., O. Vassieva, M. S. Gelfand, A. Osterman, and R. Overbeek. 2003. Bioinformatics classification and functional analysis of PhoH homologs. In Silico Biol. 33-15. [PubMed] [Google Scholar]
- 35.Kondo, R., T. Yoshida, Y. Yuki, and S. Hiroishi. 2000. DNA-DNA reassociation among a bloom-forming cyanobacterial genus, Microcystis. Int J. Syst. Evol. Microbiol. 50767-770. [DOI] [PubMed] [Google Scholar]
- 36.Koppen, A., S. Krobitsch, B. Thoms, and W. Wackernagel. 1995. Interaction with the recombination hot spot chi in vivo converts the RecBCD enzyme of Escherichia coli into a chi-independent recombinase by inactivation of the RecD subunit. Proc. Natl. Acad. Sci. USA 926249-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindell, D., J. D. Jaffe, Z. I. Johnson, G. M. Church, and S. W. Chisholm. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 43886-89. [DOI] [PubMed] [Google Scholar]
- 38.Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm. 2004. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 10111013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35324-340. [DOI] [PubMed] [Google Scholar]
- 40.Loessner, M. J., and S. Scherer. 1995. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J. Bacteriol. 1776601-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKintosh, C., K. A. Beattie, S. Klumpp, P. Cohen, and G. A. Codd. 1990. Cyanobacterial microcystin-LR is potent and specific inhibitor of protein phosphatase 1 and 2A from both mammals and higher plants. FEBS Lett. 264187-192. [DOI] [PubMed] [Google Scholar]
- 43.Mann, N. H. 2005. Phages of cyanobacteria, p. 517-533. In R. L. Calendar (ed.), The bacteriophages, 2nd, ed. Oxford University Press, New York, NY.
- 44.Mann, N. H., M. R. Clokie, A. Millard, A. Cook, W. H. Wilson, P. J. Wheatley, A. Letarov, and H. M. Krisch. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 1873188-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424741. [DOI] [PubMed] [Google Scholar]
- 46.Mesyanzhinov, V. V., J. Robben, B. Grymonprez, V. A. Kostyuchenko, M. V. Bourkaltseva, N. N. Sykilinda, V. N. Krylov, and G. Volckaert. 2002. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 3171-19. [DOI] [PubMed] [Google Scholar]
- 47.Millard, A., M. R. Clokie, D. A. Shub, and N. H. Mann. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. USA 10111007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 6786-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mlouka, A., K. Comte, A. M. Castets, C. Bouchier, and N. Tandeau de Marsac. 2004. The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J. Bacteriol. 1862355-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosig, G., J. Gewin, A. Luder, N. Colowick, and D. Vo. 2001. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc. Natl. Acad. Sci. USA 988306-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297105-107. [DOI] [PubMed] [Google Scholar]
- 52.Nagasaki, K., Y. Tomaru, K. Nakanishi, N. Hata, N. Katanozaka, and M. Yamaguchi. 2004. Dynamics of Heterocapsa circularisquama (Dinophyceae) and its viruses in Ago Bay, Japan. Aquat. Microb. Ecol. 34219-226. [Google Scholar]
- 53.Nishiwaki-Matsushima, R., T. Ohta, S. Nishiwaki, M. Suganuma, K. Kohyama, T. Ishikawa, W. W. Carmichael, and H. Fujiki. 1992. Liver tumor promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J. Cancer Res. Clin. Oncol. 118420-424. [DOI] [PubMed] [Google Scholar]
- 54.Nishizawa, T., M. Asayama, K. Fujii, K. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. (Tokyo) 126520-529. [DOI] [PubMed] [Google Scholar]
- 55.Nishizawa, T., A. Ueda, M. Asayama, K. Fujii, K. Harada, K. Ochi, and M. Shirai. 2000. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. (Tokyo) 127779-789. [DOI] [PubMed] [Google Scholar]
- 56.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 1862862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliver, R. L., and G. G. Ganf. 2000. Fresh water blooms, p. 149-194. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 58.Otsuka, S., S. Suda, R. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 17215-21. [DOI] [PubMed] [Google Scholar]
- 59.Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kutzing ex Lemmermann 1907 under the rules of the Bacteriological Code. Int. J. Syst. Evol Microbiol. 51873-879. [DOI] [PubMed] [Google Scholar]
- 60.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul, J. H., S. J. Williamson, A. Long, R. N. Authement, D. John, A. M. Segall, F. L. Rohwer, M. Androlewicz, and S. Patterson. 2005. Complete genome sequence of phiHSIC, a pseudotemperate marine phage of Listonella pelagia. Appl. Environ. Microbiol. 713311-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pope, W. H., P. R. Weigele, J. Chang, M. L. Pedulla, M. E. Ford, J. M. Houtz, W. Jiang, W. Chiu, G. F. Hatfull, R. W. Hendrix, and J. King. 2007. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J. Mol. Biol. 368966-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Borner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohwer, F., and R. Edwards. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 1844529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakaguchi, Y., T. Hayashi, K. Kurokawa, K. Nakayama, K. Oshima, Y. Fujinaga, M. Ohnishi, E. Ohtsubo, M. Hattori, and K. Oguma. 2005. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc. Natl. Acad. Sci. USA 10217472-17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shearwin, K. E., A. M. Brumby, and J. B. Egan. 1998. The Tum protein of coliphage 186 is an antirepressor. J. Biol. Chem. 2735708-5715. [DOI] [PubMed] [Google Scholar]
- 68.Stothard, P., and D. S. Wishart. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21537-539. [DOI] [PubMed] [Google Scholar]
- 69.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan, M. B., D. Lindell, J. A. Lee, L. R. Thompson, J. P. Bielawski, and S. W. Chisholm. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suttle, C. A. 2005. Viruses in the sea. Nature 437356-361. [DOI] [PubMed] [Google Scholar]
- 72.Tillett, D., E. Dittmann, M. Erhard, H. von Dohren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7753-764. [DOI] [PubMed] [Google Scholar]
- 73.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ Microbiol. 672810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomaru, Y., K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Quantitative and qualitative impacts of viral infection on a Heterosigma akashiwo (Raphidophyceae) bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34227-238. [Google Scholar]
- 75.Tucker, S., and P. Pollard. 2005. Identification of cyanophage Ma-LBP and infection of the cyanobacterium Microcystis aeruginosa from an Australian subtropical lake by the virus. Appl. Environ Microbiol. 71629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.UniProt Consortium. 2007. The Universal Protein Resource (UniProt). Nucleic Acids Res. 35D193-D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 1826472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weigele, P. R., W. H. Pope, M. L. Pedulla, J. M. Houtz, A. L. Smith, J. F. Conway, J. King, G. F. Hatfull, J. G. Lawrence, and R. W. Hendrix. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 91675-1695. [DOI] [PubMed] [Google Scholar]
- 79.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, L. Y. Geer, Y. Kapustin, O. Khovayko, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, V. Miller, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, R. L. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2007. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35D5-D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida, T., M. Maki, H. Okamoto, and S. Hiroishi. 2005. Coordination of DNA replication and cell division in cyanobacteria Microcystis aeruginosa. FEMS Microbiol. Lett. 251149-154. [DOI] [PubMed] [Google Scholar]
- 81.Yoshida, T., Y. Takashima, Y. Tomaru, Y. Shirai, Y. Takao, S. Hiroishi, and K. Nagasaki. 2006. Isolation and characterization of a cyanophage infecting the toxic cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 721239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshizawa, S., R. Matsushima, M. F. Watanabe, K. Harada, A. Ichihara, W. W. Carmichael, and H. Fujiki. 1990. Inhibition of protein phosphatases by microcystins and nodularin associated with hepatotoxicity. J. Cancer Res. Clin. Oncol. 116609-614. [DOI] [PubMed] [Google Scholar]