Environmental Epigenetics and Asthma: Current Concepts and Call for Studies (original) (raw)
Abstract
Recent studies suggest that epigenetic regulation (heritable changes in gene expression that occur in the absence of alterations in DNA sequences) may in part mediate the complex gene-by-environment interactions that can lead to asthma. The variable natural history of asthma (i.e., incidence and remission of symptoms) may be a result of epigenetic changes, such as DNA methylation, covalent histone modifications, microRNA changes, and chromatin alterations, after early or later environmental exposures. Findings from multiple epidemiologic and experimental studies indicate that asthma risk may be modified by epigenetic regulation. One study suggested that the transmission of asthma risk may occur across multiple generations. Experimental studies provide substantial in vitro data indicating that DNA methylation of genes critical to T-helper cell differentiation may induce polarization toward or away from an allergic phenotype. Despite this initial progress, fundamental questions remain that need to be addressed by well-designed research studies. Data generated from controlled experiments using in vivo models and/or clinical specimens collected after environmental exposure monitoring are limited. Importantly, cohort-driven epigenetic research has the potential to address key questions, such as those concerning the influence of timing of exposure, dose of exposure, diet, and ethnicity on susceptibility to asthma development. There is immense promise that the study of environmental epigenetics will help us understand a theoretically preventable environmental disease.
Keywords: epigenetics, asthma, DNA methylation, gene–environment interactions
Asthma has long been recognized as a complex genetic disease mediated by exposures to a variety of environmental triggers. The greater concordance of asthma among monozygotic twins when compared with dizygotic twins demonstrates that both genetic and environmental influences are important to asthma pathogenesis (reviewed in Reference 1). By 2007, several dozen publications provided evidence of specific gene-by-environment interactions for this disease (Table 1), and many more can be anticipated. Yet, the clinical presentation of asthma is heterogeneous. Some individuals are affected as young children, others as adults. Furthermore, some develop asthma or wheezing illness as a result of occupational exposures, whereas others may become affected after exposure to urban air pollution (2, 3). Traditional models for gene-by-environment interactions proposed in research studies have not always matched the observations that physicians make in the clinic.
TABLE 1.
REPRESENTATIVE RESEARCH ON GENE-BY-ENVIRONMENT INTERACTIONS IN ASTHMA OR ATOPY
| Genes | Environmental Exposure | First author, year (reference no.) |
|---|---|---|
| Genomewide linkage | Active smoke | Dizier, 2007 (72) |
| Colilla, 2003 (73) | ||
| CD14 | Choudhry, 2005 (74) | |
| β2-Adrenergic receptor | Wang, 2001 (75) | |
| Genome screen | Passive smoke | Meyers, 2005 (76) |
| IL-1R | Ramadas, 2007 (77) | |
| GSTM1 | In utero smoke | Gilliland, 2002 (78) |
| TNF | Ozone | Li, 2006 (79) |
| Genomewide linkage | Allergens | Blumenthal, 2006 (80) |
| CD14 | Farm | Leynaert (81) |
| Eder, 2005 (82) | ||
| LeVan, 2005 (83) | ||
| Toll-like receptor 2 | Eder, 2004 (84) | |
| Innate immunity receptors | Ege, 2006 (85) | |
| CD14 | Endotoxin | Simpson, 2006 (86) |
| Zambelli-Weiner, 2005 (87) | ||
| Williams, 2006 (88) | ||
| β2-Adrenergic receptor | Obesity | Barr, 2001 (89) |
| IL4RA, CD-14, and IL-13 | Diisocyanate | Bernstein, 2006 (90) |
| IL-4 | Helicobacter pylori infection | Pessi, 2005 (91) |
In 1992, the Barker hypothesis postulated that organs undergo developmental programming in utero that predetermines subsequent physiologic and metabolic adaptations during adult life. Early (including fetal) insults after nutritional and/or environmental exposures lead to a greater propensity to later disease (4–6). Early support for this hypothesis included evidence that severe fetal malnutrition was associated with an increased risk for multiple health problems throughout adulthood (5–7). Subsequent epidemiologic and experimental studies have found associations between the effects of a variety of prenatal environmental exposures, including allergens, antibiotics, and tobacco smoke, on disorders such as allergies, diabetes, neurodegenerative diseases, and cardiovascular diseases (5, 7, 8). This research inspired the fetal and early origins of adult disease hypothesis that proposes that prenatal or early postnatal environmental exposures influence developmental plasticity and result in altered programming. This programming is responsible for lasting functional changes of organs that lead to the development of a variety of complex diseases (reviewed in Reference 9).
Epigenetics is the study of heritable changes in gene expression that occur without directly altering the DNA sequence. One mechanism is DNA methylation, the covalent addition of a methyl group to a cytosine residue in a CpG site (i.e., where a cytosine lies next to guanine in the DNA sequence). CpG sites generally are clustered in high frequency near gene promoters and these regions are referred to as CpG islands. The methylation states of CpG islands in turn may affect gene activity and expression. Another epigenetic mechanism responsible for modulation of gene expression is post-translational modification of histones, including, but not limited to, acetylation, methylation, phosphorylation, and ubiquitylation (10, 11). Concurrent, multiple modifications of various histones create a complex pattern, often referred to as the histone code. These modifications permit transitions between chromatin states and alterations in transcriptional activity. DNA methylation usually works hand in hand with histone modifications to activate or silence genes by influencing chromatin structure and stability and therefore its accessibility by transcriptional factors (12). Both DNA methylation and histone modification are heritable from one cell generation to the next (13). As described by Callinan and Feinberg (12), potentially hundreds of methylated cytosines in multiple genes and dozens of post-translational chromatin modifications can arise. Epigenetic alterations are believed to occur predominantly prenatally and shortly after birth. However, recent evidence suggests they can occur during later periods (see below), influencing gene expression differentially throughout the lifespan. As such, epigenetic regulation provides an attractive mechanistic explanation, in addition to gene-by-environment interactions, for some of the molecular events linking early exposures with later disease.
Early seminal work performed by Cooney and colleagues, and by Waterland and Jirtle, demonstrated that dietary methyl supplementation during pregnancy with folic acid, vitamin B12, and other agents influenced the heritable phenotype of the agouti mice offspring (i.e., coat color distribution) via increased CpG methylation of the region upstream of the agouti gene (14, 15). Subsequent research demonstrated that the early postnatal diet induced epigenetic regulation of the imprinted gene lgf2, associated with several human cancers, in murine models (16, 17). Additional environmental exposures implicated in epigenetic regulation include xenobiotic chemicals, endocrine disruptors, maternal stress and nurturing, and low-dose radiation (reviewed in References 9 and 18). Importantly, epigenetic regulation, particularly DNA methylation, also appears to be inherited transgenerationally such that parental (or grandparental) exposures or phenotypes impact gene expression in the offspring (or offspring's offspring) without changing the DNA sequence. This phenomenon has been demonstrated for the kinked tail (AxinFu allele) and agouti-viable yellow (Avy) allele, and for associations between endocrine disrupters and male infertility, hormone-dependent cancer risk, and obesity (18).
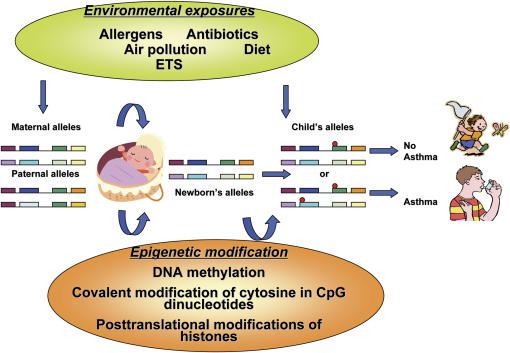
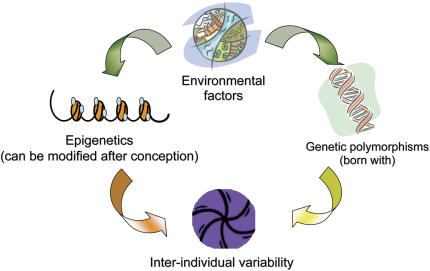
Arguably, the variable natural history of asthma (i.e., incidence and remission of symptoms) may be a result of epigenetic regulation after early or later environmental exposures (Figures 1 and 2). A large body of research has implicated specific time periods when individuals seem to be more susceptible to the effects of environmental exposures and other asthma triggers. These include prenatal development, early childhood, and adolescence (19–21). During these time periods, epigenetic modifications may be more likely to develop. In other work, adult rats underwent reversal of stable epigenetic programming associated with glucocorticoid receptor expression and hypothalamic–pituitary–adrenal and behavioral responses to stress after exposure to histone deacetylase (HDAC) inhibitors or methionine (i.e., donor of methyl groups for DNA methylation) (22). This study raises the question of whether adulthood, a period when asthma often remits (23), may also be a period when critical epigenetic changes can be deprogrammed.
Figure 1.
Environmental epigenetics and asthma. Red circles refer to sites of epigenetic changes. ETS = environmental tobacco smoke.
Figure 2.
Epigenetic and genetic interactions influence interindividual asthma risk.
This perspective will describe evidence addressing the hypothesis that asthma risk may be influenced by environmentally induced epigenetic changes. Several epidemiologic studies, including one that suggests transmission of asthma risk after maternal exposure to environmental tobacco smoke (ETS) may continue across multiple generations, will be described. Experimental studies also will be reviewed that provide substantial in vitro data demonstrating that DNA methylation of genes critical to T-helper (Th) differentiation may induce polarization toward or away from an allergic phenotype. Despite this early progress, fundamental questions remain that need to be addressed by well-designed research studies. Currently, data generated from controlled experiments using in vivo models and/or clinical specimens after environmental exposure monitoring are limited. Importantly, cohort-driven epigenetic research has the potential to address key questions, such as those concerning the influence of timing of exposure, dose of exposure, diet, and ethnicity, on susceptibility to asthma development.
PRENATAL ENVIRONMENTAL EXPOSURES, GENE INTERACTIONS, AND ASTHMA RISK
A preponderance of evidence from multiple large prospective studies indicates that prenatal exposure to ETS is associated with impaired respiratory function, transient wheeze, asthma, and/or respiratory infections in infants, young children, and adolescents (24, 25). It is evident that prenatal exposure to ETS alters airway structure, because greater distances between alveolar attachments in intraparenchymal airways have been measured among exposed infants who died of sudden infant death compared with unexposed infants (26). This body of work provides the most convincing evidence that prenatal environmental exposures can influence the risk for subsequent asthma. In addition to ETS, low maternal intake of foods containing vitamin E and zinc (27), or use of antibiotics during pregnancy (28), may increase the risk for childhood asthma. In contrast, maternal intake of probiotics, and higher levels of fruits, vegetables, and oily fish during pregnancy, may decrease the risk (29, 30). A prospective study from the Columbia Center for Children's Environmental Health showed that, at age 2 years, more difficulty breathing and probable asthma was reported among children jointly exposed to prenatal polycyclic aromatic hydrocarbons (PAHs) and postnatal ETS (31).
Experimental animal models also have shown that intrauterine exposure to airborne pollutants may increase the risk for respiratory disease in offspring. Hamada and colleagues developed a mouse model that demonstrated that exposure during pregnancy to residual oil fly ash (extracted from a precipitator unit that removes particulate contaminates of an oil-fired power plant) led to a greater susceptibility to an asthmalike phenotype in the offspring mice, including airway hyperresponsiveness and inflammation, such as eosinophil infiltration and goblet cell hyperplasia (32). In other experimental studies, prenatal exposure to endotoxin and probiotic bacteria protected mice against an asthmalike phenotype (33, 34). Most recently, prenatal exposure of mice to either diesel exhaust particles or control “inert” titanium dioxide led to greater airway inflammation and hyperresponsiveness in the offspring (35).
PARENT-OF-ORIGIN EFFECT
Another set of studies suggested that inheritance of particular genetic polymorphisms from the mother is more likely to be associated with asthma in the child than inheritance from the father. This parent-of-origin effect points to a significant role of the maternal prenatal environment and gene interactions on later asthma risk in the offspring. As examples, polymorphisms in the β-chain of the high-affinity receptor for IgE (FcɛRI-β) demonstrated stronger associations with positive allergy skin prick tests and greater allergen-specific IgE levels when they were inherited from the mother (36). In two separate cohorts, the HLA-G −964G allele was overexpressed in children with bronchial hyperresponsiveness if the mother was affected by bronchial hyperresponsiveness (37). The North Staffordshire cohort of subjects with mild asthma found that maternal, but not paternal, transmission of the glutathione-S-transferase P1 (GSTP1) Val105/Val105 and Ala114/Val114 genotypes was associated with greater lung function (i.e., FEV1/FVC) in children. The phenotype was not more frequent after maternal inheritance of GSTP1 Ile105/Ile105, Ile105/Val105, or Ala114/Ala114 (38). Moreover, maternal, but not paternal, history of asthma was associated with asthma among children younger than 5 years (39). Maternal, but not paternal, IgE was positively associated with elevated IgE levels in cord blood and at age 6 months (40). The preferential association of children's outcomes with maternal history of asthma or elevated IgE implies that the disease phenotype during pregnancy also may mediate the fetal response to environmental triggers and presumably the offspring's subsequent asthma risk.
MULTIGENERATIONAL TRANSMISSION OF ASTHMA
Recent data also suggest that the risk of ETS exposure on asthma may be transmitted across two generations. The Children's Health Study conducted a nested case-control study of 338 children with asthma who had been diagnosed by age 5 years versus 570 control subjects who were matched on in utero exposure to maternal smoking within grade, sex, and community of residence. Maternal and household smoking histories were obtained by telephone interviews. Grandmaternal smoking during the mother's fetal period may have been associated with a greater asthma risk in the grandchildren, independent of maternal smoking (odds ratio [OR], 1.8; 95% confidence interval [CI], 1.0–3.3). The risk for asthma in grandchildren was heightened if both the grandmother and mother smoked during pregnancy (OR, 2.6; 95% CI, 1.6–4.5) (41). Although much of the data were collected retrospectively, the findings lend support to the possibility that environmental exposures may alter asthma risk across generations.
EARLY POSTNATAL ENVIRONMENTAL EXPOSURES AND BOTH EARLY- AND LATE-ONSET ASTHMA RISK
In addition to prenatal exposures, multiple cohort studies suggest that early postnatal exposures modify the risk for developing later childhood or adult-onset asthma. As an example, exposure to dust mite allergen during infancy may be an important determinant for later childhood asthma (42). Exposure to dog or cat allergen is associated with protection from later childhood wheeze in some (43), but not all (44), cohort studies. Exposure during infancy to indoor combustion-related pollutants has been associated with later childhood sensitization to dust mite and a reduction in FEV1 (45). Despite these associations between early postnatal environmental exposures and later disease, direct links to epigenetic mechanisms have not been established to date.
The links between early environmental exposure and adult-onset asthma are even more indirect, but several studies suggest possible relationships. Very low birth weight and prematurity, two disease phenotypes that have been associated with prenatal exposure to pollutants such as ETS and PAHs (46, 47), were associated with reduced FEF25–75% (forced expiratory flow, midexpiratory phase) FEV1/FVC, chronic cough, wheeze, and asthma during subsequent adolescence (48). Two other studies reported that respiratory infections during infancy were associated independently with a greater incidence of chronic obstructive lung disease (49, 50). Combined, both studies point to an enduring effect of adverse environmental conditions experienced during fetal development (i.e., during growth of airways) and shortly afterward (during expansion of alveoli). Therefore, exposures during either of these critical periods for lung development appear to affect the risk for developing asthma at later life stages, including during childhood and later adulthood.
PROPOSED EPIGENETIC MECHANISMS
The traditional view that interindividual risk for asthma, like other complex diseases, is determined solely by interactions between genetic polymorphisms and environmental exposures needs to be reconciled with new findings suggesting that epigenetic mechanisms also may contribute. These mechanisms include genomic imprinting, histone modification, altered DNA methylation of regulatory sequences in Th and other genes, and regulation by microRNA (miRNA), which may change asthma risk after conception via environmentally mediated epigenetic disruption of gene expression (18). In genomic imprinting, unequal expression of the maternal and paternal alleles occurs, presumably due to reversible modification of gene activity in association with the sex of the parent. Differential DNA methylation of promoter regions of reprogrammable genes may be an important mechanism in establishing the imprint (reviewed in Reference 51). For example, infections incurred during pregnancy have been hypothesized to transmit transgenerationally the imprints of infections and inflammation, reduce the offspring's ability to withstand environmental pathogens, and in turn impact later morbidity and mortality (52). Direct mechanistic links to asthma pathogenesis still need to be determined.
Another proposed mechanism is induction of histone modifications, a process that is reversible and may be associated with chromatin remodeling and gene transcription. Oxidant-generating systems and proinflammatory mediators, some of which are implicated in asthma, may regulate histone acetylation (53). As an example, exposure to H2O2 (a reactive oxygen species) caused an increase in histone acetyltransferase (HAT) activity that promoted acetylation and induced chromatin remodeling (53). In other work, untreated subjects with asthma possessed greater levels of HAT and reduced levels of HDACs in bronchial biopsies, with levels that reversed after treatment with inhaled steroids (54). In addition, endotoxin exposure, associated with protection from developing atopy or asthma in several studies (55, 56), binds histones. It may disrupt chromatin remodeling as well and participate in gene silencing. The evidence is the finding in a septicemia cell model in which endotoxin exposure altered nuclear factor-κB binding and chromatin remodeling of the proinflammatory gene IL-1β promoter nucleosome (57).
Methylation of DNA and resulting changes in chromatin structure have been shown to initiate the process by which the Th cells lose their plasticity and differentiate productively toward the Th1 versus the proallergic Th2 pattern of cytokine gene expression. Demethylation of sites at the proximal promoter and conserved intronic regulatory element (CIRE) in the first intron of the proallergic IL-4 gene, and hypermethylation of sites in the counterregulatory IFN-γ promoter, all result in greater IL-4 production and Th2 differentiation (58–61). Methylation of a highly conserved DNaseI-hypersensitive region at the 3′ end of IL-4 is associated with Th1 differentiation (58). Interestingly, many CpG sites found in Th genes are highly conserved across species, including CpG−53 and CpG−190 of the IFN-γ promoter (61, 62). CpG−53 resides in a proximal activator protein 1 (AP1)-binding site, and when methylated, can change transcription factor binding (63). In addition, methylation of CpG−53 in the IFN-γ promoter was associated with Th2 polarization after inhibition of cAMP response element binding protein (CREB) and activating transcription factor 2 (ATF2)/c-Jun binding to the CpG-containing AP1 site (61). Therefore, mounting evidence from in vitro experimental studies suggests that the development of a polarized Th2 phenotype associated with atopy may result from altered DNA methylation at genes regulating Th differentiation. Moreover, recent work by our group demonstrated that inhaled diesel exposure and intranasal Aspergillus fumigatus induced hypermethylation at the CpG−45, CpG−53, and CpG−205 sites of the IFN-γ promoter and hypomethylation at CpG−408 of the IL-4 promoter in mice. Altered methylation of both gene promoters was correlated significantly with changes in IgE levels, suggesting that inhaled environmental exposures can induce methylation of Th genes and IgE regulation in vivo (64). In addition, ETS has been shown to induce gene hypermethylation in a gene associated with lung cancer (i.e., p16 [INK4a] tumor suppressor gene involved in cell cycle control) (65). Whether these observations occur in association with clinical asthma still needs to be elucidated.
A rapidly emerging area of epigenetics research relates to the role played by miRNA in regulating asthma-related gene expression. Since the first miRNA, Lin 4, was discovered in 1993 (66), over 300 miRNAs have been identified in humans. Each miRNA may regulate up to 200 target genes by blocking the translation of the target protein (67). These molecules are believed to regulate up to one-third of all human genes by promoting the degradation of target messenger RNA. Aberrant expression of miRNA has been shown to contribute to the pathogenesis of many human diseases and may serve as valuable diagnostic or prognostic disease markers (68). However, studies relevant to asthma or asthma risk are still lacking, except for a recent report demonstrating that a single nucleotide polymorphism at the 3′ untranslated region of HLA-G, an asthma-susceptibility gene, affects the binding of three miRNAs to this gene (69, 70). This report raises the exciting possibility of a gene-by-epigenetics interaction. Perhaps future discoveries will implicate specific environmental exposures that might disrupt miRNA expression patterns in cells and tissues relevant to asthma pathogenesis.
WHAT ARE THE KEY RESEARCH QUESTIONS?
The first important question for epigenetic studies is to determine whether any in vivo environmental exposure can induce epigenetic alterations that underlie asthma. The clinical presentation of asthma is mediated by exposure to allergens, air pollutants, endotoxin, viruses, and ETS. Which of these are important? What are the effects of exposure dose or diet (i.e., methyl donors)?
Second, when are the time periods during fetal development and afterward when individuals are more susceptible to epigenetic modifications that influence the clinical course of asthma? How do they differ by environmental exposure, and how do they explain the induction or remission of asthma that is observed at different time points and ages? Can epigenetic changes be reversed?
Third, how do genetics and epigenetics interact? For example, how do polymorphisms in asthma-related genes, many of which have been identified (Table 1), interact with epigenetic mechanisms to confer heightened susceptibility to environmental influences?
Fourth, can epigenetic changes provide measurable biomarkers that may predict later asthma and greater need for immediate intervention? Can one identify an epigenome that is susceptible to, for example, air pollution–related asthma, and enact specific interventions to reduce asthma risk?
POTENTIAL APPROACHES TO FUTURE EPIGENETIC STUDIES
Both in vivo experimental animal models and cohort-driven epigenetic research have great potential to answer many of the above questions. Mouse models may provide the experimental controls needed to examine the main effects of a variety of airborne exposures, including allergens and air pollutants, on selected candidate asthma genes. The modifying effects of diet enriched with methyl donors or HDAC inhibitors also can be tested. In addition, mice of different genetic background and those with unique transgenes could illuminate further these issues.
More exciting may be the potential to apply newly developing technologies to epidemiologic studies. Longitudinal cohort studies that feature repeated measures of specific environmental exposures, collection of biological specimens over time, and comprehensive clinical outcomes assessment have enormous potential to add a wealth of information on specific environmental triggers and critical time windows of susceptibility. In partnership with global, unbiased screening methods, such as methylation-sensitive restriction fingerprinting, restriction landmark genomic scanning, methylation of CpG island amplification–representational difference analysis, and methylation target microarrays to identify candidate regions, as well as validation assays to reassess genes of interest (71), epigenomes susceptible to ETS-induced asthma, for example, may be identifiable in the near future.
It is unlikely that any individual study will be able to account for all of the potentially confounding factors, limiting one's ability to examine epigenetic pathways in observational studies. These include environment-by-environment interactions, traditional gene-by-environment interactions, effects of birth order, and other host factors such as obesity. Despite these limitations, there is great promise that the study of environmental epigenetics will help us understand a theoretically preventable environmental disease.
Acknowledgments
The authors thank Drs. Frederica Perera, Julie Herbstman, and Matthew Perzanowski for their input.
Supported by National Institutes of Health grants ES013063, ES013163, ES015905, ES013071, ES015584, ES05008, ES006096, and ES013071.
Originally Published in Press as DOI: 10.1164/rccm.200710-1511PP on January 10, 2008
Conflict of Interest Statement: Neither author has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Los H, Postmus PE, Boomsma DI. Asthma genetics and intermediate phenotypes: a review from twin studies. Twin Res 2001;4:81–93. [DOI] [PubMed] [Google Scholar]
- 2.Venn AJ, Lewis SA, Cooper M, Hubbard R, Britton J. Living near a main road and the risk of wheezing illness in children. Am J Respir Crit Care Med 2001;164:2177–2180. [DOI] [PubMed] [Google Scholar]
- 3.Hoppin JA, Umbach DM, London SJ, Alavanja MCR, Sandler DP. Diesel exhaust, solvents, and other occupational exposures as risk factors for wheeze among farmers. Am J Respir Crit Care Med 2004;169:1308–1313. [DOI] [PubMed] [Google Scholar]
- 4.Lau C, Rogers JM. Embryonic and fetal programming of physiological disorders in adulthood. Birth Defects Research C Embryo Today 2004;72:300–312. [DOI] [PubMed] [Google Scholar]
- 5.Hales C, Barker D. The thrifty phenotype hypothesis. Br Med Bull 2001;60:5–20. [DOI] [PubMed] [Google Scholar]
- 6.Hales C, Barker D, Clark P, Cox L, Fall C, Osmond C, Winter P. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 1991;303:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKeever T, Lewis SA, Smith C, Hubbard R. The importance of prenatal exposures on the development of allergic disease: a birth cohort study using the West Midlands General Practice Database. Am J Respir Crit Care Med 2002;166:827–832. [DOI] [PubMed] [Google Scholar]
- 8.Mone S, Gillman MW, Miller TL, Herman EH, Lipshultz SE. Effects of environmental exposures on the cardiovascular system: prenatal period through adolescence. Pediatrics 2004;113:1058–1069. [PubMed] [Google Scholar]
- 9.Dolinoy D, Weidman J, Jirtle R. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 2007;23:297–307. [DOI] [PubMed] [Google Scholar]
- 10.Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev 2006;16:125–136. [DOI] [PubMed] [Google Scholar]
- 11.Krebs JE. Moving marks: dynamic histone modifications in yeast. Mol Biosyst 2007;3:590–597. [DOI] [PubMed] [Google Scholar]
- 12.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet 2006;15:R95–R101. [DOI] [PubMed] [Google Scholar]
- 13.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 2004;15:595–605. [DOI] [PubMed] [Google Scholar]
- 14.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 2002;132:2393S–2400S. [DOI] [PubMed] [Google Scholar]
- 15.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg A, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143–153. [DOI] [PubMed] [Google Scholar]
- 17.Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol 1999;154:635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang W, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord 2007;8:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilliland FD, Berhane K, Li Y-F, Rappaport EB, Peters JM. Effects of early onset asthma and in utero exposure to maternal smoking on childhood lung function. Am J Respir Crit Care Med 2003;167:917–924. [DOI] [PubMed] [Google Scholar]
- 20.Kurukulaaratchy RJ, Matthews S, Arshad SH. Does environment mediate earlier onset of the persistent childhood asthma phenotype? Pediatrics 2004;113:345–350. [DOI] [PubMed] [Google Scholar]
- 21.Mandhane PJ, Greene JM, Cowan JO, Taylor DR, Sears MR. Sex differences in factors associated with childhood- and adolescent-onset wheeze. Am J Respir Crit Care Med 2005;172:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver I, Meaney M, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA 2006;103:3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marco RD, Locatelli F, Cerveri I, Bugiani M, Marinoni A, Giammanco G. Incidence and remission of asthma: a retrospective study on the natural history of asthma in Italy. J Allergy Clin Immunol 2002;110:228–235. [DOI] [PubMed] [Google Scholar]
- 24.Magnusson L, Olesen A, Wennborg H, Olsen J. Wheezing, asthma, hayfever, and atopic eczema in childhood following exposure to tobacco smoke in fetal life. Clin Exp Allergy 2005;35:1550–1556. [DOI] [PubMed] [Google Scholar]
- 25.Alati R, Al-Mamun A, O'Callaghan M, Najman JM, Williams GM. In utero and postnatal maternal smoking and asthma in adolescence. Epidemiology 2006;17:138–144. [DOI] [PubMed] [Google Scholar]
- 26.Elliot JG, Carroll NG, James AL, Robinson PJ. Airway alveolar attachment points and exposure to cigarette smoke in utero. Am J Respir Crit Care Med 2003;167:45–49. [DOI] [PubMed] [Google Scholar]
- 27.Devereux G, Turner SW, Craig LC, McNeill G, Martindale S, Harbour PJ, Helms PJ, Seaton A. Low maternal vitamin E intake during pregnancy is associated with asthma in 5-year-old children. Am J Respir Crit Care Med 2006;174:499–507. [DOI] [PubMed] [Google Scholar]
- 28.Jedrychowski W, Galas A, Whyatt R, Perera F. The prenatal use of antibiotics and the development of allergic disease in one year old infants: a preliminary study. Int J Occup Med Environ Health 2006;19:70–76. [DOI] [PubMed] [Google Scholar]
- 29.Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, Tuure T, Kuitunen M. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol 2007;119:192–198. [DOI] [PubMed] [Google Scholar]
- 30.Fitzsimon N, Fallon U, O'Mahony D, Loftus BG, Bury G, Murphy AW, Kelleher CC. Mothers' dietary patterns during pregnancy and risk of asthma symptoms in children at 3 years. Ir Med J 2007;100(Suppl):27–32. [PubMed] [Google Scholar]
- 31.Miller RL, Garfinkel R, Horton M, Camann D, Perera F, Whyatt RM, Kinney PL. Polycyclic aromatic hydrocarbons, environmental tobacco smoke, and respiratory symptoms in an inner-city birth cohort. Chest 2004;126:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada K, Suzaki Y, Leme A, Ito T, Miyamoto K, Kobzik L, Kimura H. Exposure of pregnant mice to an air pollutant aerosol increases asthma susceptibility in offspring. J Toxicol Environ Health 2007;70:688–695. [DOI] [PubMed] [Google Scholar]
- 33.Blumer N, Herz U, Wegmann M, Renz H. Prenatal lipopolysaccharide-exposure prevents allergic sensitization and airway inflammation, but not airway responsiveness in a murine model of experimental asthma. Clin Exp Allergy 2005;35:397–402. [DOI] [PubMed] [Google Scholar]
- 34.Blumer N, Sel S, Virna S, Patrascan C, Zimmermann S, Herz U, Renz H, Garn H. Perinatal maternal application of Lactobacillus rhamnosus GG suppresses allergic airway inflammation in mouse offspring. Clin Exp Allergy 2007;37:348–357. [DOI] [PubMed] [Google Scholar]
- 35.Fedulov AV, Leme A, Yang Z, Dahl M, Lim R, Mariani TJ, Kobzik L. Pulmonary exposure to particles during pregnancy causes increased neonatal asthma susceptibility. Am J Respir Cell Mol Biol 2008;38:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traherne JA, Hill MR, Hysi P, D'Amato M, Broxholme J, Mott R, Moffatt MF, Cookson WO. LD mapping of maternally and non-maternally derived alleles and atopy in Fc epsilon RI-beta. Hum Mol Genet 2003;12:2577–2585. [DOI] [PubMed] [Google Scholar]
- 37.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet 2005;763:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll WD, Lenney W, Child F, Strange RC, Jones PW, Fryer AA. Maternal glutathione s-transferase gstp1 genotype is a specific predictor of phenotype in children with asthma. Pediatr Allergy Immunol 2005;16:32–39. [DOI] [PubMed] [Google Scholar]
- 39.Litonjua A, Carey VJ, Burge HA, Weiss ST, Gold DR. Parental history and the risk for childhood asthma: does mother confer more risk than father? Am J Respir Crit Care Med 1998;158:176–181. [DOI] [PubMed] [Google Scholar]
- 40.Liu CA, Wang CL, Chuang H, Ou CY, Hsu TY, Land KD. Prenatal prediction of infant atopy by maternal but not paternal total IgE levels. J Allergy Clin Immunol 2003;112:899–904. [DOI] [PubMed] [Google Scholar]
- 41.Li Y-F, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 2005;127:1232–1241. [DOI] [PubMed] [Google Scholar]
- 42.Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ. Exposure to house-dust mite allergen (Der p i) and the development of asthma in childhood: a prospective study. N Engl J Med 1990;323:502–507. [DOI] [PubMed] [Google Scholar]
- 43.Remes ST, Castro-Rodriguez JA, Holberg CJ, Martinez FD, Wright AL. Dog exposure in infancy decreases the subsequent risk of frequent wheeze but not of atopy. J Allergy Clin Immunol 2001;108:509–515. [DOI] [PubMed] [Google Scholar]
- 44.Lau S, Illi S, Platts-Mills TA, Riposo D, Nickel R, Grüber C, Niggemann B, Wahn U. Longitudinal study on the relationship between cat allergen and endotoxin exposure, sensitization, cat-specific IgG and development of asthma in childhood–report of the German Multicentre Allergy Study (MAS 90). Allergy 2005;60:766–773. [DOI] [PubMed] [Google Scholar]
- 45.Ponsonby AL, Dwyer T, Kemp A, Couper D, Cochrane J, Carmichael A. A prospective study of the association between home gas appliance use during infancy and subsequent dust mite sensitization and lung function in childhood. Clin Exp Allergy 2001;31:1544–1552. [DOI] [PubMed] [Google Scholar]
- 46.Perera F, Rauh V, Tsai W-Y, Kinney P, Camann D, Barr D, Bernert T, Garfinkel R, Tu Y-h, Diaz D, et al. Effects of transplacental exposure to environmental pollutants on birth outcomes in a multiethnic population. Environ Health Perspect 2003;111:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Windham GC, Hopkins B, Fenster L, Swan SH. Prenatal active or passive tobacco smoke exposure and the risk of preterm delivery or low birth weight. Epidemiology 2000;11:427–433. [DOI] [PubMed] [Google Scholar]
- 48.Anand D, Stevenson CJ, West CR, Pharoah PO. Lung function and respiratory health in adolescents of very low birth weight. Arch Dis Child 2003;88:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker DJP, Godfrey KM, Fall C, Osmond C, Winter PD, Shaheen SO. Relation of birth weight and childhood respiratory infection to adult lung function and death from chronic obstructive airways disease. BMJ 1991;303:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaheen SO, Barker DJ, Shiell AW, Crocker FJ, Wield GA, Holgat ST. The relationship between pneumonia in early childhood and impaired lung function in late adult life. Am J Respir Crit Care Med 1994;149:616–619. [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y-h, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet 2004;5:479–510. [DOI] [PubMed] [Google Scholar]
- 52.Finch CE, Crimmins EM. Inflammatory exposure and historical changes in human life-spans. Science 2004;305:1736–1739. [DOI] [PubMed] [Google Scholar]
- 53.Rahman I. Oxidative stress, chromatin remodeling and gene transcription in inflammation and chronic lung diseases. J Biochem Mol Biol 2003;36:95–109. [DOI] [PubMed] [Google Scholar]
- 54.Ito K, Caramori G, Lim S, Oates T, Chung K, Barnes P, Adcock I. Expression and activity of histone deacetylases in human asthmatic airway. Am J Respir Crit Care Med 2002;166:392–396. [DOI] [PubMed] [Google Scholar]
- 55.Perzanowski MS, Miller RL, Thorne PS, Barr RG, Divjan A, Sheares BJ, Garfinkel RS, Perera FP, Goldstein IF, Chew GL. Endotoxin in inner-city homes and the association with atopy and wheeze in the first two years of life. J Allergy Clin Immunol 2006;117:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, Maisch S, Carr D, Gerlach F, Bufe A, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med 2002;347:869–877. [DOI] [PubMed] [Google Scholar]
- 57.Chan C, Li L, McCall CE, Yoza BK. Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB transactivation at the IL-1beta promoter. J Immunol 2005;175:461–468. [DOI] [PubMed] [Google Scholar]
- 58.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 2002;16:649–660. [DOI] [PubMed] [Google Scholar]
- 59.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 1998;9:765–775. [DOI] [PubMed] [Google Scholar]
- 60.Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, et al. A critical control element for interleukin-4 memory expression in t helper lymphocytes. J Biol Chem 2005;280:28177–28185. [DOI] [PubMed] [Google Scholar]
- 61.Jones B, Chen J. Inhibition of IFN-g transcription by site-specific methylation during T helper cell development. EMBO J 2006;25:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-g promoter at CPG and non-CPG sites underlie differences in IFN-g gene expression between human neonatal and adult CD45ro- T cells. J Immunol 2002;168:2820–2827. [DOI] [PubMed] [Google Scholar]
- 63.Young H, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard J, Penix L, Wilson C, Melvin A, McGurn M, et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol 1994;153:3603–3610. [PubMed] [Google Scholar]
- 64.Liu J, Ballaney M, Al-Alem U, Quan C, Jin X, Perera F, Chen LC, Miller RL. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci 2008;102:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Digel W, Lübbert M. DNA methylation disturbances as novel therapeutic target in lung cancer: preclinical and clinical results. Crit Rev Oncol Hematol 2005;55:1–11. [DOI] [PubMed] [Google Scholar]
- 66.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene LIN-4 encodes small RNAs with antisense complementarity to LIN-14. Cell 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 67.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, Piedade ID, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500. [DOI] [PubMed] [Google Scholar]
- 68.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189–198. [DOI] [PubMed] [Google Scholar]
- 69.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J 2005;19:681–693. [DOI] [PubMed] [Google Scholar]
- 70.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 2007;81:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ho S-m, Tang W-y. Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod Toxicol 2007;23:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dizier M, Bouzigon E, Guilloud-Bataille M, Siroux V, Lemainque A, Boland A, Lathrop M, Demenais F. Evidence for gene × smoking exposure interactions in a genome-wide linkage screen of asthma and bronchial hyper-responsiveness in EGEA families. Eur J Hum Genet 2007;15:810–815. [DOI] [PubMed] [Google Scholar]
- 73.Colilla S, Nicolae D, Pluzhnikov A, Blumenthal M, Beaty T, Bleecker E, Lange E, Rich S, Meyers D, Ober C, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol 2003;111:840–846. [DOI] [PubMed] [Google Scholar]
- 74.Choudhry SP, Avila PC, Nazario S, Ung J, Kho J, Rodriguez-Santana R, Csal J, Tsai HJ, Torres A, Ziv E, et al. CD14 tobacco gene–environment interaction modifies asthma severity and immunoglobulin E levels in Latinos with asthma. Am J Respir Crit Care Med 2005;172:173–182. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Chen C, Niu T, Wu D, Yang J, Wang B, Fang Z, Yandava C, Drazen J, Weiss S, et al. Association of asthma with β2-adrenergic receptor gene polymorphism and cigarette smoking. Am J Respir Crit Care Med 2001;163:1404–1449. [DOI] [PubMed] [Google Scholar]
- 76.Meyers D, Postma D, Stine O, Koppelman G, Ampleford E, Jongepier H, Howard T, Bleecker E. Genome screen for asthma and bronchial hyperresponsiveness: interactions with passive smoke exposure. J Allergy Clin Immunol 2005;115:1169–1175. [DOI] [PubMed] [Google Scholar]
- 77.Ramadas R, Sadeghnejad A, Karmaus W, Arshad S, Matthews S, Huebner M, Kim D, Ewart S. Interleukin-1R antagonist gene and pre-natal smoke exposure are associated with childhood asthma. Eur Respir J 2007;29:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gilliland FD, Li Y-F, Dubeau L, Berhane K, Avol E, McConnell R, Gauderman WJ, Peters JM. Effects of glutathione S-transferase M1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med 2002;166:457–463. [DOI] [PubMed] [Google Scholar]
- 79.Li YF, Gauderman WJ, Avol E, Dubeau L, Gilliland FD. Associations of tumor necrosis factor g-308a with childhood asthma and wheezing. Am J Respir Crit Care Med 2006;173:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blumenthal M, Langefeld C, Barnes K, Ober C, Meyers D, King R, Beaty T, Beck S, Bleecker E, Rich S. A genome-wide search for quantitative trait loci contributing to variation in seasonal pollen reactivity. J Allergy Clin Immunol 2006;117:79–85. [DOI] [PubMed] [Google Scholar]
- 81.Leynaert B, Guilloud-Bataille M, Soussan D, Benessiano J, Guenegou A, Pin I, Neukirch F. Association between farm exposure and atopy, according to the CD14 C-159t polymorphism. J Allergy Clin Immunol 2006;118:658–665. [DOI] [PubMed] [Google Scholar]
- 82.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD. Opposite effects of CD 14/-260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol 2005;116:601–607. [DOI] [PubMed] [Google Scholar]
- 83.LeVan T, Essen SV, Romberger D, Lambert G, Martinez F, Vasquez M, Merchant J. Polymorphisms in the CD14 gene associated with pulmonary function in farmers. Am J Respir Crit Care Med 2005;171:773–779. [DOI] [PubMed] [Google Scholar]
- 84.Eder W, Klimecki W, Yu L, Mutius Ev, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol 2004;113:482–488. [DOI] [PubMed] [Google Scholar]
- 85.Ege MJ, Bieli C, Frei R, Strien RTv, Riedler J, Ublagger E, Schram-Bijkerk D, Brunekreef B, Hage Mv, Scheynius A, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol 2006;117:817–823. [DOI] [PubMed] [Google Scholar]
- 86.Simpson A, John S, Jury F, Niven R, Woodcock A, Ollier WE, Custovic A. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med 2006;174:386–392. [DOI] [PubMed] [Google Scholar]
- 87.Zambelli-Weiner A, Ehrlich E, Stockton ML, Grant AV, Zhang S, Levett PN, Beaty TH, Barnes KC. Evaluation of the CD14/-260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. J Allergy Clin Immunol 2005;115:1203–1209. [DOI] [PubMed] [Google Scholar]
- 88.Williams L, McPhee R, Ownby D, Peterson E, James M, Zoratti E, Johnson C. Gene-environment interactions with CD14 c-260t and their relationship to total serum IgE levels in adults. J Allergy Clin Immunol 2006;118:851–857. [DOI] [PubMed] [Google Scholar]
- 89.Barr RG, Cooper DM, Speizer FE, Drazen JM, Camargo CA. Beta(2)-adrenoceptor polymorphism and body mass index are associated with adult-onset asthma in sedentary but not active women. Chest 2001;120:1474–1479. [DOI] [PubMed] [Google Scholar]
- 90.Bernstein D, Wang N, Campo P, Chakraborty R, Smith A, Cartier A, Boulet L, Malo J, Yucesoy B, Luster M, et al. Diisocyanate asthma and gene-environment interactions with IL4ra, CD-14, and IL-13 genes. Ann Allergy Asthma Immunol 2006;97:800–806. [DOI] [PubMed] [Google Scholar]
- 91.Pessi T, Virta M, Adjers K, Karjalainen J, Rautelin H, Kosunen TU, Hurme M. Genetic and environmental factors in the immunopathogenesis of atopy: interaction of Helicobacter pylori infection and IL4 genetics. Int Arch Allergy Immunol 2005;137:282–288. [DOI] [PubMed] [Google Scholar]