Long-Range Communication between the Silencers of HMR (original) (raw)
Abstract
Gene regulation involves long-range communication between silencers, enhancers, and promoters. In Saccharomyces cerevisiae, silencers flank transcriptionally repressed genes to mediate regional silencing. Silencers recruit the Sir proteins, which then spread along chromatin to encompass the entire silenced domain. In this report we have employed a boundary trap assay, an enhancer activity assay, chromatin immunoprecipitations, and chromosome conformation capture analyses to demonstrate that the two HMR silencer elements are in close proximity and functionally communicate with one another in vivo. We further show that silencing is necessary for these long-range interactions, and we present models for Sir-mediated silencing based upon these results.
Gene activation and gene repression are central to the proper development and differentiation of organisms. DNA elements such as promoters, enhancers, and silencers play a central role in eukaryotic gene regulation. These elements are separated from each other by several kilobase pairs of DNA but are able to communicate with one another to regulate the activation or repression of genes. The exact mechanism by which distally located elements communicate with one another is not clear and is one of the key questions in gene regulation. Long-range communication between distantly located elements in chromosomes is thought to occur by one of two principal mechanisms (8). One class of models postulate that a signal emanating from a distal regulatory element spreads along the DNA fiber until it encounters a proximal regulatory element. A second class of models postulate that distal and proximal regulatory elements interact with one another directly, with the intervening DNA forming a loop. Both mechanisms must function within the context of the global chromosome structure, which appears to be composed of large chromosome loops that attach to a proteinaceous superstructure (11). The nucleus appears to be divided further into distinct chromatin compartments, with heterochromatic domains being present in regions near the nuclear periphery while euchromatic domains are found mainly in the interior of the nucleus, although a significant portion of euchromatin is located near nuclear pores.
It has been suggested that the functionally and structurally defined chromatin domains may be coincident (36). Enhancers and locus control regions (LCRs) are long-range regulatory elements that activate promoters in a distance- and orientation-independent manner, and recent studies indicate that enhancers and LCRs often cluster together in three-dimensional space to form an “active chromatin hub” (23, 49, 58, 59). Similarly, in yeast the promoters and terminators of genes are in close proximity to one another (3, 48) and tethered to the nuclear pore (10, 52). The consequence of this spatial organization is that the DNA between these regulatory elements is looped out. It is thought that the formation of these nuclear substructures aids in transcription activation.
Silencers are negative regulatory elements composed of binding sites for various factors that act collectively in the establishment and stable inheritance of a repressed state. Like enhancers, silencers repress promoters in a distance- and orientation-independent manner (35). Silencers flank the silenced HML and HMR mating-type loci in yeast, while at telomeres the terminal repeated TG1-3 sequences serve as silencers. These silencers recruit the Sir proteins, Sir2p, Sir3p, and Sir4p, which then spread across several kilobase pairs of DNA via interactions with histones. Thus, our current understanding of Sir-mediated repression is that it is an example of long-range effects mediated via transmission along the DNA fiber rather than direct long-range interactions between the silencers (60).
DNA elements that restrict the action of long-distance regulatory elements, such as silencers and enhancers, are generically called insulators. Insulators located between an enhancer and a promoter (called enhancer blockers) disrupt enhancer-promoter communication and prevent the enhancer from activating that promoter, while insulators located between a silencer and a promoter (called barriers) block the silencer from repressing the promoter. Numerous models have been proposed to explain how insulators function to block long-range communication. Some models postulate that insulators act as decoys, forming nonproductive interactions with distal regulatory complexes, or sequester these complexes in specific regions of the nucleus, while other models suggest that insulators function locally by disrupting the propagation of a specific chromatin domain (60).
In this paper we present evidence demonstrating that while silencers function via recruitment and transmission of Sir proteins along the DNA, they also directly communicate with each other. Functional analyses of silencer-mediated repression suggest that silencer elements communicate with one another in mediating repression in the nucleus. Our studies also show that DNA fragments containing silencers (separated by several kilobase pairs of DNA) are in close spatial proximity in the nucleus and likely form chromatin loops. Interestingly, this long-range communication was lost in mutants of the Sir proteins. Our results suggest that silenced domains are formed by the spreading of repressor proteins from silencers that interact with one another enabling, compaction of the chromatin fiber and stable repression. These results are similar to the long-range interactions between LCRs and promoters and suggest conservation in the mechanism by which genes are activated and repressed.
MATERIALS AND METHODS
The genotypes of the strains, the oligonucleotides, and the exact sequences of the various integrations generated and used in this study will be provided upon request.
Yeast strains.
Yeast genomic integrations were performed by homologous recombination and gene replacement, using PCR products or DNA fragments derived from plasmids. Yeast transformations used the lithium acetate method (34). PCR amplifications were carried out with Expand high-fidelity DNA polymerase, and integrations were confirmed by PCR and sequencing analysis. SIR2, SIR3, PPR1, and ADE2 genes were deleted from the start to the stop codon and replaced with HIS3 or kanMX markers. Deletion of _MAT_α was obtained by replacing MAT_α_2 and MAT_α_1 sequences (SGD coordinates 199731 to 200964) with the kanMX cassette.
The ADE2 gene flanked by Gal4p binding sites (Gbs), present at HML, in strain KIY54 (32) was PCR amplified with appropriate primers and integrated at the HMR locus in a _sir2_Δ strain (JRY4576) or in an _HMR_Δ_I sir4_Δ strain (ROY926). Sequencing of the PCR products indicated that a single Gal4p binding site, which is contrary to published results (32), flanked ADE2. The PCR product was integrated in the HMR_a_2 coding region (SGD coordinates 293212 to 293410) with the ADE2 promoter close to the HMR-E silencer. Strains with the integrated ADE2 gene (ROY2729 [_MAT_α HMR::_Gbs-ADE2-Gbs sir2_Δ] and ROY2914 [_MAT_α _HMR-E-Gbs-ADE2-Gbs-HMR_Δ_I sir4_Δ]) were crossed with a W303 wild-type strain to obtain ROY2770 and ROY3001, respectively.
The HMRa1 coding region in ROY2729 and ROY2914 was replaced by the URA3 coding region by homologous recombination, and transformants were crossed with an _ade2_Δ::kanMX strain to obtain ROY3182 and ROY3194, respectively.
Plasmid pJR1270 contains an EcoRI-HindIII fragment with the HMR locus where the HMR-I silencer has been deleted. This fragment contains two SpeI sites. The plasmid was partially digested with SpeI, end filled, and religated to obtain pRO698, which contains only one SpeI site 290 bp upstream of the ARS element present at HMR-E. A pair of oligonucleotides with four Gbs flanked by SpeI sites were annealed, digested, and cloned into the SpeI site in pRO698 to produce pRO700. The EcoRI-BglII fragment from pRO700 was used to replace the HMR region in strain ROY2585 (HMRa2::_URA3 sir2_Δ), to give strain ROY3285 (_Gbs-HMR sir2_Δ).
The EcoRI-HindIII fragment in pRO700 was used to replace the HMR region in ROY2585 to produce ROY3283 (Gbs-_HMR_Δ_I sir2_Δ). The URA3 cassette was integrated between HMR-I or HMRΔI and the tRNA gene (SGD coordinates 295070 to 295281) in strains JRY4566 (W303 _sir2_Δ), ROY3285, ROY3550 (_HMR_Δ_I sir2_Δ), and ROY3283, and the transformants were crossed with a ppr1 Δ::kanMX strain to obtain ROY3495 to -3489, -3683, -3699, -3680, and -3697. ROY3495 and -3497 were crossed with a Gal4-TAP-tagged strain to obtain ROY4371 and -4372, respectively.
The tRNA gene was deleted and the URA3 gene integrated in strain JRY4566 and ROY3285 by PCR-mediated gene replacement. Transformants were then crossed with a _ppr1_Δ strain to obtain ROY3688, -3703, -3686, and -3701.
Plasmid pJR1571 contains an EcoRI-HindIII fragment comprising the HMR locus. Oligonucleotides with three new Sau3A sites were used to PCR amplify the XbaI-EcoNI fragment of the MATa2 gene. The PCR product was cloned into the XbaI-EcoNI fragment of HMRa2 in plasmid pJR1571. The EcoRI-HindIII fragment of the new construct (pOS154) was used to replace the HMR locus in strain ROY2800 (HMRa2::URA3), and transformants were crossed with _mat_αΔ::kanMX, _sir3_Δ::HIS3, and _hml_Δ::TRP1 strains to obtain ROY4064 and ROY4065.
Plasmids.
Plasmids RO590 and RO635 contained the full-length SAS2 or NUP2 coding regions fused in frame to the Gal4 DNA binding domain (GBD), with transcription driven by the ADH1 promoter in the pGBK-RC-TRP1 base plasmid (pGBD) (47).
Serial dilutions.
Yeast cells were grown overnight at 30°C in 5 ml YPAD or Hartwell's complete (HC)) medium without tryptophan to allow maintenance of the plasmids. Cells were diluted to an _A_600 of 1.0 unit/ml in HC-trp medium and serially diluted 5- or 10-fold. Using a cell spotter, approximately 3 μl of each serial dilution was placed onto properly supplemented HC plates to assay for ADE2 and URA3 expression, or onto properly supplemented YMD plates previously spread with 1.0 _A_600 unit of mating lawn (strain JRY19a) diluted in 300 μl of YPD, for the mating assays. For the mating assays, selection for plasmids was maintained. The plates were incubated at 30°C and photographed. Cells grown in limiting amounts of adenine were kept at 4°C for an additional 2 days for development of the color prior to photography.
ChIP.
Quantitative chromatin immunoprecipitation (ChIP) analysis was performed as previously described (46), with minor modifications. The program for the PCR was as follows: 95°C for 3 min (1 cycle), and 95°C for 1 s, 52°C for 30 s, and 72°C for 1 min (45 cycles). The fold enrichment was calculated using the formula 2_CT_(IP) − CT(input) as described previously (40a) and was normalized to the telomeric probe. For Rap1p immunoprecipitation, polyclonal antibodies (Y300; Santa Cruz Biotechnology, Inc.) were used, while antibodies against H4K16Ac were purchased from Upstate.
3C.
The chromosome conformation capture (3C) analyses of yeast strains were performed exactly as described previously (18) with a few specific changes. Each strain in each experiment was cross-linked for 0, 5, 10, and 20 min, and each sample was independently processed and analyzed. The restriction enzyme used for the digestion was Sau3A, and the digestion buffers were as recommended by the manufacturer of the enzyme. All primers used were tested with un-cross-linked/ligated DNA, and only primers with equal amplification efficiencies were used for the 3C analyses.
Fluorescence analysis.
Two diploid strains were constructed to visualize the relative positions of HMR and the nucleolus. The strains differ only by the absence of the tRNA barrier at HMR. The lac operator array is telomere proximal to HMR-I. Both strains contain Sik1p fused to red fluorescent protein (RFP) (a nucleolar marker), express _lac_-green fluorescent protein (_lac_-GFP), and contain a galactose inducible R recombinase.
To examine the relative positions of HMR with respect to the nucleolus, the cells were first grown in SC-trp medium containing dextrose. Cells were fixed 2 h with paraformaldehyde and mounted on microscope slides containing agar plugs. Parallel Z stacks of cells were obtained using both rhodamine and GFP filters to visualize the nucleolus and HMR, respectively (17 sequential images separated by 0.2 μm). The two landmarks were considered colocalized if the corresponding fluorescence signals fully overlapped within the same plane or in adjacent planes. The landmarks were considered to touch if contact (but not overlap) was seen between them within a plane or adjacent planes. The landmarks were considered to be fully separated if no contact was observed or if image planes lacking fluorescence separated fluorescent foci in different planes. Multiple fields of cells were examined for each of the two trials. Cell morphology was used to estimate the cell cycle stage of each cell examined. However, the same general trends were observed in G1, S, and G2 phases, so these data were pooled (G1 and S phase cells were well represented, whereas there were considerably fewer G2 cells).
RESULTS
Silencers flank silenced genes at HML and HMR. At HML, silencing initiates from both silencers, but at HMR silencing initiates only at the HMR-E silencer. Previous work from our lab showed that tethering proteins with barrier activity near HMR-E blocked the spread of silencing from the HMR-E silencer if the HMR-I silencer was absent (21). However the barrier could be bypassed if a second silencer (HMR-I) was positioned downstream of the barrier (references 21 and 47 and data not shown). One explanation for this phenomenon is that silencing nucleates at HMR-I, as well as HMR-E. However, functional data clearly indicates that HMR-I only augments the activity of HMR-E and does not possess an autonomous silencing activity (1, 7, 50).
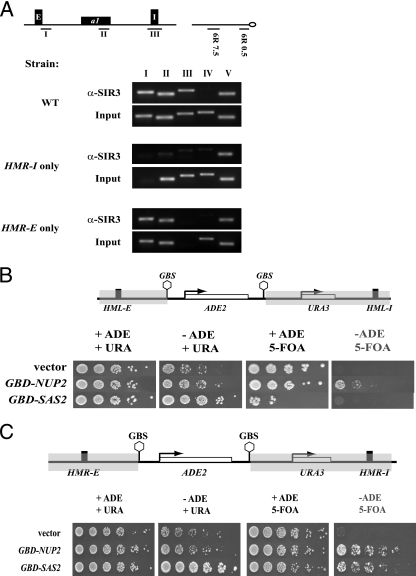
Since our analyses suggested that silencing at HMR might be initiating at HMR-I, we reasoned that if HMR-I was a silencer, then it should be able to recruit at least some Sir proteins in the absence of the HMR-E silencer. To directly test this possibility, we used ChIP to examine the binding of Sir3p near HMR-E and HMR-I in a variety of HMR variants. As expected, Sir3p localized to the two silencers in the wild-type strain (Fig. 1A). Deleting HMR-I did not affect Sir3p levels at the HMR-E silencer, but deleting HMR-E resulted in loss of Sir3p localization from the HMR locus but not the telomeres. In the absence of HMR-E, the levels of Sir3p at HMR were equivalent to those observed at the negative control, the TEL6R 7.5kb probe, where Sir3p has not been found previously. Therefore, these results, at this level of sensitivity, demonstrate that HMR-I does not recruit Sir proteins in the absence of HMR-E and are consistent with previous functional results (1, 7, 50).
FIG. 1.
Boundary trap assays at HMR and HML. (A) HMR-I does not recruit Sir3p. ChIP was used to map the presence of Sir3p in strains with mutant silencers at HMR, and the immunoprecipitated samples were analyzed by PCR. The locations of the PCR probes are shown in the schematic diagram. WT, wild type. (B) At HML, only Nup2 allows discontinuous silencing. Strains with a boundary trap construct at HML were transformed with _TRP1_-containing plasmids constitutively expressing the chimeric protein Gbd-Sas2p (pRO590) or Gbd-Nup2p (pRO635) or the vector. Cells were grown in liquid YM medium (HC-trp), and expression of the ADE2 and URA3 genes was monitored by serial dilutions on HC-trp plates lacking or containing adenine, uracil, and 5-FOA as indicated. The plates were photographed after 2 days. The panel labeled in gray allows differentiation between “true barrier” proteins and “desilencing” proteins. (C) At HMR, both Nup2 and Sas2p allow discontinuous silencing. Strains with a boundary trap construct at HMR were transformed with _TRP1_-containing vector or with TRP1 plasmids constitutively expressing Gbd-Nup2p or Gbd-Sas2p. Cells were grown overnight in liquid HC-trp, and serial dilutions were spotted on appropriate plates. Cells were spotted on HC-trp plates lacking adenine or containing 30 μg per ml of adenine and allowed to grow at 30°C prior to photography. To assay for stable repression of URA3, cells were spotted onto HC-trp plates containing 5-FOA and lacking adenine or containing 30 μg per ml of adenine and allowed to grow at 30°C prior to photography. The panel labeled in gray allows differentiation between “true barrier” proteins and “desilencing” proteins.
These results lead to an alternative explanation that HMR-E “communicates” with HMR-I to facilitate silencing at a distance. To explore this paradoxical phenomenon further, we monitored silencing of a dual reporter system known as the boundary trap assay that was developed and used to investigate insulator proteins at HML (33). In this assay, the silenced locus was modified and the mating-type genes were replaced with ADE2 and URA3 genes. Gal4 binding sites flank the ADE2 gene, whereas a second reporter, URA3, is not flanked by these sites and resides adjacent to the I silencer (Fig. 1B and C). The assay monitors the ability of a protein tethered to the Gal4p binding sites to insulate the ADE2 gene from repression but not the neighboring URA3 gene.
The dual reporter system was first used at HML. Unlike at HMR, at HML both silencers are able to independently initiate silencing (Fig. 1B). We tested the behavior of Gal4-Nup2p and Gal4-Sas2p. Nuclear pore proteins such as Nup2p were claimed to be “true barrier” proteins that can insulate the ADE2 gene while maintaining the neighboring URA3 gene in a silenced state (33). We also tested Gal4-Sas2p, since acetyltransferases are believed to function by a “desilencing” mechanism. We measured expression of ADE2 by growth on medium lacking adenine, and we measured expression of URA3 by growth on medium containing 5-fluoroorotic acid (5-FOA). Cells expressing URA3 convert 5-FOA to a toxic metabolite and die. We used these assays because they are far more sensitive to changes in the expression levels than Northern blots. Furthermore, these assays allow us to determine the mitotic stability of these epigenetic states. Our results (Fig. 1B) are consistent with previously published data (33). Gal4-Nup2p insulated ADE2 from repression while allowing URA3 to be stably repressed in a small percentage of cells. On the other hand, Gal4-Sas2p derepressed both ADE2 and URA3, presumably by disrupting silencing across the entire silenced domain.
We next constructed a dual reporter system at HMR that was similar to the system at HML, placing the ADE2 gene near HMR-E and the URA3 gene near HMR-I (Fig. 1C). The strain was transformed with Gal4-Nup2p, Gal4-Sas2p, or vector alone. The cell growth assays in Fig. 1C clearly show that Gal4-Nup2 functions again as a true barrier, producing colonies of cells in which ADE2 was active but URA3 was stably repressed. However, unlike the situation at HML, Gal4-Sas2p derepressed ADE2 expression while permitting URA3 repression at HMR. Thus, Gal4-Sas2p also functions as a “true barrier” at HMR. Importantly, the ability of cells to form colonies on medium lacking adenine but containing 5-FOA indicates that the “discontinuous” silenced state that is established in these cells is stably inherited for several generations, enabling these cells to form colonies. A dual reporter system containing ADE2 and MATa1 yielded similar results, suggesting that this effect was not reporter specific (data not shown). Furthermore, the fact that Sas2p, a bona fide histone acetyltransferase which is expected to behave as a “desilencer,” can function as a “true barrier” protein indicates that the molecular underpinnings for these definitions will need to be reconsidered.
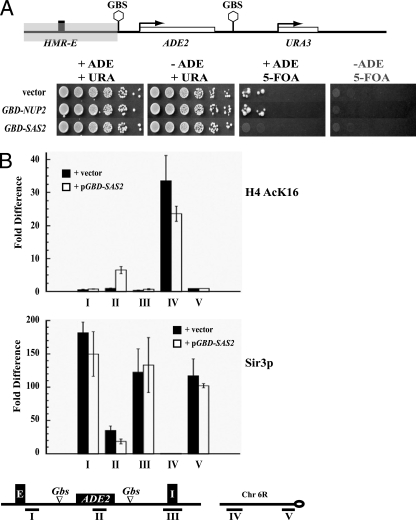
We next determined whether the generation of the discontinuous silenced state required HMR-I. We deleted the HMR-I silencer from the boundary trap strain at HMR and analyzed the ability of these strains to grow on medium lacking adenine but containing 5-FOA. Our results (Fig. 2A) showed that silencing of the URA3 gene in the dual reporter system at HMR required HMR-I. When this silencer was removed, no colonies formed on the plates lacking adenine and containing 5-FOA.
FIG. 2.
Discontinuous silencing at HMR. (A) HMR-I is necessary for discontinuous silencing. Strains with a boundary trap construct at HMR but lacking the HMR-I silencer were transformed with _TRP1_-containing vector or with TRP1 plasmids constitutively expressing Gbd-Nup2p or Gbd-Sas2p. The strains were assayed as described for Fig. 1C. (B) Mapping the distribution of acetylated histones and Sir3p in the boundary trap constructs. Strains constitutively expressing Gbd-Sas2p or vector alone were grown in HC-trp medium, selecting for the plasmids (the medium was supplemented with 90 μg/ml adenine). ChIP with antibodies against H4K16Ac or Sir3p were performed exactly as previously described (46). The graphs depict the enrichment of the immunoprecipitated sample over the input normalized to a telomeric probe. Enrichment and standard errors were computed from at least two independent cross-linked samples and three independent immunoprecipitation experiments. Localization of the PCR probes is depicted in the schematic.
This result demonstrates that the Sir proteins recruited at HMR-E can transpose across an active domain only when a silencer is present on either side of this domain. These results with the dual reporter systems are concordant with our earlier studies of single tethered barrier proteins (47). Silencing adjacent to HMR-I requires the HMR-I silencer if a barrier blocks the action of HMR-E.
To explore the discontinuous silencing phenomenon at a molecular level, we mapped the distribution of Sir3p and H4AcK16 across the HMR domain in the presence and absence of Gal4-Sas2p. We chose to analyze these two proteins since they are markers of active and inactive chromatin (46). In the absence of Gal4-Sas2p, there is no acetylation at either silencer or the ADE2 gene (Fig. 2B). When Sas2p is recruited to sites flanking the ADE2 gene, there is no detectable acetylation at the HMR-E and HMR-I silencers but there is a significant increase in H4K16 acetylation at the ADE2 gene, consistent with the observation that ADE2 is active in these cells.
On the other hand, Sir3p was present at the two silencers, in both the presence and absence of Gal4-Sas2p, but was reduced at the ADE2 gene when Sas2p was tethered at the Gal4p binding sites flanking the ADE2 gene. Thus, tethered Gal4-Sas2p does not block the normal function of the two silencers, and the growth phenotypes observed in Fig. 1 are indeed due to discontinuous silenced domains that initiate from HMR-E.
Interestingly, we consistently see increased levels of Sir3p at both the HMR-E and HMR-I silencers compared with the ADE2 gene, in the presence or absence of Gal4-Sas2p. Our results demonstrate that HMR-I alone cannot recruit Sir proteins, but in the presence of HMR-E it is able to stably maintain elevated levels of Sir proteins. While the elevated levels of Sir3p at HMR-E can be explained by the fact that HMR-E recruits the Sir proteins and initiates silencing, the reason for the elevated levels at HMR-I were unexpected and not initially obvious. One possibility is that the increased levels of Sir proteins at HMR-I may be due to the two silencers being in close proximity to one another.
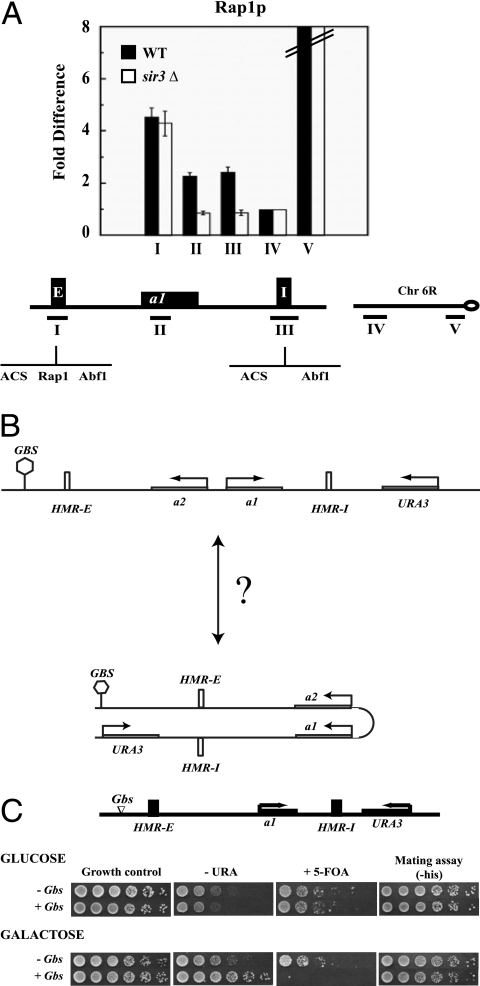
Rap1p localizes to HMR-I.
Our data indicate that HMR-E functionally communicates with HMR-I, resulting in a discontinuous silenced domain, but they do not specify how this might occur. One possibility is that the two silencers reside in close proximity to one another.
To confirm these long-range interactions, we asked whether a DNA-bound protein at one end of the domain was in close proximity to the other end of the domain, similar to the experiments used to show long-range interactions in Drosophila (5). There is a single binding site for Rap1p at the HMR-E silencer, where the protein has been shown to bind (53). No Rap1p sites are known to exist at HMR-I. We used ChIP to map the presence of Rap1p at HMR, as well as at loci on chromosome 6R (Fig. 3A). While we did not observe significant binding of Rap1p to the telomere 6R 7.5-kb probe, the quantitative analyses showed that Rap1p was present immediately adjacent to telomere 6R and at HMR. At HMR we observed Rap1p binding to HMR-E, the silenced MATa1 gene at HMR and at HMR-I. This result was obtained with two different commercially available antibodies (data not shown), validating the presence of Rap1p at HMR silencers.
FIG. 3.
Functional long-range communication at HMR. (A) Mapping Rap1p at HMR. Antibodies against the Rap1p C terminus were used to map the distribution of Rap1p across the HMR locus in a wild-type (WT) strain and a _sir3_Δ strain. Quantitative ChIPs were performed as described for Fig. 2. The PCR probes used are shown in the schematic diagram. Error bars indicate standard errors. (B) Schematic representation of the enhancer construct at HMR. The locations of Gal4p binding sites and the URA3 gene are shown. (C) “Enhancer” activity at HMR. Strains with URA3 located downstream of HMR-I containing no Gal4 binding sites (-Gbs) or four Gbs upstream of HMR-E (+Gbs), were grown in 5 ml YPD overnight. Cells were washed, and fivefold serial dilutions were prepared. Properly supplemented YM plates containing 2% galactose (YMG) or 2% glucose (YMD) as a carbon source were used to induce or to repress expression of Gal4p, respectively. To assay for expression of URA3, cells were spotted onto YMG or YMD plates lacking or containing uracil or 5-FOA and photographed. To assay for expression of MATa1 at HMR, cells were spotted onto properly supplemented YMG or YMD plates with mating lawns.
We next determined if loss of Sir3p affected the distribution of Rap1p. Loss of Sir3p did not lead to any decrease in the amount of Rap1p at HMR-E, but there was a complete loss of Rap1p from HMR-I. These results are consistent with the observation that HMR-E was in close proximity to HMR-I, al though it is also possible that despite Rap1p being a sequence-specific DNA binding protein, it spread along the silenced chromatin through interactions with the Sir proteins.
“Enhancer” activity at HMR.
To investigate the spatial localization of the silencers relative to each other, we decided to develop an “enhancer assay” (Fig. 3B). The assay is premised on the assumption that when an upstream activation sequence (UAS) is brought in close spatial proximity to the promoter of a repressed gene, it will activate that gene. We placed Gal4p binding sites upstream of HMR-E and placed a repressed reporter gene (URA3) several kilobase pairs (4 kb) downstream from the Gal4p binding sites on the distal side of HMR, between HMR-I and the tRNA barrier. Transcription of the reporter was directed toward the Gal4p binding sites (Fig. 3B). At this location, the URA3 gene was subjected to repression by Sir-mediated silent chromatin, and in the absence of the Sir proteins, URA3 was active in both glucose and galactose (data not shown). Interestingly, stable repression of URA3 located downstream of HMR-I was dependent upon the HMR-I silencer, because in the absence of this silencer, the reporter was no longer stably silenced (data not shown).
We next tested whether binding of Gal4p to its sites upstream of HMR-E could disrupt the silencing of the URA3 gene located downstream of HMR-I. When cells were grown in glucose, Gal4p was not activated and URA3 remained silenced in strains that contained or lacked Gal4p binding sites (Fig. 3C, top). When these cells were grown in medium containing galactose, however, URA3 was activated and cells were able to grow in medium lacking uracil and not in medium containing 5-FOA (Fig. 3C, bottom). This result was observed only in strains that contained Gal4p binding sites, indicating that Gal4 binding upstream of HMR mediated the galactose-dependent URA3 induction.
One possibility is that Gal4p was disrupting silencer function. If Gal4p was disrupting silencing across the entire domain, then the MATa1 gene located in the silenced region should also be activated. We therefore monitored expression of the MATa1 gene located between the two silencers by performing a mating assay and selecting for diploids. The appearance of diploids (Fig. 3C) demonstrated that at this level of sensitivity, MATa1 was silenced.
We also investigated whether Gal4p binding upstream of HMR was disrupting Sir3p binding at HMR by quantitative ChIP. If this was the case, then in strains containing Gal4p binding sites grown in galactose, one might expect to see a reduction in the levels of Sir3p at HMR. We mapped the levels of Sir3p by ChIP across the entire HMR domain in strains grown in galactose with and without Gal4p binding sites. Our analyses showed only a slight change in the levels of Sir3p at the two silencers, with no discernible change at the MATa1 gene located between the two silencers in the presence or absence of Gal4p (data not shown). These results are consistent with our mating assays (Fig. 3C) showing that silencing did not significantly change at HMR.
These results demonstrate “enhancer” function of a gene across a silenced domain. They also demonstrate that a protein bound to a UAS located several kilobase pairs from the promoter of a gene could derepress that gene in yeast. This result is highly unusual, since activation over such long distances has not been observed in yeast (20). The simplest explanation for these results is that the UAS was in close spatial proximity to the promoter of the reporter gene in the nucleus, which then alleviated silencing of the reporter. However one cannot rule out other possibilities due to the inherent limitations of these assays.
Loss of the barrier does not affect long-range communications.
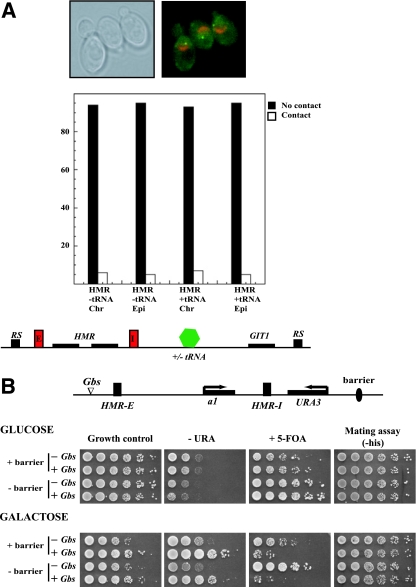
Our results suggested that the two silencers might be in close proximity to one another; we were interested in determining the DNA elements and factors that affected this localization. In chicken cells the globin insulator helps tether the globin domain to the nucleolus and aids in the formation of chromatin loops (61, 62). In yeast, tRNA genes are dispersed throughout the genome, but in situ hybridization demonstrated that the genes are clustered adjacent to the nucleolus (30). One of the HMR barriers is a tRNA gene (21), and it is therefore possible that tethering of the HMR barriers to the nucleolus might be the mechanism by which the two silencers were brought in close proximity to one another. A prediction of this model would be that the HMR locus would reside adjacent to the nucleolus and deletion of the barrier would result in a concomitant loss of this localization.
To determine whether the tRNA barrier adjacent to HMR associated with the nucleolus, we used a cytological approach. A lac operator array was incorporated adjacent to HMR (inserted approximately 4 kb from HMR) in a strain that expressed _lac_-GFP, as well as Sik1-RFP. _lac_-GFP binds the lac operator array to create a bright green spot of fluorescence (marking HMR), whereas Sik1p, a nucleolar protein, imparts red fluorescence to the perinuclear crescent-shaped nucleolus. Stacks of fluorescent images along the Z axis using both GFP and rhodamine (red) filters were collected to determine the relative positions of HMR and the nucleolus. Colocalization was defined as full overlap of the green and red signals within the same or adjacent image planes. Over 200 cells were examined in at least three independent trials. The data in Fig. 4A show that in over 90% of the cases, HMR and the nucleolus did not contact one another. The low level of coincident colocalization is similar to that found for other noninteracting chromatin landmarks (9, 12). Similar results were found when HMR and the adjacent barrier were liberated from the chromosome by site-specific recombination to form an extrachromosomal DNA circle. These results indicated that HMR and the associated boundary did not reside at the nucleolus. Furthermore, deleting the barrier did not alter the localization of HMR in the nucleus (Fig. 4A). It is therefore unlikely that the mechanism by which silencers are brought in close proximity is via tethering to the nucleolus.
FIG. 4.
Long-range communication and barrier function. (A) HMR does not colocalize with the nucleolus. Fluorescence analysis was performed with strains expressing Sik1-RFP and _lac_-GFP. A lac operator array placed approximately 4 kb from HMR, adjacent to the promoter of the GIT1 gene, allowed us to map the localization of the silenced domain relative to the nucleolus. The GFP and RFP signals were monitored in strains containing or lacking the tRNA barrier. Colocalization was defined as full overlap of the green and red signals within the same or adjacent image planes. Over 200 cells were examined in two independent trials. Colocalization was monitored when the HMR domain was present on a chromosome as well as on an episome (following recombination). A representative picture of the cells is shown above the graphs. (B) Loss of the tRNA barrier does not affect long-range activation at HMR. Strains containing or lacking the tRNA barrier without or with four Gal4p binding sites upstream of HMR-E and with URA3 located downstream of HMR-I were grown overnight. Properly supplemented YM plates containing 2% glucose or galactose as carbon source were used to induce expression of Gal4p. To assay for expression of URA3, cells were spotted onto YMG plates lacking or containing uracil or 5-FOA and photographed. To assay for expression of MATa1 at HMR, cells were spotted onto properly supplemented YMG plates with mating lawns.
Tethering of insulators to nuclear superstructures has been proposed to be important for insulation. In Drosophila, the Su(Hw) insulators cluster in the nucleus, forming insulator bodies (28), while in yeast, nuclear pore proteins localize to the silenced chromatin (10), and models suggest that tethering of insulators to the pores, forming a chromatin loop, is the mechanism by which chromatin domains are organized and maintained (32).
Therefore it was still possible that the barrier insulator elements at HMR were important for the observed long-range communication between the two silencers, albeit not by tethering to the nucleolus. If barrier elements were necessary for organizing chromatin domains into loops, then loss of a barrier should result in loss or diminution of long-range communications. Using the enhancer assay, we investigated the role of the HMR tRNA barrier in this process. We generated two strains lacking the tRNA barrier and possessing URA3 immediately downstream of HMR-I. One strain contained Gal4p binding sites located upstream of HMR-E, while the second strain lacked these binding sites. Monitoring the expression of URA3 in these two strains in galactose showed that loss of the tRNA barrier did not adversely affect the long-range communication between the two ends of the silenced chromatin domain (Fig. 4B).
It has been suggested that chromatin loops are formed by the attachment of barrier elements to the nuclear pore via Nup2p (32). Analyses of Nup2p mutants indicate that Nup2p is necessary for robust tRNA barrier function, but loss of Nup2p did not affect the long-range communication between the two silencers (G. Ruben and R. T. Kamakaka, unpublished data).
HMR-E and HMR-I are in close spatial proximity.
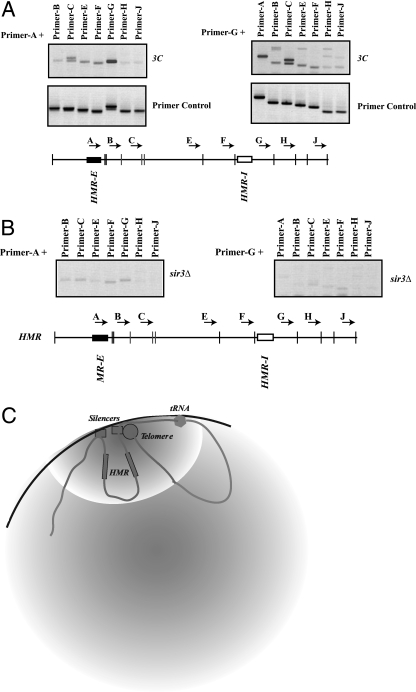
All of our analyses described thus far suggested that the two silencers were in close spatial proximity to one another. We therefore directly analyzed the spatial relationships at the native HMR locus in the yeast nucleus by using the 3C method, which was developed to investigate the three-dimensional relationships between DNA elements (15, 18). Cells were briefly treated with formaldehyde to cross-link DNA to proteins, followed by cleavage with a specific restriction enzyme; we digested the DNA with Sau3A since this enzyme generated small fragments (55). The fragments were then diluted and ligated, such that ligations were primarily between cross-linked DNA fragments. The fragments that ligated to one another were identified using PCR with specific pairs of primers. Using this method, the cross-linking frequency between two restriction fragments is expected to be roughly proportional to their proximity to one another in the nucleus.
The DNA between the HMR-E and HMR-I silencers does not contain many Sau3A sites. To improve the resolution of our analyses, we introduced three additional Sau3A sites within this region (in the MATa2 gene). Furthermore, since regions at HMR are homologous to regions at _HML_α and MAT, we performed this analysis with a strain in which these two loci were deleted. All of the primers that we used in this analysis were oriented in the same direction. Therefore, a PCR product can arise only after restriction fragments were digested and religated. This eliminated PCR products that might arise from incomplete digestion of the cross-linked samples.
We initially analyzed the ligations with a fixed oligonucleotide located in a Sau3A fragment containing the HMR-E silencer (primer A) with restriction fragments that encompassed the silenced domain and beyond to determine which fragments were in close proximity to the reference fragment. Our analyses showed that the HMR-E fragment ligated most frequently to a single Sau3A fragment (amplified with primer G) containing the HMR-I silencer (Fig. 5A).
FIG. 5.
Spatial organization at HMR. (A) 3C analysis of HMR. Wild-type strains (lacking HML and MAT) were cross-linked, digested with Sau3A, and ligated, and the ligation products were analyzed by PCR (labeled 3C). Reference primer A or G was used along with the other primers across the domain. The location and orientation of the primers are shown schematically. The primer control panel involved intermolecular ligations and PCR analyses of un-cross-linked DNA. (B) Loss of Sir3p affects long-range interactions. Strains with a deletion of the SIR3 gene were analyzed with the 3C assay as described above. (C) Schematic representation of the HMR domain in the yeast nucleus.
To ensure that the PCR amplification efficiencies between different primer pairs were comparable, we digested plasmid DNA containing the HMR locus with Sau3A in the absence of cross-linking, followed by ligation under conditions that favored intermolecular ligation. PCR analyses indicated that all of the primer pairs were approximately equally efficient in amplifying the ligated products (Fig. 5A). We confirmed the equivalent PCR amplification efficiencies of the primers used by twofold serial dilutions of the un-cross-linked (intermolecular) ligation reaction prior to PCR (data not shown). These data showed that the differences in amplification observed across the HMR domain, in the nucleus, were due to differences in cross-linking/ligation of various fragments to the reference fragment (HMR-E) and not due to differences in PCR amplification. As an additional control, we purified the PCR products obtained from the cross-linked nuclear samples and sequenced them to unambiguously determine the identity of the ligated fragments (data not shown).
We next used the primer in the Sau3A fragment containing HMR-I (primer G) as the reference primer and assayed the proximity of this fragment to other fragments across HMR. Our results, shown in Fig. 5A, revealed that the HMR-I silencer-containing fragment ligated most frequently to the HMR-E silencer-containing fragment (primer A) as well as to a fragment downstream of HMR-E (primer C) that harbored the end of the MATa2 gene. From these data we inferred that HMR-I was in close proximity to HMR-E and the 3′ end of the MATa2 gene. It is possible that HMR-I is in close proximity with both fragments simultaneously or exchanges rapidly between these two fragments.
The 3C method was initially used to demonstrate the proximity between various centromeres in yeast (18). We also tested this interaction with our cross-linked samples. Consistent with previously published data, we found that EcoRI fragments at Cen IV (primer 14) ligated only to fragments at Cen III (primer 6) and not to other chromosome III EcoRI fragments (primers 5 and 7) (data not shown).
Long-range interactions require silencing.
Our results suggest that long-range communication between the two HMR silencers was not a fortuitous result of the clustering or long-range interactions between the barrier elements that flank the two silencers at HMR. Silencing at HMR utilizes the Sir proteins that interact with the chromatin to mediate silencing. We therefore investigated whether the communication between the two silencers was a result of silencing. We determined whether the long-range interaction between the two silencers was disrupted in a _sir3_Δ mutant by 3C analyses. Our results showed that in the absence of this repressor, the extent of ligation between the _HMR-E_- and _HMR-I_-containing Sau3A fragments was dramatically reduced (Fig. 5B), suggesting that these long-range interactions required the Sir proteins.
Since HMR-E no longer ligated to HMR-I in a _sir3_Δ mutant, to ensure that the cross-linking and ligation were normal in this sample, we examined the ligation between the centromeric fragments as controls, since Sir3p is not present at yeast centromeres and Sir3p mutations have not been shown to affect centromere function. Our analysis of centromeric chromatin demonstrated that indeed CEN III remained in close proximity to CEN IV in the absence of Sir3p (data not shown).
Our results in their totality demonstrate that silencers separated over several kilobase pairs of DNA functionally and structurally interact with one another. We have begun to identify the determinants necessary for these interactions and show that the Sir proteins are necessary for this long-range communications between silencers.
DISCUSSION
The control of eukaryotic gene expression involves communication between regulatory elements that are often separated by great distances. There are now numerous examples of distal enhancers and LCRs that contact the genes they activate (reviewed in reference 60). More recent studies have even found interactions between regulatory elements and genes that reside on entirely different chromosomes (39, 54). In this report, for the first time, we show long-range interactions between the silencers that flank the HMR locus in yeast, and we identify the determinants required for these interactions. Interestingly, we have shown that deletion of the tRNA insulator element or the nuclear pore protein Nup2p did not affect functional long-range communication between the two silencers but that loss of Sir3p did result in diminution of these interactions, suggesting that silencing itself may be important for organizing this chromatin domain.
Silencers and the mechanisms for silencing.
Numerous studies have shown that HMR-E is sufficient to nucleate silencing at HMR. HMR-I cooperates with HMR-E, but it cannot initiate silencing on its own (42, 50). Therefore, HMR-I is analogous to proto-silencers that have been found at telomeres and HML (6, 13). At telomeres, proto-silencers and proteins with barrier activity are interspersed in the subtelomeric blocks to yield domains of discontinuous silencing (24, 25, 38), and these functional assays have led to models where the proto-silencer elements might interact with one another, but direct long-range interactions at telomeric loci have not been demonstrated using the 3C technique. Long-range interactions between the proto-silencers and terminal telomeric sequences, which function as silencers, may indeed be occurring, similar to what we observe at HMR. The distribution of terminally bound Rap1 and Ku at telomeres is consistent with telomere loop formation (43, 56), and enhancer assays like the one we have employed in this study have also suggested looping within silent chromatin at telomeres (16). However, this is the first report to unambiguously describe long-range interactions and the formation of chromatin loops at an internal silenced locus in yeast.
Placement of the dual reporter constructs of the boundary trap assay at the silent mating-type loci created discontinuous silencing states. At HMR, Sir3p was found at both silencers but was consistently reduced at the insulated reporter gene in between. In agreement, the H4K16Ac mark for active chromatin was found in a reciprocal pattern. How can a discontinuous state be created when HMR-I does not function on its own? One possibility is that both silencers are held in close proximity so that HMR-I shares the nucleation activity of the more potent HMR-E silencer. In this case, silent chromatin would nucleate at both silencers and spread from both until encountering synthetic (or natural) barriers. Thus, when a pair of barrier proteins is situated between HMR-E and HMR-I, a domain of active chromatin will reside between domains of silent chromatin.
An alternative possibility is that the insulated ADE2 construct counteracts silent chromatin that has spread from a sole nucleation point at HMR-E. In this scenario, ADE2 activation would occur following a cell cycle event, such as DNA replication, that compromises silencing efficiency (4). Silent chromatin would persist on both sides of the activated domain because the silencers stabilized the existing repressed state (2, 13, 14, 42, 50). However, it is hard to visualize how this mechanism lends itself to stable inheritance (which we observe in our assays). An alternative model that combines these two scenarios is possible where binding of Sir proteins to HMR-E facilitates interactions between HMR-E and HMR-I. Once HMR-I is brought in proximity to HMR-E, it can also nucleate silencing, which then would spread from both silencers.
Sir3p and long-range repression.
The long-range communication between the two silencer fragments is dependent upon Sir3p. Sir3p is a structural repressor protein that binds the histones in nucleosomes to mediate repression. In vitro binding studies with oligonucleosomes have shown that Sir3p oligomers bind multiple chromatin fragments and “cross-links” nucleosomal arrays (27, 29, 40). It is therefore possible that Sir3p binding to chromatin cross-links the silenced domain and the resulting compaction brings distal sites together. The dependence on Sir3p is reminiscent of long-range repression in Drosophila, where Polycomb-mediated repression involves interactions between chromatin memory module elements and distally repressed promoters via the association of Polycomb group proteins (17, 44). We note, however, that Sir3p is not sufficient to hold HMR loci on sister chromatids together (12). Instead, silent chromatin recruits cohesin, which mediates pairing of the twin silent chromatin domains. Additional factors could similarly facilitate interactions between distal silent chromatin segments within the same chromatin fiber.
Regulatory elements important for long-range communication.
An interaction specifically between the two silencer-containing DNA fragments raises the question of which DNA elements, if any, are required. Our results with the boundary trap system demonstrate that HMR-I is necessary. Preliminary data using the 3C technique also suggest that the silencer elements are necessary. ORC and Abf1p bind HMR-I and might be involved in mediating these long-range interactions. Rap1p might also aid long-range communication. When bound to two sites on naked DNA, the protein induces loop formation (31). Further experiments will be necessary to dissect the roles of these elements and proteins in long-range communication.
In an alternative scenario, the two silencers could be brought in close proximity by insulator elements that flank the silenced domain. It has been suggested that chromatin loops are formed through association of insulator elements with the nuclear pores (32, 33, 52). It is therefore possible that the barrier elements flanking HMR (21, 22) or the hypersensitive sites at this locus (45) associate with nuclear pores or active chromatin hubs and cluster in the nucleus, the consequence of which would be to align HMR-E and HMR-I in close proximity. However, we have shown that deleting the barrier or Nup2p, which interacts with the barrier, had little effect on long-range communication between the silencers. It therefore seems unlikely that chromatin barriers play a major role in the observed interactions between HMR-E and HMR-I.
Sir spreading, long-range interactions, and silencing.
While we have shown that the HMR silencers are in close proximity to one another, it is unclear whether this long-range communication between silencers has any functional significance at the native locus. It is possible that the two silencers are fortuitously brought in close proximity to one another simply by the compaction of chromatin. However, data show that the HMR-I silencer is important for stabilizing the repressed state (50). In its absence, the silent state is reduced and silencing is weakened in a population of cells. Importantly, our data with the boundary trap assay showed that HMR-I becomes an essential silencing element when propagation of silent chromatin from HMR-E is blocked. One possibility, as described above, is that an interaction between the silencers permits the robust nucleation activity of HMR-E to act locally via a spreading mechanism as well as distally at HMR-I via long-range interactions. Another possibility (the two are not mutually exclusive) is that silencers associate with one another and with telomeres to bring both ends of the silenced domain into a nuclear compartment that favors silencing (Fig. 5C). HMR resides at the nuclear periphery and frequently colocalizes with the highly concentrated foci of Sir proteins associated with telomeres (26, 57). Consistent with this is the demonstration that telomere 3L is in proximity to the silenced HML locus (37). The clustering of regulatory elements in the nucleus is a recurring theme utilized by a variety of elements (60) to robustly activate and repress genes. It is thought to increase the local concentrations of proteins and thus improve the probability that these elements will function efficiently (19).
The role of the Sir proteins in this scenario would be to stabilize the long-range interactions that arise due to tethering of the silencers. This would be consistent with in vitro evidence showing that the Sir proteins preferentially bind two DNA fragments (27, 29) and with our 3C data showing that the long-range interactions are lost in the absence of Sir3p. This role for the Sir proteins is also consistent with the observation that loss of silencing affects telomere-telomere interactions and formation of telomeric foci in yeast.
Besides playing a role in tethering the HMR domain to telomeric foci, the role of the silencers would also be to nucleate the spread of Sir proteins (41, 51). Our quantitative analysis demonstrating increased concentrations of Sir proteins at silencers with reduced levels of these proteins the further one traverses from the silencer would be consistent with this role for the silencers, though other models are equally possible.
The mechanism by which long-range silencing occurs may be more complicated than previously anticipated. Interactions between silencer elements mediated by the repressor proteins may compete with interactions between enhancer and promoters to affect the three-dimensional organization and functional status of the nucleus. Further studies should help us to understand the significance of long-range interactions in chromatin domain organization and gene regulation.
Acknowledgments
We thank Genevieve Fourel, David Donze, Grant Hartzog, Michael Lichten, Orna Cohen-Fix, Masaya Oki, and Catherine Fox for comments on the manuscript; C. Fox and U. Laemmli for specific strains; and Riza Ysla for technical assistance.
This work was supported by grants from the NIH to R.T.K. (GM078068) and M.R.G. (GM51402).
Footnotes
▿
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Abraham, J., J. Feldman, K. A. Nasmyth, J. N. Strathern, A. J. Klar, J. R. Broach, and J. B. Hicks. 1983. Sites required for position-effect regulation of mating-type information in yeast. Cold Spring Harbor Symp. Quant. Biol. 47989-998. [DOI] [PubMed] [Google Scholar]
- 2.Ansari, A., and M. R. Gartenberg. 1999. Persistence of an alternate chromatin structure at silenced loci in vitro. Proc. Natl. Acad. Sci. USA 96343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, A., and M. Hampsey. 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 192969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., and D. E. Gottschling. 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 81133-1146. [DOI] [PubMed] [Google Scholar]
- 5.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boscheron, C., L. Maillet, S. Marcand, M. Tsai-Pflugfelder, S. M. Gasser, and E. Gilson. 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 152184-2195. [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz, and K. Nasmyth. 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 4141-48. [DOI] [PubMed] [Google Scholar]
- 8.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 132465-2477. [DOI] [PubMed] [Google Scholar]
- 9.Bystricky, K., P. Heun, L. Gehlen, J. Langowski, and S. M. Gasser. 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA 10116495-16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117427-439. [DOI] [PubMed] [Google Scholar]
- 11.Chambeyron, S., and W. A. Bickmore. 2004. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16256-262. [DOI] [PubMed] [Google Scholar]
- 12.Chang, C. R., C. S. Wu, Y. Hom, and M. R. Gartenberg. 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 193031-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, T. H., and M. R. Gartenberg. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14452-463. [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, T. H., Y. C. Li, and M. R. Gartenberg. 1998. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc. Natl. Acad. Sci. USA 955521-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, K. E., M. P. Kladde, and M. A. Seyfred. 1993. Interaction between transcription regulatory regions of prolactin chromatin. Science 261203-206. [DOI] [PubMed] [Google Scholar]
- 16.de Bruin, D., Z. Zaman, R. A. Liberatore, and M. Ptashne. 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409109-113. [DOI] [PubMed] [Google Scholar]
- 17.Dejardin, J., and G. Cavalli. 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 23857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 2951306-1311. [DOI] [PubMed] [Google Scholar]
- 19.Dillon, N., and R. Festenstein. 2002. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18252-258. [DOI] [PubMed] [Google Scholar]
- 20.Dobi, K. C., and F. Winston. 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 275575-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donze, D., C. R. Adams, J. Rine, and R. T. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 182485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fourel, G., E. Lebrun, and E. Gilson. 2002. Protosilencers as building blocks for heterochromatin. Bioessays 24828-835. [DOI] [PubMed] [Google Scholar]
- 25.Fourel, G., E. Revardel, C. E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 182522-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gartenberg, M. R., F. R. Neumann, T. Laroche, M. Blaszczyk, and S. M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119955-967. [DOI] [PubMed] [Google Scholar]
- 27.Georgel, P. T., M. A. Palacios DeBeer, G. Pietz, C. A. Fox, and J. C. Hansen. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 988584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 61025-1035. [DOI] [PubMed] [Google Scholar]
- 29.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 204522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haeusler, R. A., and D. R. Engelke. 2004. Genome organization in three dimensions: thinking outside the line. Cell Cycle 3273-275. [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann, J. F., T. Laroche, A. H. Brand, and S. M. Gasser. 1989. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell 57725-737. [DOI] [PubMed] [Google Scholar]
- 32.Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U. K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109551-562. [DOI] [PubMed] [Google Scholar]
- 33.Ishii, K., and U. K. Laemmli. 2003. Structural and dynamic functions establish chromatin domains. Mol. Cell 11237-248. [DOI] [PubMed] [Google Scholar]
- 34.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamakaka, R. T. 1997. Silencers and locus control regions: opposite sides of the same coin. Trends Biochem. Sci. 22124-128. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli, U. K., E. Kas, L. Poljak, and Y. Adachi. 1992. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2275-285. [DOI] [PubMed] [Google Scholar]
- 37.Lebrun, E., G. Fourel, P. A. Defossez, and E. Gilson. 2003. A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol. Cell. Biol. 231498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebrun, E., E. Revardel, C. Boscheron, R. Li, E. Gilson, and G. Fourel. 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312269-272. [DOI] [PubMed] [Google Scholar]
- 40.Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz, and D. Moazed. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121515-527. [DOI] [PubMed] [Google Scholar]
- 40a.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 202224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, K., M. A. Vega-Palas, and M. Grunstein. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 161528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahoney, D. J., R. Marquardt, G. J. Shei, A. B. Rose, and J. R. Broach. 1991. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5605-615. [DOI] [PubMed] [Google Scholar]
- 43.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97621-633. [DOI] [PubMed] [Google Scholar]
- 44.Maurange, C., and R. Paro. 2002. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 162672-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasmyth, K. A. 1982. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell 30567-578. [DOI] [PubMed] [Google Scholar]
- 46.Oki, M., and R. T. Kamakaka. 2005. Barrier function at HMR. Mol. Cell 19707-716. [DOI] [PubMed] [Google Scholar]
- 47.Oki, M., L. Valenzuela, T. Chiba, T. Ito, and R. T. Kamakaka. 2004. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol. Cell. Biol. 241956-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Sullivan, J. M., S. M. Tan-Wong, A. Morillon, B. Lee, J. Coles, J. Mellor, and N. J. Proudfoot. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 361014-1018. [DOI] [PubMed] [Google Scholar]
- 49.Patrinos, G. P., M. de Krom, E. de Boer, A. Langeveld, A. M. Imam, J. Strouboulis, W. de Laat, and F. G. Grosveld. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 181495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivier, D. H., J. L. Ekena, and J. Rine. 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 132207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel, and U. K. Laemmli. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21379-391. [DOI] [PubMed] [Google Scholar]
- 53.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10408-412. [DOI] [PubMed] [Google Scholar]
- 54.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435637-645. [DOI] [PubMed] [Google Scholar]
- 55.Splinter, E., F. Grosveld, and W. de Laat. 2004. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 375493-507. [DOI] [PubMed] [Google Scholar]
- 56.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1183-93. [DOI] [PubMed] [Google Scholar]
- 57.Taddei, A., F. Hediger, F. R. Neumann, C. Bauer, and S. M. Gasser. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 231301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 101453-1465. [DOI] [PubMed] [Google Scholar]
- 59.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17453-462. [DOI] [PubMed] [Google Scholar]
- 60.Valenzuela, L., and R. T. Kamakaka. 2006. Chromatin insulators. Annu. Rev. Genet. 40107-138. [DOI] [PubMed] [Google Scholar]
- 61.Yusufzai, T. M., and G. Felsenfeld. 2004. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 1018620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13291-298. [DOI] [PubMed] [Google Scholar]