Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane (original) (raw)
Abstract
Endocytosis in yeast requires actin and clathrin. Live cell imaging has previously shown that massive actin polymerization occurs concomitant with a slow 200-nm inward movement of the endocytic coat (Kaksonen, M., Y. Sun, and D.G. Drubin. 2003. Cell. 115:475–487). However, the nature of the primary endocytic profile in yeast and how clathrin and actin cooperate to generate an endocytic vesicle is unknown. In this study, we analyze the distribution of nine different proteins involved in endocytic uptake along plasma membrane invaginations using immunoelectron microscopy. We find that the primary endocytic profiles are tubular invaginations of up to 50 nm in diameter and 180 nm in length, which accumulate the endocytic coat components at the tip. Interestingly, significant actin labeling is only observed on invaginations longer than 50 nm, suggesting that initial membrane bending occurs before initiation of the slow inward movement. We also find that in the longest profiles, actin and the myosin-I Myo5p form two distinct structures that might be implicated in vesicle fission.
Introduction
Genetic analysis of the endocytic uptake in yeast identified >50 proteins involved in the process, including clathrin, actin, and numerous proteins that control actin dynamics (Engqvist-Goldstein and Drubin, 2003). Whether clathrin and actin cooperated within the same pathway in yeast and the sequence of events driving formation of the primary endocytic vesicles remained completely open questions until recent development of live cell fluorescence microscopy applied to this organism (Kaksonen et al., 2003). This technique showed that once a cortical site is marked by the clathrin coat, recruitment of all other endocytic proteins, including those controlling actin dynamics, follows in a highly reproducible, ordered, and linear manner (Kaksonen et al., 2003, 2005; Jonsdottir and Li, 2004; Newpher et al., 2005; Toshima et al., 2006; Sun et al., 2006, 2007). Based on differences in protein function and dynamics at the endocytic cortical patch, four molecular modules have been defined (Kaksonen et al., 2005): (1) the endocytic coat module composed of clathrin (Chc1p/Clc1p) and several clathrin-interacting proteins such as the Hip1R homologue Sla2p, the intersectin-like proteins Sla1p and End3p, and the eps15 family member Pan1p; (2) the Wiskott-Aldrich syndrome protein (WASP)/Myo module containing the yeast WASP (Las17p) and WASP-interacting protein (Vrp1p) homologues, the unconventional myosin-I Myo5p, the Myo5p-interacting protein Bbc1p, and the yeast syndapin Bzz1p; (3) the vesicle fission module containing the yeast amphiphysins Rvs161p and Rvs167p; and (4) the actin module, which includes the Arp2/3p complex, the actin binding protein Abp1p, capping protein (Cap1/2p), and fimbrin (Sac6p).
The coat components and Las17p are the first to be observed as patches at the plasma membrane (Kaksonen et al., 2003, 2005; Newpher et al., 2005; Toshima et al., 2006). After a period of restrained motility, which can last >1 min, the coat undergoes a 200-nm slow inward movement, which takes ∼10–15 s. Shortly after the transient recruitment of the yeast amphiphysins, the coat starts disassembling as it rapidly moves into the cytosol (Kaksonen et al., 2003, 2005). The released vesicle then moves rapidly into the cell along actin cables to finally fuse with internal compartments (Kaksonen et al., 2003; Huckaba et al., 2004; Toshima et al., 2006). The unconventional myosin-I Myo5p is recruited at the endocytic patch at the onset of the slow inward movement together with Bbc1p and slightly after Bzz1p and Vrp1p. All components of the WASP/Myo module stay at the plasma membrane as the vesicle detaches, and they are thought to orchestrate Arp2/3-dependent actin polymerization at sites of endocytosis (Sun et al., 2006). Finally, coincident with initiation of the slow inward movement and with the onset of massive actin polymerization is also the recruitment of the actin module, which remains associated with the endocytic vesicle as it moves into the cytosol (Kaksonen et al., 2003, 2005). Based on the time of recruitment of the yeast amphiphysins, it has been proposed that the slow inward movement represents the initial invagination of the plasma membrane (Kaksonen et al., 2006). However, the precise point when vesicle fission occurs has not been determined. Furthermore, it is not really understood how clathrin and actin machineries cooperate to drive membrane invagination and vesicle fission.
Fluorescence microscopy does not offer sufficient resolution to define the nature of the primary endocytic profiles and to dissect the architecture of the molecular complexes during initial membrane bending. In contrast to mammalian cells, the ultrastructural studies on the early stages of endocytic budding in yeast have been scarce and difficult to interpret (Mulholland et al., 1994, 1999). In this study, we use immunoelectron microscopy to analyze plasma membrane–associated profiles decorated for components of the different endocytic modules. With the live cell imaging data in hand, the ultrastructural analysis indicates that the primary endocytic profiles in yeast are tubular invaginations of up to 50 nm in diameter and 180 nm in length. Analysis of the distribution of immunogold labeling nine different proteins required for the endocytic uptake shows that they segregate along endocytic invaginations. The data provide essential information to model the molecular mechanism driving endocytic uptake in yeast and unveil interesting similarities and differences with the analogous process in mammals.
Results
Yeast proteins involved in endocytic uptake label tubular invaginations of up to 50 nm in diameter and 180 nm in length
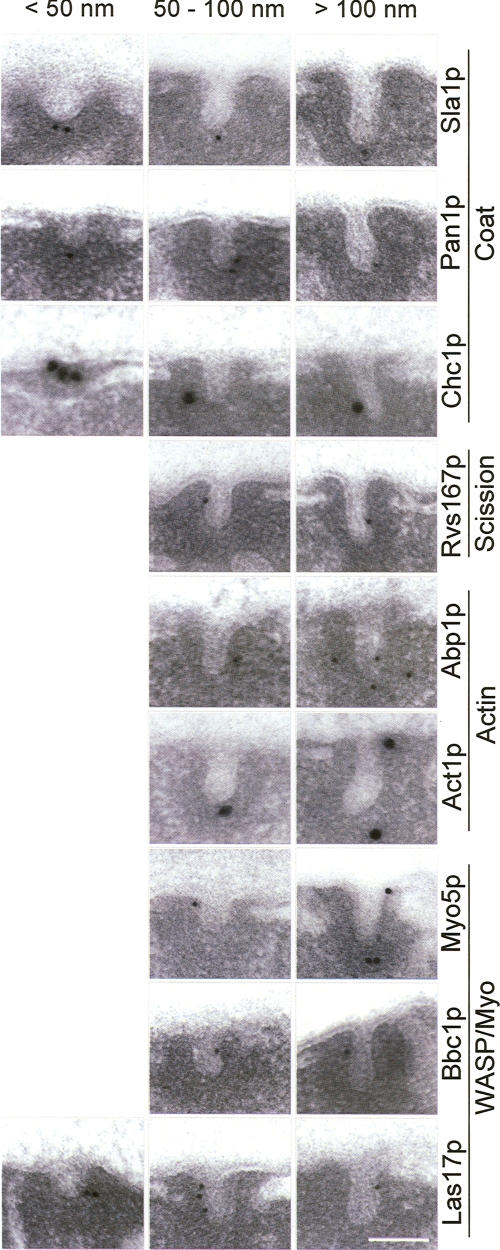
The actual point when vesicle scission from the plasma membrane occurs during the 200-nm coat slow inward movement has not been defined. Small vesicles could form at the plasma membrane that then slowly move through a dense cortex. Alternatively, the endocytic coat could progress into the cytosol at the tip of tubular profiles while still attached to the plasma membrane. To define the nature of the primary endocytic profiles in yeast, we used immunoelectron microscopy to analyze the morphology of membrane profiles located within 200 nm from the cell surface (plasma membrane invaginations and vesicles), which could be immunolabeled with gold particles directed against the endocytic coat components. Because the coat proteins are recruited first to the sites of endocytosis and only disassemble once the fast inward movement starts, we reasoned that labeling against these components should decorate the incipient endocytic profiles at all maturation stages. Ultrathin sections from chemically fixed yeast cells expressing HA-tagged versions of Sla1p or Pan1p were prepared and labeled using a rat monoclonal antibody against HA or a mixture of monoclonal antibodies against clathrin (Chc1p) and the adequate colloidal gold-conjugated secondary antibodies. Micrographs of membrane profiles decorated with gold against the coat were collected, and their length (invagination length [IL]) and diameter were measured. This type of analysis demonstrated that all components of the endocytic coat associate with tubular invaginations with diameters varying between 35 and 50 nm and lengths up to 180 nm, which were often surrounded by an electron-dense material (Fig. 1 and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200708060/DC1). Interestingly, no significant labeling associated with small vesicles close to the plasma membrane was observed. These observations were consistent with the hypothesis proposing that the endocytic coat progresses inside the cell in association with tubular profiles before vesicle scission.
Figure 1.
Proteins involved in the endocytic uptake associate with plasma membrane invaginations of about 50 nm in diameter and up to 180 nm in length. Representative electron micrographs of ultrathin sections from yeast showing plasma membrane–associated invaginations shorter than 50 nm, between 50 nm and 100 nm, and longer than 100 nm decorated with gold particles against the indicated proteins. Sections of yeast strains expressing HA-tagged Sla1p (HR2634/pMIG692), Pan1p (ScMIG946), Rvs167p (ScMIG995), Abp1p (ScMIG723), Myo5p (ScMIG994), Bbc1p (ScMIG903), and Las17p (ScMIG516) were decorated with a rat monoclonal anti-HA antibody and the appropriate 10-nm gold-conjugated secondary IgGs. Chc1p and actin (Act1p) were localized on ultrathin sections of ScMIG946 using a mixture of mouse monoclonal anti-Chc1p antibodies or a mouse monoclonal anti-actin antibody, respectively, and the appropriate 20-nm gold-conjugated secondary IgGs. Bar, 100 nm.
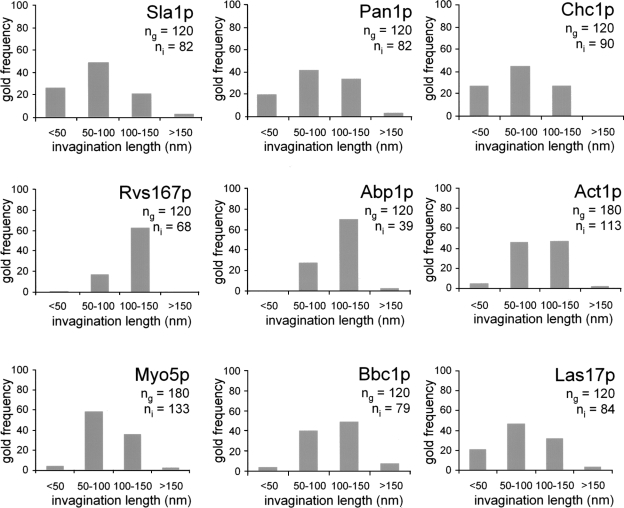
To investigate whether the plasma membrane–associated tubular profiles were actually endocytic invaginations that grow into the cytosol during the slow inward movement, we examined the presence of Abp1p and Rvs167p on similar invaginations. Abp1p is recruited to the cell cortex at the beginning of the slow inward movement, whereas the yeast amphiphysins are the last proteins arriving before initiation of the fast inward movement (Kaksonen et al., 2005). Micrographs from yeast cells expressing HA-tagged versions of these proteins were prepared and analyzed as described in the previous paragraph. Consistent with our hypothesis, gold decorating Abp1p and Rvs167p again appeared near tubular profiles similar to those labeled for Sla1p, Pan1p, and Chc1p (Fig. 1). Interestingly, however, and in contrast to gold decorating the coat, no significant labeling for Abp1p and Rvs167p was detected in association with profiles shorter than 50 nm (Fig. 2). Only one gold particle out of 120 decorating Rvs167p and none decorating Abp1p appeared to be associated with short profiles, whereas >20% of the gold labeling the endocytic coat were found near this kind of profile (IL ≤ 50 nm; Fig. 2). These data suggested that initial membrane bending might occur before assembly of the actin module, previous to initiation of the slow inward movement and massive actin polymerization. Consistently, only 5% of the particles labeling actin (Act1p) were associated with invaginations shorter than 50 nm (Fig. 2). Also in striking agreement with this hypothesis was the distribution of gold particles decorating different proteins of the WASP/Myo module. Again, 19% of the labeling against Las17p, which is recruited to the endocytic patch before initiation of the slow inward movement, was found on invaginations shorter than 50 nm. In contrast, <5% of the gold particles decorating Bbc1p and Myo5p, which arrive at the cortical patch at the onset of the slow inward movement, were found associated with this type of profile. Consistent with the relatively similar dynamics of the coat components and Las17p at the endocytic patch, protein comparison for the length of the labeled invaginations (IL) using a generalized linear model demonstrated that Las17p was more similar to Pan1p than to the members of the WASP/Myo module (Table I).
Figure 2.
Shallow plasma membrane invaginations are almost devoid of endocytic proteins recruited during the slow inward movement. Frequency of gold decorating the indicated proteins binned according to the length of the associated invagination (IL; 50-nm bins). Gold-labeled ultrathin sections were prepared as described in Fig. 1, and the length of the associated plasma membrane invagination was measured. The number of gold particles associated with invaginations of lengths within the indicated ranges were counted and normalized against the total number of gold particles recorded for each protein (n g). The total number of invaginations (n i) associated with the recorded gold particles is indicated.
Table I. Protein comparison for the IL parameter using a generalized linear model.
| Protein | Sla1p | Pan1p | Chc1p | Abp1p | Act1p | Rvs167p | Myo5p | Bbc1p | Las17p |
|---|---|---|---|---|---|---|---|---|---|
| Sla1p | 1.0000 | 0.0263 | 0.8629 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0156 |
| Pan1p | 1.0000 | 0.0168 | <0.0001 | 0.0002 | <0.0001 | 0.0128 | <0.0001 | 0.8414 | |
| Chc1p | 1.0000 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0096 | ||
| Abp1p | 1.0000 | 0.0019 | 0.3842 | <0.0001 | 0.2383 | <0.0001 | |||
| Act1p | 1.0000 | 0.0318 | 0.1600 | 0.0710 | 0.0005 | ||||
| Rvs167p | 1.0000 | 0.0006 | 0.7568 | <0.0001 | |||||
| Myo5p | 1.0000 | 0.0021 | 0.0228 | ||||||
| Bbc1p | 1.0000 | <0.0001 | |||||||
| Las17p | 1.0000 |
The observation that gold frequencies labeling the different proteins binned for the length of the associated invagination approximately recapitulated their dynamics at the endocytic patch (Fig. 2) strongly indicated that tubular invaginations of increasing length represent progressively more mature endocytic profiles.
Proteins required for endocytic uptake spatially segregate along plasma membrane invaginations
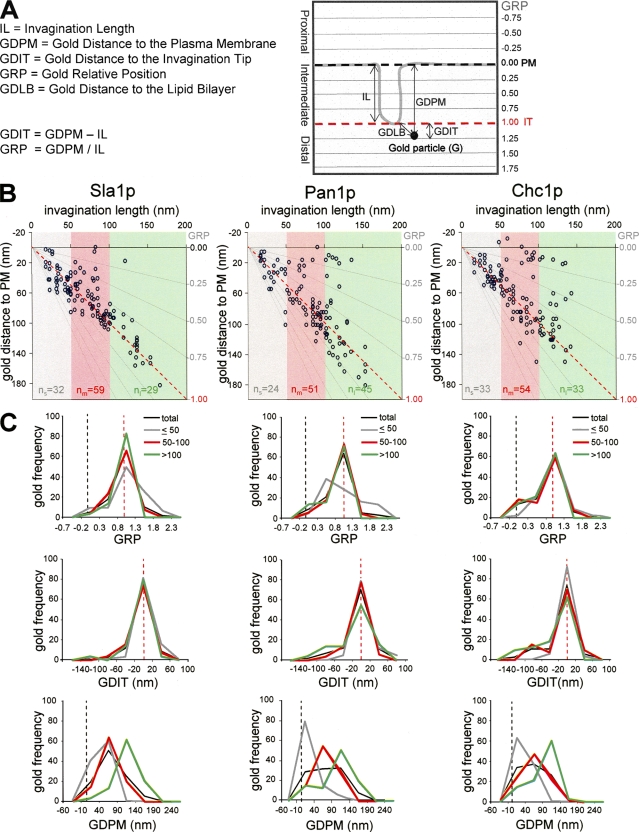
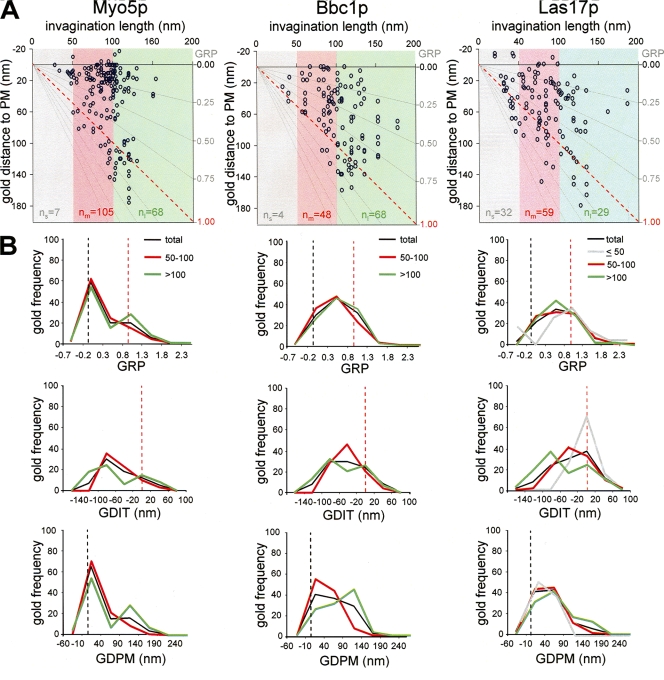
In light of the live cell fluorescence microscopy data (Kaksonen et al., 2005), our results strongly indicated that endocytic budding in yeast requires the formation of tubular invaginations that grow into the cytosol before vesicle scission. To obtain information regarding the architecture of endocytic complexes along these membrane profiles, we analyzed the distribution of gold particles labeling components of different endocytic modules. Several parameters were measured for every recorded immunogold. In addition to the length of the labeled invagination (IL), the distance from the gold particle to the basal plasma membrane (gold distance to plasma membrane [GDPM]), the distance from the gold particle to the invagination tip (gold distance to invagination tip [GDIT] = GDPM − IL), and the relative position of the gold particle with respect to the invagination (gold relative position [GRP] = GDPM/IL) were analyzed (Fig. 3 A). GRPs close to 1 described gold localized at the invagination tip, and GRPs near 0 defined gold particles situated at the base of the invagination (plasma membrane), close to the negatively curved area. GRPs >1 described gold particles located at the distal area. In addition, the minimal distance of the gold particle to the lipid bilayer (gold distance to lipid bilayer [GDLB]) was measured (Fig. 3 A). To gain information about possible rearrangements in the architecture of endocytic complexes as invaginations elongate, the GDPM, GDIT, and GRP parameters were plotted against the IL (Fig. 3 B). The distribution of these parameters was then analyzed for the entire immunogold population labeling each protein and for the subpopulations associated with short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) invaginations (Fig. 3; and see Figs. 4, 5, and 6).
Figure 3.
Endocytic coat components accumulate at the tip of tubular invaginations. (A) Scheme of an immunogold-labeled invagination showing the parameters used to characterize the particle position: invagination length (IL), gold distance to the basal plasma membrane (GDPM), gold distance to the invagination tip (GDIT = GDPM − IL), gold relative position (GRP = GDPM/IL), and minimal gold distance to the lipid bilayer (GDLB). (B) Graphs representing the GDPM for each recorded gold particle immunolabeling Sla1p, Pan1p, and Chc1p versus the IL. The x axis represents the level of the basal plasma membrane. The red dotted lines indicate the positions of the invagination tip. Gray lines converging at the xy axes crossing point define the GRP areas (left axis) as a function of the IL. Gray, red, and green areas enclose the gold subpopulations associated with short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) profiles, respectively. Sample sizes of subpopulations associated with short, intermediate, and long profiles are indicated as n s, n m, and n l, respectively. (C) Graphs representing the frequency of gold labeling Sla1p, Pan1p, and Chc1p binned according to their GRP, GDIT, or GDPM. The entire gold subpopulation (total; black) and the subpopulations associated with short (≤ 50; gray), intermediate (50–100; red) and long (>100; green) profiles were analyzed. The number of gold particles with values falling into the indicated intervals was counted and normalized with respect to the total population (n g; Fig. 2) or for the corresponding subtotals (n s, n m, and n l). Immunogold-labeled ultrathin sections were prepared as described in Fig. 1. (A and C) Black and red dotted lines indicate the positions of the basal plasma membrane (PM) and the invagination tip (IT), respectively.
Figure 4.
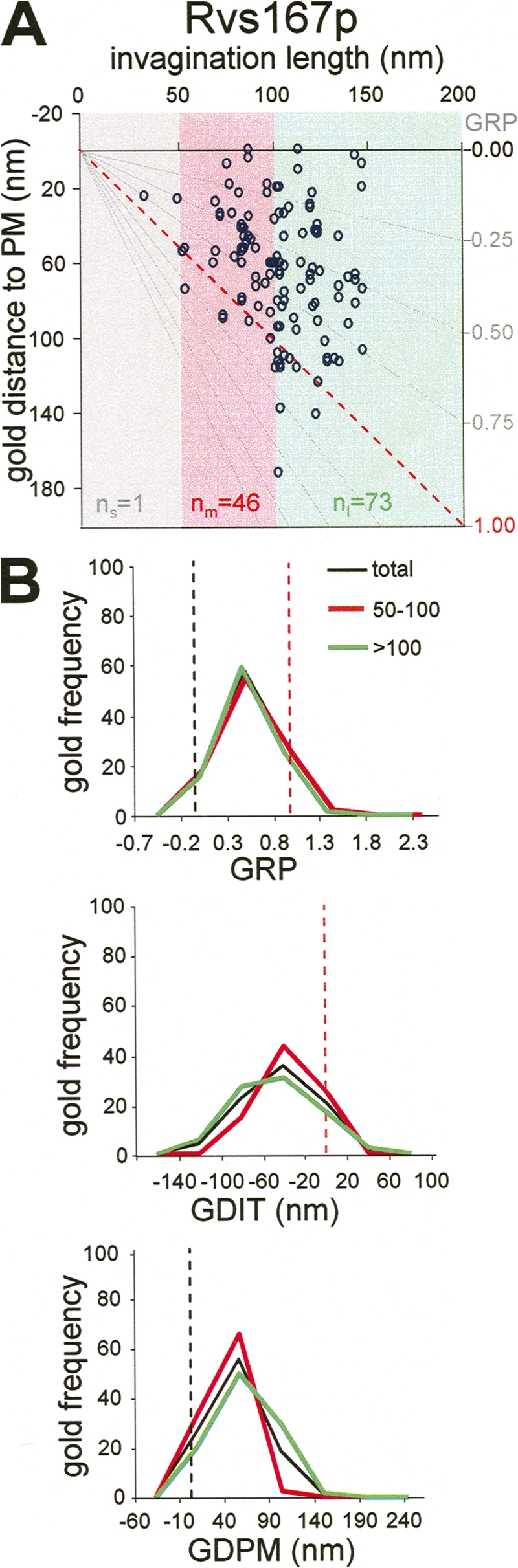
The yeast amphiphysin Rvs167p occupies a subapical area on plasma membrane–associated invaginations. (A) Graph representing the GDPM for each recorded gold particle labeling Rvs167p versus the IL. The x axis represents the level of the basal plasma membrane. The red dotted line indicates the position of the invagination tip. Gray lines converging at the xy axes crossing point define the GRP areas (left axis) as a function of the IL. Gray, red, and green areas enclose the gold subpopulations associated with short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) profiles, respectively. Sample sizes of subpopulations associated with short, intermediate, and long profiles are indicated as n s, n m, and n l, respectively. (B) Graphs representing the frequency of gold labeling Rvs167p binned according to their GRP, GDIT, or GDPM. The entire gold subpopulation (total; black) and the subpopulations associated with intermediate (50–100; red) and long (>100; green) profiles were analyzed. The number of gold particles with values falling into the indicated intervals was counted and normalized with respect to the total population (n g; Fig. 2) or for the corresponding subtotals (n m and n l). Dotted black and red lines indicate the positions of the basal plasma membrane and the invagination tip, respectively. Immunogold-labeled ultrathin sections were prepared as described in Fig. 1.
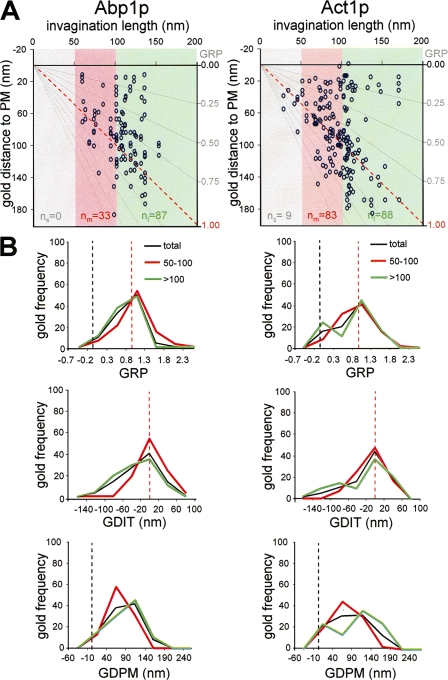
Figure 5.
Actin segregates into two distinct pools in long plasma membrane invaginations. (A) Graphs representing the GDPM for each recorded gold particle labeling Abp1p or actin (Act1p) versus the IL. The x axis represents the level of the basal plasma membrane. The red dotted lines indicate the positions of the invagination tip. Gray lines converging at the xy axes crossing point define the GRP areas (left axis) as a function of the IL. Gray, red, and green areas enclose the gold subpopulations associated with short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) profiles, respectively. Sample sizes of subpopulations associated with short, intermediate, and long profiles are indicated as n s, n m, and n l, respectively. (B) Graphs representing the frequency of gold labeling Abp1p or Act1p binned according to their GRP, GDIT, or GDPM. The entire gold subpopulation (total; black) and the subpopulations associated with intermediate (50–100; red) and long (>100; green) profiles were analyzed. The number of gold particles with values falling into the indicated intervals was counted and normalized with respect to the total population (n g; Fig. 2) or for the corresponding subtotals (n m and n l). Dotted black and red lines indicate the positions of the basal plasma membrane and the invagination tip, respectively. Immunogold-labeled ultrathin sections were prepared as described in Fig. 1.
Figure 6.
Myo5p but not Las17p or Bbc1p mimics the distribution of actin in long profiles. (A) Graphs representing the GDPM for each recorded gold particle labeling Myo5p, Bbc1p, or Las17p versus the IL. The x axis represents the level of the basal plasma membrane. The red dotted lines indicate the positions of the invagination tip. Gray lines converging at the xy axes crossing point define the GRP areas (left axis) as a function of the IL. Gray, red, and green areas enclose the gold subpopulations associated with short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) profiles, respectively. Sample sizes of subpopulations associated with short, intermediate, and long profiles are indicated as n s, n m, and n l, respectively. (B) Graphs representing the frequency of gold labeling Myo5p, Bbc1p, or Las17p binned according to their GRP, GDIT, or GDPM. The entire gold subpopulation (total; black) and the subpopulations associated with intermediate (50–100; red) and long (>100; green) profiles were analyzed for Myo5p, Bbc1p, and Las17p. In addition, the distribution for the Las17p subpopulation associated with short profiles (≤ 50 nm; gray) was included. The number of gold particles with values falling into the indicated intervals was counted and normalized with respect to the total population (n g; Fig. 2) or for the corresponding subtotals (ns, n m, and n l). Dotted black and red lines indicate the positions of the basal plasma membrane and the invagination tip, respectively. Immunogold-labeled ultrathin sections were prepared as described in Fig. 1.
Analysis of gold labeling the coat components Sla1p, Pan1p, and Chc1p indicated that they were all tightly associated with the invagination tip (Figs. 1 and 3 B). Particles labeling these proteins exhibited mean GRP values near 1, with >80% of the particles located <40 nm from the invagination tip (Fig. 3 C and Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200708060/DC1). The observation that the mean GDIT did not significantly change with the IL, whereas the mean GDPM clearly increased (Fig. 3, B and C; Table II; and Table S2), suggested that the coat travels into the cytosol at the tip of the tubular profiles. Consistent with the similar dynamics of Sla1p, Pan1p, and Chc1p at the endocytic patch (Kaksonen et al., 2005), statistical comparison of the coat components failed to demonstrate significant differences (P < 0.02) for any of the positional parameters (GRP, GDIT, GDPM, and GDLB) analyzed (Table III). All of these results strongly suggested that the endocytic coat forms a complex that moves into the cytosol at the tip of tubular profiles.
Table II. Homogeneity analysis for the GRP, GDIT, GDPM, and GDLB parameters using the Kruskal-Wallis test.
| Protein | GRP | GDIT | GDPM | GDLB |
|---|---|---|---|---|
| Sla1p | 0.706 | 0.414 | 0.000 | 0.142 |
| Pan1p | 0.086 | 0.096 | 0.001 | 0.003 |
| Chc1p | 0.172 | 0.058 | 0.000 | 0.174 |
| Abp1p | 0.004 | 0.001 | 0.109 | 0.474 |
| Act1p | 0.394 | 0.368 | 0.002 | 0.891 |
| Rvs167p | 0.907 | 0.026 | 0.001 | 0.346 |
| Myo5p | 0.086 | 0.096 | 0.001 | 0.003 |
| Bbc1p | 0.085 | 0.351 | 0.000 | 0.013 |
| Las17p | 0.500 | 0.000 | 0.025 | 0.595 |
Table III. Protein comparison for the GRP, GDIT, GDPM, and GDLB parameters using a generalized linear model.
| Protein | Sla1p | Pan1p | Chc1p | Abp1p | Act1p | Rvs167p | Myo5p | Bbc1p | Las17p |
|---|---|---|---|---|---|---|---|---|---|
| GRP | |||||||||
| Sla1p | 1.0000 | 0.5897 | 0.0373 | 0.1691 | 0.6899 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Pan1p | 1.0000 | 0.1271 | 0.3686 | 0.8670 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Chc1p | 1.0000 | 0.6080 | 0.0795 | 0.0001 | <0.0001 | <0.0001 | 0.0002 | ||
| Abp1p | 1.0000 | 0.2529 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |||
| Act1p | 1.0000 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||
| Rvs167p | 1.0000 | <0.0001 | 0.5524 | 0.7000 | |||||
| Myo5p | 1.0000 | 0.0006 | <0.0001 | ||||||
| Bbc1p | 1.0000 | 0.3216 | |||||||
| Las17p | 1.0000 | ||||||||
| GDIT | |||||||||
| Sla1p | 1.0000 | 0.7505 | 0.1386 | 0.0955 | 0.2626 | 0.0023 | 0.0004 | 0.0020 | 0.0035 |
| Pan1p | 1.0000 | 0.1548 | 0.0916 | 0.3336 | <0.0001 | <0.0001 | 0.0003 | 0.0007 | |
| Chc1p | 1.0000 | 0.7821 | 0.4514 | 0.0008 | <0.0001 | 0.0005 | 0.0025 | ||
| Abp1p | 1.0000 | 0.2347 | <0.0001 | <0.0001 | <0.0001 | 0.0005 | |||
| Act1p | 1.0000 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||
| Rvs167p | 1.0000 | 0.0074 | 0.8129 | 0.6325 | |||||
| Myo5p | 1.0000 | 0.0158 | 0.0045 | ||||||
| Bbc1p | 1.0000 | 0.4923 | |||||||
| Las17p | 1.0000 | ||||||||
| GDPM | |||||||||
| Sla1p | 1.0000 | 0.7755 | 0.0206 | 0.0619 | 0.3468 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Pan1p | 1.0000 | 0.0346 | 0.0944 | 0.5070 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Chc1p | 1.0000 | 0.0871 | 0.5058 | 0.0013 | <0.0001 | 0.0023 | 0.0041 | ||
| Abp1p | 1.0000 | 0.2281 | <0.0001 | <0.0001 | <0.0001 | 0.0002 | |||
| Act1p | 1.0000 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||
| Rvs167p | 1.0000 | 0.0005 | 0.8670 | 0.8809 | |||||
| Myo5p | 1.0000 | 0.0003 | 0.0006 | ||||||
| Bbc1p | 1.0000 | 0.9947 | |||||||
| Las17p | 1.0000 | ||||||||
| GDLB | |||||||||
| Sla1p | 1.0000 | 0.2386 | 0.2246 | 0.0001 | <0.0001 | 0.6496 | 0.4625 | 0.0642 | 0.1708 |
| Pan1p | 1.0000 | 0.2246 | 0.0030 | <0.0001 | 0.0991 | 0.5687 | 0.4734 | 0.8380 | |
| Chc1p | 1.0000 | 0.0002 | <0.0001 | 0.6888 | 0.4411 | 0.0638 | 0.1607 | ||
| Abp1p | 1.0000 | 0.0424 | <0.0001 | 0.0001 | 0.0144 | 0.0051 | |||
| Act1p | 1.0000 | <0.0001 | <0.0001 | <0.0001 | <.0001 | ||||
| Rvs167p | 1.0000 | 0.2151 | 0.0192 | 0.0657 | |||||
| Myo5p | 1.0000 | 0.1672 | 0.4266 | ||||||
| Bbc1p | 1.0000 | 0.6090 | |||||||
| Las17p | 1.0000 |
The apical distribution observed for Sla1p, Pan1p, and Chc1p seemed to be specific for the coat components. Gold labeling Rvs167p preferred a slightly negatively curved area situated immediately beneath the region where particles labeling the coat accumulated (Fig. 1 and Fig. 4 A). Statistical comparison of Rvs167p with the coat components demonstrated very significant differences for all positional parameters analyzed except for GDLB (Table III). Rvs167p labeling was found concentrated within a subapical area, with a mean GRP near 0.6 (Figs. 1 and 4 B; and Table S2). The mean GDIT for Rvs167p was close to −40 nm (Fig. 4 B and Table S2). Again, it did not significantly change as a function of IL, whereas the mean GDPM did, suggesting that the yeast amphiphysin follows the endocytic coat as it moves into the cytosol (Fig. 4 and Tables II and S2).
Analysis of the distribution of gold decorating Abp1p and actin indicated that these proteins form a loose cloud around the tip that spreads toward the base (Fig. 1 and Fig. 5). The actin and Abp1p distribution was consistent with that previously described using rapid-freeze and deep-etch electron microscopy techniques (Rodal et al., 2005). Statistical comparison of Abp1p or actin with the coat components did not show significant differences regarding the GRP, GDIT, and GDPM parameters (Table III). Similar to the coat, Abp1p and actin had GRP mean values near 1 even though the particles decorating these proteins seemed to spread more along the entire invagination (Fig. 5 and Table S2). However, in contrast to Rvs167p, Abp1p and actin exhibited very significant differences with the coat components when the GDLB parameter was used for comparison (Table III). On average, particles decorating Sla1p, Pan1p, Chc1p, and Rvs167p were found at ∼10 nm from the lipid bilayer, whereas those decorating Act1p and Abp1p were located at ∼20 nm from the membrane (Table S2).
Closer inspection of the distribution of particles labeling actin and Abp1p demonstrated some other important differences with the coat (Figs. 3 and Figs.5). In contrast to Sla1p, Pan1p, and Chc1p, the Kruskal-Wallis test showed significant divergences between the subpopulations of particles labeling Abp1p on intermediate (50 nm ≤ IL < 100 nm) and long (IL ≥ 100 nm) profiles for GRP and GDIT but not for GDPM (Table II). Analysis of the gold frequency binned for GDIT on intermediate and long profiles indicated that in contrast to the coat components, Abp1p spreads toward the profile base as the invagination elongates (Fig. 5 B). Interestingly, particles labeling actin did not exhibit the same pattern as Abp1p in the longest profiles (Fig. 5). Analysis of the gold frequency binned for GRP, GDIT, and GDPM indicated that in long profiles, actin split into two different pools associated with the base and the tip of the invagination (Figs. 1 and 5 B). The two gold subpopulations were most evident when profiles longer than 110 nm were analyzed (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708060/DC1). In the very long invaginations, ∼35% and 45% of the particles labeling actin were found associated with the invagination base (GRP = −0.2–0.3) and tip (GRP = 0.8–1.3), respectively, whereas <5% was found at the intermediate region (GRP = 0.3–0.8); Fig. S1 A). The χ2 test applied to the gold frequencies in those GRP regions for three independent gold countings demonstrated that the likelihood ratio was very small (P = 0.013; Fig. S1 B).
Finally, we investigated the distribution of gold decorating the most potent actin-nucleating promoting factors during endocytic uptake: Myo5p and Las17p (Sun et al., 2006). In addition, we analyzed the distribution of Bbc1p, a protein that has been shown to act as an inhibitor of both Las17p and Myo5p (Sun et al., 2006). Myo5p, Las17p, and Bbc1p remain at the plasma membrane as the coat and the actin modules move into the cytosol. Thus, we expected to see important differences in their distribution when compared with other endocytic modules.
Accordingly, we found that Myo5p labeling exhibited an exclusive pattern, with most particles located near the basal plasma membrane (Fig. 6). More than 65% of the gold had GDPM values inferior to 40 nm (Fig. 6 B and Table S2). Gold particles decorating Myo5p were often tightly associated with the negatively curved area (Fig. 1). Statistical comparison of the entire population of gold labeling Myo5p with those decorating the coat, actin, or fission modules showed very significant differences for GRP, GDIT, and GDPM (Table III). Interestingly, however, closer inspection of the Myo5p gold distribution in the longest profiles indicated that similar to actin, the myosin-I segregated into two distinct pools: one of them was still associated with the invagination base, and the other was associated with the tip (Figs. 1 and 6 B). In profiles longer than 110 nm, ∼55% and 35% of the particles labeling Myo5p were found at the invagination base (GRP = −0.2–0.3) or at the tip (GRP = 0.8–1.3), respectively, whereas <10% was close to the intermediate region (GRP = 0.3–0.8; Fig. S1 A). A χ2 test applied to the gold frequencies in those GRP regions for three independent gold countings demonstrated that the likelihood ratio was very small (P ≤ 0.001; Fig. S1 B).
Surprisingly, although Bbc1p and Las17p physically and functionally interact with Myo5p (Anderson et al., 1998; Geli et al., 2000; Lechler et al., 2000; Drees et al., 2001; Mochida et al., 2002), and Bbc1p and Myo5p have identical dynamics at the cortical patch (Sun et al., 2006), the distribution of gold labeling Bbc1p and Las17p was significantly different from those decorating Myo5p, specially when GRP and GDPM parameters were considered (P < 0.001; Fig. 6 and Table III). Similar to Rvs167p (Fig. 4), gold against Bbc1p and Las17p mostly occupied the invagination intermediate area, with GRP values near 0.6 regardless of the IL (Fig. 6 B, Table II, and Table S2). Consistently, protein comparison using a generalized linear model could not demonstrate significant differences between Bbc1p, Las17p, and Rvs167p for any of the positional parameters analyzed except for GDLB (particles decorating Bbc1p appeared slightly more separated from the lipid bilayer; Table III and Table S2). In contrast, statistical analysis demonstrated very significant differences (P < 0.008) with Myo5p for GRP, GDIT, and GDPM (Figs. 4 and 6, Table III, and Table S2).
Two distinct acto/myosin-I pools associate with deeply invaginated endocytic profiles
Our data strongly indicated that proteins involved in the endocytic uptake segregate along plasma membrane invaginations. One of our most striking observations, which could not be predicted from previous experiments, was that actin and Myo5p rearrange in the longest profiles to generate two acto/myosin-I structures associated with the base and tip of the endocytic invaginations.
Differences and similarities between the patterns of particles labeling Myo5p and actin as a function of IL were most obvious when subpopulations associated with profiles shorter than 80 nm or longer than 110 nm were compared (Fig. S1 and Table IV). The Kruskal-Wallis test failed to evidence statistically significant differences between the actin and Myo5p gold subpopulations associated with profiles longer than 110 nm for any positional parameters, whereas highly significant differences (P < 0.0004) could be demonstrated for all parameters in shorter profiles using similar sample sizes (Table IV). These data demonstrated that although in short profiles, the distributions of gold decorating actin and Myo5p were complementary, in the longest profiles, actin and myosin-I occupied the same invagination areas, and even they were placed at a similar distance from the lipid bilayer (Figs. 5, 6, and S1 A).
Table IV. Homogeneity analysis for the GRP, GDIT, GDPM, and GDLB parameters using the Kruskal-Wallis test for the indicated Myo5p and Act1p subpopulations.
| Protein | IL | Act1pIL ≤ 80 nm | Act1pIL ≥ 110 nm | Myo5pIL ≤ 80 nm | Myo5pIL ≥ 110 nm |
|---|---|---|---|---|---|
| GPR | |||||
| Act1p | IL ≤ 80 nm | 1.0000 | 0.0216 | <0.0001 | ND |
| Act1p | IL ≥ 110 nm | 1.0000 | ND | 0.0995 | |
| Myo5p | IL ≤ 80 nm | 1.0000 | 0.1300 | ||
| Myo5p | IL ≥ 110 nm | 1.0000 | |||
| GDIT | |||||
| Act1p | IL ≤ 80 nm | 1.0000 | 0.0375 | <0.0001 | ND |
| Act1p | IL ≥ 110 nm | 1.0000 | ND | 0.2782 | |
| Myo5p | IL ≤ 80 nm | 1.0000 | 0.1521 | ||
| Myo5p | IL ≥ 110 nm | 1.0000 | |||
| GDPM | |||||
| Act1p | IL ≤ 80 nm | 1.0000 | 0.0350 | <0.0001 | ND |
| Act1p | IL ≥ 110 nm | 1.0000 | ND | 0.0271 | |
| Myo5p | IL ≤ 80 nm | 1.0000 | <0.0001 | ||
| Myo5p | IL ≥ 110 nm | 1.0000 | |||
| GDLB | |||||
| Act1p | IL ≤ 80 nm | 1.0000 | 0.9429 | 0.0004 | ND |
| Act1p | IL ≥ 110 nm | 1.0000 | ND | 0.7070 | |
| Myo5p | IL ≤ 80 nm | 1.0000 | 0.0004 | ||
| Myo5p | IL ≥ 110 nm | 1.0000 |
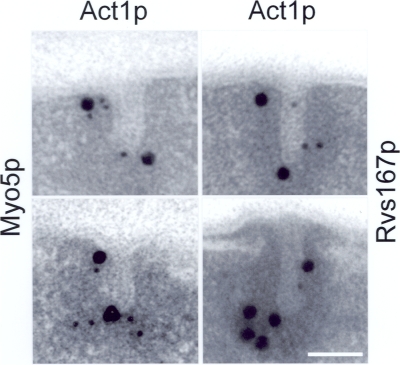
To investigate whether two distinct acto/myosin-I pools actually exist, double-labeling experiments on ultrathin sections of yeast cells expressing HA-tagged Myo5p were performed using the rat monoclonal anti-HA and the mouse monoclonal anti-actin antibodies combined with 10-nm and 20-nm gold-conjugated goat anti–rat and mouse IgGs, respectively. Tight colocalization of gold particles decorating Myo5p (10 nm) and actin (20 nm) could be observed at the base and at the tip of long invaginations (Fig. 7). Furthermore, to demonstrate that the two acto/myosin-I pools existed in bona fide endocytic invaginations previous to the fission event, double-labeling experiments were performed for HA-tagged Rvs167p and actin. The distal and proximal actin pools could also be demonstrated on long profiles that were also immunodecorated for Rvs167p at the intermediate area (Fig. 7).
Figure 7.
Two distinct acto/myosin-I pools associate with deep plasma membrane invaginations. Representative electron micrographs of ultrathin sections showing the colocalization of either Myo5p and Act1p or Rvs167p and Act1p. Strains expressing HA-tagged Myo5p (ScMIG994) or Rvs167p (ScMIG995) were immunolabeled with a rat anti-HA monoclonal antibody or the mouse monoclonal anti-actin antibody and the appropriate 10-nm (Myo5p and Rvs167p) and 20-nm (Act1p) gold-conjugated secondary IgGs, respectively. Bar, 100 nm.
Discussion
In contrast to mammalian cells, ultrastructural studies on the endocytic pathway in yeast have been scarce. An early study describing the yeast actin cytoskeleton demonstrated plasma membrane invaginations coated with actin and Abp1p that were proposed as endocytic intermediates (Mulholland et al., 1994). However, the same authors failed to localize endocytic cargo on this type of membrane invagination, leaving the definition of the primary endocytic profiles still open (Mulholland et al., 1999).
Analysis of the endocytic uptake using live cell fluorescence microscopy now provides tools to better interpret the electron microscopy results. Live cell imaging shows that assembly of the coat precedes sequential recruitment of all other proteins involved in the process (Kaksonen et al., 2003, 2005; Jonsdottir and Li, 2004; Newpher et al., 2005; Sun et al., 2006). The data also demonstrate that the coat remains associated with the vesicles until it moves into the cytosol (Kaksonen et al., 2003). Accordingly, plasma membrane invaginations labeled for the endocytic coat necessarily represent the primary endocytic profiles. Immunolabeling of Sla1p, Pan1p, and clathrin on ultrathin sections demonstrated that the primary endocytic profiles in yeast are tubular invaginations of ∼50 nm and up to 180 nm in length, which are often surrounded by an electron-dense material. Not only the gold against these proteins decorated the same type of profiles, but also, consistent with the view that Sla1p, Chc1p, and Pan1p form an endocytic coat, the distribution of particles along the membrane profiles was remarkably similar (Fig. 3, Fig. 8, and Table III). More than 95% of the gold particles labeling the endocytic coat (n = 360) were found at <50 nm from the invagination tip. Because the distance of the gold particle to the actual antigen is ∼20 nm (Hermann et al., 1996), our data suggest that the endocytic coat roughly occupies the first 30 nm from the invagination tip.
Figure 8.
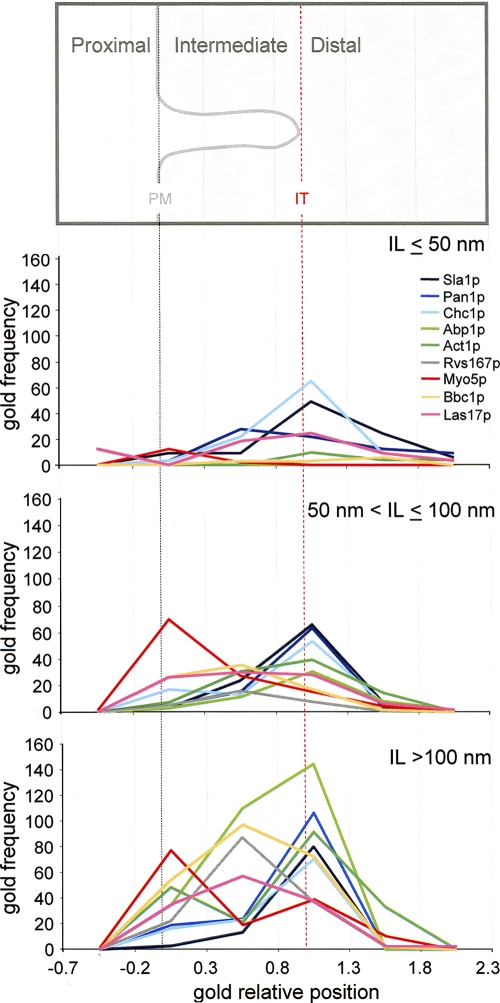
Comparative distribution of endocytic proteins along short, intermediate, and long plasma membrane–associated invaginations. The graphs represent the relative frequency of immunogold associated with the indicated proteins on short (IL ≤ 50 nm), intermediate (50 nm < IL ≤ 100 nm), and long (IL > 100 nm) profiles normalized for the Sla1p gold subpopulations associated with those profiles binned according to their GRP. Immunogold particles were binned according to their association with the indicated areas and normalized to the Sla1p n s, n m, and n l sample sizes (Fig. 3) for comparison. The number of immunogold particles decorating Act1p and Myo5p was first multiplied by the correction factor (120/180) to equal the sample size. A scheme illustrating the different invagination areas is shown in which the levels of the invagination tip (IT) and the basal plasma membrane (PM) are represented as vertical lines. Immunogold-labeled ultrathin sections were prepared as described in Fig. 1.
Live cell imaging also demonstrates that the endocytic coat travels unidirectional into the cytosol concomitant with the sequential recruitment of all other endocytic modules (Kaksonen et al., 2003). Interestingly, we found that the frequency of gold labeling the different endocytic proteins binned according to the length of the associated invagination recapitulated their temporal pattern of recruitment to the cortical patch (Figs. 2 and Figs.8; Kaksonen et al., 2003, 2005; Jonsdottir and Li, 2004; Newpher et al., 2005; Sun et al., 2006). This indicates that invaginations of increasing length represent progressively more mature endocytic profiles. Accordingly, the observation that small invaginations were practically devoid of particles decorating proteins recruited during the slow inward movement implies that initial membrane bending occurs at some point during the immotile phase and, thus, before massive actin polymerization. Initial membrane curvature in yeast might require the epsin N-terminal homology domains of some coat components and/or clathrin polymerizing in a scaffold, as proposed for clathrin-dependent budding in mammalian cells (Hurley and Wendland, 2002; Hinrichsen et al., 2006).
A molecular model was recently proposed to explain actin-driven membrane invagination during endocytic uptake in yeast (Kaksonen et al., 2006). According to the model, the strongest actin-nucleating promoting factors, Myo5p and Las17p, would concentrate at the rim of preassembled endocytic coated pits. Attachment of actin filaments to the endocytic coat together with continuous nucleation of actin at the plasma membrane would then push the pit inwards and lead to the formation of a tubular invagination. Our observations (1) that the primary endocytic profiles are tubular invaginations that accumulate the coat components at the tip, (2) that a slightly bended coated pit is probably formed before initiation of the slow inward movement, (3) that actin forms a loose cloud around the endocytic coat, and (4) that Myo5p accumulates at the invagination base are in striking agreement with the proposed model.
However, in the context of that model, it was unexpected to find that Las17p labeling did not concentrate at the base of the invagination but rather distributed along the intermediate area (Figs. 1, Figs.6, and Figs.8). Furthermore, we found that the distribution of Las17p on invaginations longer than 50 nm very much resembled that of Bbc1p (Figs. 6 and Figs.8 and Table III), suggesting that the nucleating activity of Las17p might be switched off by association with Bbc1p at this stage (Rodal et al., 2003; Sun et al., 2006). Accordingly, Myo5p has been shown to be the primary Arp2/3p activator during the slow inward movement (Sun et al., 2006).
Las17p-induced actin polymerization could play a major role previous to Bbc1p recruitment. An interesting possibility is that Las17p promotes initial assembly of a disordered actin network on the surface of the coated pit, which is then linked to the endocytic coat and serves as an anchoring scaffold for a more productive burst of actin polymerization organized by Myo5p at the profile base. Consistent with a role of Las17p controlling the onset of the slow inward movement rather than in promoting actin polymerization at this stage, it was shown that deletion of the Las17p domains required for its nucleating activity does not abolish but rather delays membrane invagination (Sun et al., 2006).
Our ultrastructural experiments now clarify some similarities and differences between the clathrin-dependent endocytic uptake in mammalians and yeast. Clathrin-dependent endocytosis in higher eukaryotes involves the formation of a spherical constricted pit stabilized by a clathrin lattice. In yeast, although a partially curved endocytic coat is probably assembled, clathrin polymerization fails to drive constriction of the incipient pit. Instead, an actin-based mechanism that involves the formation of a tubular profile is in charge of completing budding. The structure of the yeast clathrin might preclude its assembly into a lattice or provide insufficient force to bend the lipid bilayer. Alternatively, the thickness or the rigidity of the yeast plasma membrane might prevent closure of the incipient coat. Interestingly, actin polymerization and myosin-I recruitment occur on a subset of forming clathrin-coated pits in mammalian cells (Merrifield et al., 2002; Krendel et al., 2007). The lipid composition of the donor membrane and/or the nature of the endocytosed cargo might also define in mammalians the local stiffness of the lipid bilayer and, thus, the necessity for an actin-based mechanism to help membrane bending or vesicle scission.
In mammalian cells, scission of clathrin-coated buds requires the GTPase dynamin, which may work as a regulatory enzyme and/or as a molecular motor that constricts the vesicle neck (Song and Schmid, 2003). Strikingly, dynamin does not seem to be required for endocytic uptake in yeast (Gammie et al., 1995; Yu and Cai, 2004). Our study now provides important information regarding a possible alternative mechanism for vesicle fission. Statistical analysis of the distribution of gold particle labeling actin and Myo5p and double-labeling experiments demonstrated that in long profiles, actin and myosin-I form two distinct structures separated by the scission machinery (Fig. 8). A cooperative role for myosin-I with the fission machinery is consistent with our own unpublished observations demonstrating synthetic growth defects when combining mutations in the yeast amphiphysins and myosins-I (unpublished data) and also with the observation that a point mutation in the myosin-I lipid-binding domain causes accumulation of long plasma membrane–associated invaginated profiles in yeast (Jonsdottir and Li, 2004). The basal acto/myosin-I structure might form a ringlike structure that either stiffens the base of the endocytic profile or acts as a contractile ring in a way similar to what has been proposed for dynamin (Song and Schmid, 2003). The observations that actin might adopt a spiral-like arrangement around plasma membrane invaginations (Mulholland et al., 1994) and that myosin-I can cross-link and/or slide actin filaments (Pollard et al., 1991) support these hypotheses. The distal myosin-I pool could anchor the incipient endocytic profile to preformed actin cables or to internal compartments (Toshima et al., 2006) to provide the tension necessary for scission (Roux et al., 2006).
Characterization of the distribution of other proteins required for the endocytic uptake and analysis of the ultrastructure of the endocytic profiles in mutants with internalization defects are now required to unveil the mechanism of primary endocytic budding in yeast.
Materials and methods
Yeast strains and plasmid construction
The yeast strains used are listed in Table V. HA tag was fused at the C terminus of each protein by homologous recombination in the gene as described previously (Wach et al., 1997). PCRs were performed using a DNA polymerase with proof reading activity (Vent polymerase; New England Biolabs, Inc.). Oligonucleotides were synthesized by MWG-Biotech AG. pMIG692 is a centromeric shuttle vector based on pYCplac111 containing a C-terminal HA-tagged SLA1 gene under its own promoter and the selectable marker HIS3MX. It was constructed by homologous recombination in yeast. Unless otherwise mentioned, strains without plasmid were grown in complete yeast peptone dextrose medium, and strains with plasmid were grown on synthetic dextrose complete medium without leucin at 26°C (Guthrie and Fink, 1991). Transformation of yeast was accomplished by the lithium acetate method (Ito et al., 1983).
Table V. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| RH2881 | _MAT_a his3 leu2 ura3 trp1 bar1-1 | H. Riezmana |
| RH2634 | _MAT_a his3 leu2 ura3 bar1-1 sla1Δ∷URA3 | H. Riezmana |
| ScMIG516 | _MAT_a his3 leu2 ura3 trp1 bar1-1 LAS17-3HA∷TRP1 | This study |
| ScMIG946 | _MAT_a his3 leu2 ura3 trp1 bar1-1 PAN1-3HA∷TRP1 | This study |
| ScMIG903 | _MAT_a his3Δ1 ura3Δ0 leu2Δ10 met15Δ0 bar1Δ∷URA3 BBC1-3HA∷HIS3MX | This study |
| ScMIG100 | _MAT_a his3Δ1 ura3Δ0 leu2Δ10 met15Δ0 bar1Δ∷URA3 | This study |
| ScMIG723 | _MAT_a his3Δ1 ura3Δ0 leu2Δ10 met15Δ0 bar1Δ∷URA3 ABP1-3HA∷HIS3MX | This study |
| ScMIG994 | _MAT_a his3Δ1 ura3Δ0 leu2Δ10 met15Δ0 bar1Δ∷URA3 MYO5-3HA∷HIS3MX | This study |
| ScMIG995 | _MAT_a his3Δ1 ura3Δ0 leu2Δ10 met15Δ0 bar1Δ∷URA3 RVS167-3HA∷HIS3MX | This study |
Preparation of yeast ultrathin sections and immunolabeling
Cells were grown in yeast peptone dextrose medium to 4–5 × 106 cells/ml and harvested over a disposable Stericup 0.22-μm filter unit with a vacuum of −20 to −15 Hg, leaving behind 5 ml of media. 25 ml of 1.2× fixative solution was immediately added to obtain final concentrations of 0.04 M KPO4, pH 6.6, 0.6 M sorbitol, 4% formaldehyde (from a 16% ethanol-free ultrapure electron microscopy–grade solution; Polysciences, Inc.), 0.4% glutaraldehyde (25% electron microscopy–grade solution; Fluka), 1 mM MgCl2, 0.5 mM EGTA (glycol-bis[2-aminoethylether]-N,N,_N_′,_N_′–tetraacetic acid), 10 mM NaF, and 10 mM KF. Cells were then transferred to a 50-ml Falcon tube (BD Biosciences), and fixation was continued overnight at 4°C with rolling. Subsequent steps (metaperiodate and ammonium chloride treatments, dehydration, infiltration, embedding, and sectioning) were performed as described previously (Mulholland et al., 1994) without further modifications. For immunolabeling, ultrathin sections were incubated for 15 min in blocking buffer (10 mM KPO4, pH 7.5, 150 mM NaCl, 2% BSA, and 0.05% Tween 20), transferred to a 25-μl drop of primary antibody in blocking buffer for 3 h, and washed over 30 min in washing buffer (10 mM KPO4 buffer, pH 7.5, 150 mM NaCl, and 0.05% Tween 20). After blocking for 15 min, grids were incubated with the corresponding gold-conjugated secondary antibody for 60 min and were washed first for 30 min in washing buffer and then for a further 30 min in washing buffer without Tween 20. All steps of immunolabeling were performed at room temperature. Grids were then washed in double-distilled water and poststained with uranyl acetate (2% in water) over 60 min and lead citrate for 30 s as described previously (Reynolds, 1963). For double immunolabeling, ultrathin sections were incubated with a mixture of the two first antibodies for 3 h, washed as described for the single immunolabeling, incubated with a mixture of the corresponding 1:50 diluted secondary antibodies, and conjugated to gold particles of different sizes. 8 μg/ml anti-HA rat monoclonal antibody (3F10; Roche), 200 μg/ml mouse anti-actin monoclonal antibody (C4; Chemicon International), and a mixture of six mouse anti-Chc1p monoclonal antibodies diluted 1:5 (Lemmon et al., 1988) were used as primary antibodies for the detection of HA-tagged proteins, actin, and clathrin, respectively (the antibodies against clathrin were gifts from S.K. Lemmon, University of Miami, Miami, FL). 20-nm gold-conjugated goat anti–mouse IgG (EM.GMHL20) and 10-nm gold-conjugated anti–rat IgG (EM.GAT10; BB International) were used as secondary antibodies (diluted 1:50). Ultrathin sections were examined using a transmission electron microscope (model 1010; JEOL) at 50 Kv accelerating voltage. Micrographs of the yeast plasma membrane invaginations were acquired at 100,000× magnification with a CCD camera (MegaView III; Olympus) and image acquisition analySIS software (Olympus). Adjustments of image size, brightness, and contrast were performed on Photoshop 5.0 (Adobe). The IL was measured on immunoelectron micrographs as the distance from a reference line that defines the basal plasma membrane to the invagination tip. The GDPM was measured as the minimal distance from the basal plasma membrane to the center of the gold particle. The GDLB was measured as the minimal distance from the gold center to the lipid bilayer. The distance measurements on micrographs were performed by Photoshop 5.0.2 ruler.
The specificity of the anti-HA labeling on plasma membrane invaginations was assessed for the less abundant endocytic protein Las17p at the endocytic patch and for the most transiently recruited one, Rvs167p (Sun et al., 2006). 45% of the plasma membrane invaginations (n i = 200) on ultrathin sections of the strain expressing HA-tagged Las17p (ScMIG516) were labeled with immunogold particles versus 12.5% of the plasma membrane invaginations (n i = 200) on ultrathin sections of the isogenic wild type (ScMIG228). For Rvs167p, 61% of the plasma membrane invaginations (n i = 200) on ultrathin sections of the strain expressing HA-tagged Rvs167p (ScMIG995) were labeled with immunogold particles versus 9.5% of the plasma membrane invaginations (n i = 200) on ultrathin sections of the isogenic wild type (ScMIG100).
Statistical analysis of the immunogold distribution
All statistical analyses of the immunogold distributions were performed with a statistical package for Windows (version 15.0; SPSS). A nonparametric Kruskal-Wallis test (Kruskal and Wallis, 1952) was used for comparison of the immunogold subpopulation associated with the intermediate (50 nm ≤ IL < 100 nm) and long (IL ≥ 100 nm) profiles within the same protein and for comparison of the Act1p and Myo5p distributions in short (IL ≤ 80 nm) and very long (IL ≥ 110 nm) profiles. A generalized linear model (McCullagh and Nelder, 1989) was applied for comparison of proteins using the IL, GRP, GDPM, and GDIT parameters. A χ2 test (Cochran, 1954) was used to analyze the likelihood ratio between the frequencies of gold particles decorating Myo5p or actin on the invagination basal, intermediate, and apical regions.
Online supplemental material
Fig. S1 shows statistical analysis of the distribution of gold particles labeling actin and Myo5p on invaginations shorter than 80 nm or longer than 110 nm. Table S1 shows the descriptive statistics for the IL parameter. Table S2 shows the descriptive statistics for the GRP, GDIT, GDPM, and GDLB parameters. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708060/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We thank E. Coll, G. Martínez, and C. López-Iglesias (University of Barcelona) for electron microscopy technical support, M. Pons and M. Cairó for technical assistance, A. Espinal and A. Blasco (Universidad Autónoma de Barcelona) for statistical analysis, H. Girao, S. Lemmon, T. Newpher, and B. Grosshans for critical reading of the manuscript, and S.K. Lemmon for providing material.
This work was supported by a grant from the Ministerio de Educación y Ciencia (MEC; grant BUF2005-04089). H. Grotsch is a recipient of a predoctoral fellowship from the Generalitat de Catalunya. F.-Z. Idrissi is a recipient of the Ramón y Cajal postdoctoral contract from the MEC. I.M. Fernandez-Golbano is a recipient of a predoctoral fellowship from the MEC.
Abbreviations used in this paper: GDIT, gold distance to invagination tip; GDLB, gold distance to lipid bilayer; GDPM, gold distance to plasma membrane; GRP, gold relative position; IL, invagination length; WASP, Wiskott-Aldrich syndrome protein.
References
- Anderson, B.L., I. Boldogh, M. Evangelista, C. Boone, L.A. Greene, and L.A. Pon. 1998. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J. Cell Biol. 141:1357–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran, W.G. 1954. Some methods for strengthening the common χ2 tests. Biometrics. 10:417–451. [Google Scholar]
- Drees, B.L., B. Sundin, E. Brazeau, J.P. Caviston, G.C. Chen, W. Guo, K.G. Kozminski, M.W. Lau, J.J. Moskow, A. Tong, et al. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., and D.G. Drubin. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19:287–332. [DOI] [PubMed] [Google Scholar]
- Gammie, A.E., L.J. Kurihara, R.B. Vallee, and M.D. Rose. 1995. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130:553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli, M.I., R. Lombardi, B. Schmelzl, and H. Riezman. 2000. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 19:4281–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology (Methods in Enzymology). Vol. 194. Academic Press Inc., San Diego, CA. 933 pp. [PubMed]
- Hermann, R., P. Walther, and M. Muller. 1996. Immunogold labeling in scanning electron microscopy. Histochem. Cell Biol. 106:31–39. [DOI] [PubMed] [Google Scholar]
- Hinrichsen, L., A. Meyerholz, S. Groos, and E.J. Ungewickell. 2006. Bending a membrane: how clathrin affects budding. Proc. Natl. Acad. Sci. USA. 103:8715–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba, T.M., A.C. Gay, L.F. Pantalena, H.C. Yang, and L.A. Pon. 2004. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J.H., and B. Wendland. 2002. Endocytosis: driving membranes around the bend. Cell. 111:143–146. [DOI] [PubMed] [Google Scholar]
- Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir, G.A., and R. Li. 2004. Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr. Biol. 14:1604–1609. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Y. Sun, and D.G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 115:475–487. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., C.P. Toret, and D.G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 123:305–320. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., C.P. Toret, and D.G. Drubin. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7:404–414. [DOI] [PubMed] [Google Scholar]
- Krendel, M., E.K. Osterweil, and M.S. Mooseker. 2007. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 581:644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal, W.H., and W.A. Wallis. 1952. Use of ranks in one-criterion variance analysis. J. Amer. Statis. Assoc. 47:583–621. [Google Scholar]
- Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, S., V.P. Lemmon, and E.W. Jones. 1988. Characterization of yeast clathrin and anticlathrin heavy-chain monoclonal antibodies. J. Cell. Biochem. 36:329–340. [DOI] [PubMed] [Google Scholar]
- McCullagh, P., and J.A. Nelder. 1989. Generalized Linear Models. Second edition. Chapman and Hall, New York. 511 pp.
- Merrifield, C.J., M.E. Feldman, L. Wan, and W. Almers. 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4:691–698. [DOI] [PubMed] [Google Scholar]
- Mochida, J., T. Yamamoto, K. Fujimura-Kamada, and K. Tanaka. 2002. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics. 160:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland, J., D. Preuss, A. Moon, A. Wong, D. Drubin, and D. Botstein. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland, J., J. Konopka, B. Singer-Kruger, M. Zerial, and D. Botstein. 1999. Visualization of receptor-mediated endocytosis in yeast. Mol. Biol. Cell. 10:799–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher, T.M., R.P. Smith, V. Lemmon, and S.K. Lemmon. 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 9:87–98. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., S.K. Doberstein, and H.G. Zot. 1991. Myosin-I. Annu. Rev. Physiol. 53:653–681. [DOI] [PubMed] [Google Scholar]
- Reynolds, E.S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal, A.A., A.L. Manning, B.L. Goode, and D.G. Drubin. 2003. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr. Biol. 13:1000–1008. [DOI] [PubMed] [Google Scholar]
- Rodal, A.A., L. Kozubowski, B.L. Goode, D.G. Drubin, and J.H. Hartwig. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell. 16:372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, A., K. Uyhazi, A. Frost, and P. De Camilli. 2006. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 441:528–531. [DOI] [PubMed] [Google Scholar]
- Song, B.D., and S.L. Schmid. 2003. A molecular motor or a regulator? Dynamin's in a class of its own. Biochemistry. 42:1369–1376. [DOI] [PubMed] [Google Scholar]
- Sun, Y., A.C. Martin, and D.G. Drubin. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 11:33–46. [DOI] [PubMed] [Google Scholar]
- Sun, Y., S. Carroll, M. Kaksonen, J.Y. Toshima, and D.G. Drubin. 2007. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J. Cell Biol. 177:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima, J.Y., J. Toshima, M. Kaksonen, A.C. Martin, D.S. King, and D.G. Drubin. 2006. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc. Natl. Acad. Sci. USA. 103:5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- Yu, X., and M. Cai. 2004. The yeast dynamin-related GTPase Vps1p functions in the organization of the actin cytoskeleton via interaction with Sla1p. J. Cell Sci. 117:3839–3853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]