Increased Sporulation Rate of Epidemic Clostridium difficile Type 027/NAP1 (original) (raw)
Abstract
Clostridium difficile PCR ribotype 027 comprised 0.2% of a collection of Swedish isolates in 1997-2001 (3 of 1,325 isolates). These isolates had lower moxifloxacin MICs than the epidemic type 027 isolates, but they had the same tcdC sequence and toxin yield. Type 027 produced 3- to 13-fold more toxin than did major Swedish types. One epidemic strain (027/NAP1a) sporulated more than did other type 027 isolates, a feature that should contribute to its survival and spread.
A strain of Clostridium difficile (PCR ribotype 027 and pulsed-field gel electrophoresis type NAP1 [027-NAP1]) producing high levels of toxin has in recent years been associated with increased infection rates, outbreaks, and severe disease (10, 12, 15, 16, 17, 18, 19, 22, 25, 26, 31, 34). Its high toxin yield may, in part, be caused by a frameshift mutation in tcdC (5), encoding a negative regulator of TcdR and part of the C. difficile pathogenicity locus (20, 21). Type 027 has been associated with the use of fluoroquinolones, especially moxifloxacin and gatifloxacin (3, 4, 8, 27, 32), that promote C. difficile growth and toxin production in an animal model (1). The role of these factors for the worldwide expansion of type 027/NAP1 is obscure. We compared historical and epidemic isolates of PCR ribotype 027 with respect to antibiotic susceptibility, tcdC sequence, toxin yield, S-layer, and sporulation.
C. difficile isolates were obtained from the following: a collection of 1,325 strains isolated in 1997-2001 in central and south Sweden (23, 29, 30, 35, 36, 37); the recent U.S.-Canadian epidemic (US1067 and US1165, representing PCR ribotype 027/pulsed-field gel electrophoresis types NAP1a and NAP1b, respectively); and the Culture Collection, University of Göteborg (CCUG), Göteborg, Sweden (CCUG 19125 [VPI 10463], CCUG 37783, and CCUG 20309 [8864]). For PCR ribotyping, see reference 30. Antibiotic susceptibility was determined by Etest (AB Biodisk, Solna, Sweden) using IsoSensitest agar (Oxoid Ltd, Basingstoke, United Kingdom) supplemented with 5% defibrinated horse blood and 20 μg/ml of β-NAD (Swedish Reference Group for Antibiotics [www.srga.org]). The isolation of bacterial DNA, PCR, and sequencing were performed as described previously (24) by using primers for tcdC (28). For growth experiments, overnight cultures were diluted 106-fold into triplicate tubes containing peptone-yeast without cysteine or glucose that were further grown for 48 h (13, 14). Sampling, separation of intra- and extracellular fractions, sonication, and toxin measurements (by enzyme immunoassay) were performed as described previously (13, 14). Vegetative and sporulated cells were scored by microscopy by using a Bürker chamber; 10 squares containing 5 to 15 cells were counted per isolate, and the values were averaged. Two-dimensional gel electrophoresis was performed as described in reference 13; duplicate 24-h intracellular protein samples of US1067, US1165, and T-378 and a single sample of Ö99-1670 were focused on 180-mm linear immobilized pH gradient strips (pH 4 to 7; Amersham Biosciences, Uppsala, Sweden). For identification, protein spots were excised from Coomassie-stained gels and processed using the Montage in-gel digestion kit (Millipore, Billerica, MA), together with a vacuum manifold unit. Two microliters of eluted peptides was loaded on an anchor chip plate (Bruker Daltonics, Inc., Bremen, Germany) and covered by 1 μl of α-cyano-4-hydoxycinnamic acid. Peptide mass mapping of tryptic peptides was performed by a Bruker Daltonics Reflex IV matrix-assisted laser desorption ionization-time of flight apparatus equipped with a nitrogen laser (337.1 nm) and operated in a reflective positive mode. Spectrum calibration was performed by internal use of trypsin fragments of 842.510, 1,045.564, and 2,211.105 Da and the external use of a 1,000- to 4,000-Da peptide calibration standard (Bruker Daltonics, Inc.). Identified peptide masses were analyzed in the Mascot search engine at www.matrixscience.com.
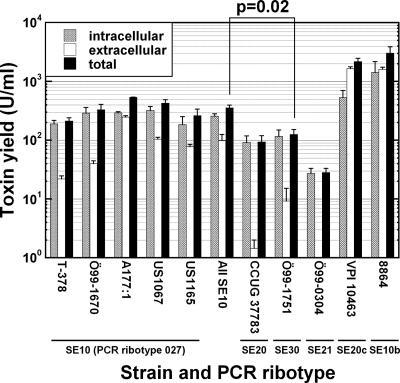
By using two C. difficile PCR ribotype 027 reference strains, three type 027 isolates were identified in the Swedish national database; these isolates represented 0.2% of strains collected in 1997-2001. (PCR ribotype 027 corresponded to type SE10 according to the Swedish nomenclature.) The type 027-infected patients had developed moderate to severe _Clostridium difficile_-associated diarrhea (CDAD) (Table 1), and like other historical 027 isolates (22), the Swedish ones had lower MICs for moxifloxacin compared to the MICs for the recent epidemic type 027/NAP1 (Table 2). In addition, the MICs for metronidazole were about threefold lower in the Swedish isolates. The tcdC sequence was identical in the type 027 isolates Ö99-1670, US1067, and US1165, including the characteristic 18-bp deletion and the frameshift mutation at position 117, while isolates representing the major Swedish types SE20 and SE30 had a wild-type tcdC allele (see Fig. S1 in the supplemental material). The toxin yield was similar in historical and epidemic type 027 isolates but 3- to 13-fold higher than the yield in isolates representing the common types SE20, SE30, and SE21 (Fig. 1).
TABLE 1.
Clinical data for Swedish patients infected with C. difficile PCR ribotype 027
| Strain | Year | Age (yrs) | No. of stools per day | Temp (°C) | No. of leukocytes (1,000/mm3) | Underlying diseasea | Antibiotic treatmentb |
|---|---|---|---|---|---|---|---|
| T-378 | 1997 | 53 | >10 | >39 | 17.9 | None | Penicillin V |
| Ö99-1670 | 1999 | 85 | 5-10 | 38 | 31.9 | Lung cancer | Amoxicillin |
| A177:1 | 2001 | 92 | 5-10 | >38 | 31.6 | None | Piperacillin-tazobactam metronidazole/ciprofloxacin |
TABLE 2.
MICs (μg/ml) of antibiotics for C. difficile isolates by using Etest
| Antibiotic | MIC for strain | |||||
|---|---|---|---|---|---|---|
| T-378a | Ö99-1670a | A177:1a | US1067b | US1165b | 8864c | |
| Ciprofloxacin | >32 | >32 | >32 | >32 | >32 | >32 |
| Levofloxacin | >32 | >32 | >32 | >32 | >32 | >32 |
| Moxifloxacin | 0.5 | 2 | 1.0 | >32 | >32 | 1.0 |
| Clindamycin | 4 | 8 | 4 | 4 | 2 | 4 |
| Metronidazoled | 0.053 (3) | 0.125 (3) | 0.084 (3) | 0.33 (3) | 0.25 (3) | 0.19 (2) |
| Vancomycin | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | 1.0 |
FIG. 1.
Toxin yields of 24-h C. difficile cultures. Isolates T-378, Ö99-1670, A177:1, US1067, and US1165 are type SE10, i.e., PCR ribotype 027. Isolates representing major Swedish PCR ribotypes, CCUG 37783 (SE20, i.e., PCR ribotype 001), Ö99-1751 (SE30), Ö99-0304 (SE21), and two high-level toxin producing reference strains, VPI 10463 (SE20c) and 8864 (SE10b), were included for comparison. Values are averages of duplicate cultures, and bars indicate standard errors. Statistics were calculated using logarithmic average values of total toxin for “All SE10,” CCUG 37783, Ö99-1751, Ö99-0304, VPI 10463, and 8864 by using analysis of variance and Bonferroni post hoc compensation for multiple comparisons (the P value for the comparison of “All SE10” and strain Ö99-1751 is shown).
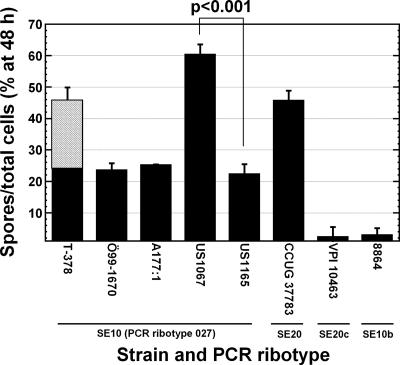
Three of the type 027 isolates had sporulation frequencies of about 25% at 48 h (Fig. 2). T-378 was morphologically different from the other 027 isolates, showing elongated doublet cells usually containing a single spore (not shown). Thus, the actual sporulation frequency per unit cell was about twofold lower than the scored one, i.e., closer to 25% than to 45% (Fig. 2). Strain US1067 (NAP1a) sporulated more (60%; P < 0.001) (Fig. 2) and had a smaller cell and colony size compared to the case for the other type 027 isolates. Excluding the low-sporulating strains VPI 10463 and 8864, US1067 also had a 20% higher optical density at 24 h than those of all other isolates (P < 0.001) (data not shown), i.e., consistent with characteristics of the epidemic 027 type (34). Despite the differences in sporulation frequency and morphology, the expressed protein patterns of US1067, US1165, and Ö99-1670 were similar (see Fig S2 in the supplemental material). However, T-378 expressed an additional S-layer (see Fig. S2 and Table S1 in the supplemental material), possibly causing its unique morphology.
FIG. 2.
Sporulation frequencies of C. difficile cultures. For further explanation of strains, see the legend for Fig. 1. Bars indicate standard errors. Statistics were calculated by using analysis of variance and Bonferroni post hoc compensation for multiple comparisons. The P value for the comparison of US1067 and US1165 is indicated. As T-378 formed doublet cells, the total frequency scored (filled and hatched areas) and an estimate of the true sporulation frequency per unit cell (filled area) are shown. For details, see the text.
The “hypervirulence” of C. difficile 027/NAP1 (16, 25, 26) has been ascribed to its higher (about 20-fold) toxin yield in vitro compared to those of toxinotype 0 strains (34), caused by loss of TcdC function (5, 21). However, the moderate threefold-higher toxin yield of type 027 (defective tcdC) compared to those of types SE30/SE20 (wild-type tcdC) indicates that other factors may also affect toxin production. For example, the nutritional sensor CodY may further affect the range of toxin levels (6). The fact that strains VPI 10463 and 8864 yielded few spores but superior amounts of toxin during stationary phase was in accord with the inverse relation between spore and toxin yield generally found among C. difficile isolates (2). No clinical isolate of the same PCR ribotype as that of strain 8864 and only two isolates sharing type and characteristics with VPI 10463 were found in our national database, suggesting that high-level toxin producers with low sporulation capacities have poor transmission rates. Although antibiotics that promote growth and toxin production by resistant C. difficile in vivo are major risk factors for developing CDAD (1, 7, 9, 11, 33), our data showed that certain PCR ribotype 027 strains have different morphologies and growth characteristics as well as high capacities for both toxin and spore production, features that may contribute to disease severity, therapy failure, relapse, and spread.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Michel Warny for providing strains US1067 and US1165 and Mats Andersson for matrix-assisted laser desorption ionization-time of flight analyses.
Footnotes
▿
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Adams, D. A., M. M. Riggs, and C. J. Donskey. 2007. Effect of fluoroquinolone treatment on growth of and toxin production by epidemic and nonepidemic Clostridium difficile strains in the cecal contents of mice. Antimicrob. Agents Chemother. 512674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Åkerlund, T., B. Svenungsson, A. Lagergren, and L. G. Burman. 2006. Correlation of disease severity with fecal toxin levels in patients with _Clostridium difficile_-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J. Clin. Microbiol. 44353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biller, P., B. Shank, L. Lind, M. Brennan, L. Tkatch, G. Killgore, A. Thompson, and L. C. McDonald. 2007. Moxifloxacin therapy as a risk factor for _Clostridium difficile_-associated disease during an outbreak: attempts to control a new epidemic strain. Infect. Control Hosp. Epidemiol. 28198-201. [DOI] [PubMed] [Google Scholar]
- 4.Bourgault, A. M., F. Lamothe, V. G. Loo, L. Poirier, and the CDAD-CSI Study Group. 2006. In vitro susceptibility of Clostridium difficile clinical isolates from a multi-institutional outbreak in Southern Québec, Canada. Antimicrob. Agents Chemother. 503473-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curry, S. R., J. W. Marsh, C. A. Muto, M. M. O'Leary, A. W. Pasculle, and L. H. Harrison. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 7.Freeman, J., S. D. Baines, K. Saxton, and W. H. Wilcox. 2007. Effect of metronidazole on growth and toxin production by epidemic Clostridium difficile PCR ribotypes 001 and 027 in a human gut model. J. Antimicrob. Chemother. 6083-91. [DOI] [PubMed] [Google Scholar]
- 8.Gaynes, R., D. Rimland, E. Killum, H. K. Lowery, T. M. Johnson II, G. Killgore, and F. C. Tenover. 2004. Outbreak of Clostridium difficile infection in a long-term facility: association with gatifloxacin use. Clin. Infect. Dis. 38640-645. [DOI] [PubMed] [Google Scholar]
- 9.Gerding, D. N., S. Johnson, L. R. Peterson, M. E. Mulligan, and J. Silva, Jr. 1995. _Clostridium difficile_-associated diarrhea and colitis. Infect. Control Hosp. Epidemiol. 16459-477. [DOI] [PubMed] [Google Scholar]
- 10.Hubert, B., V. G. Loo, A. M. Bourgault, L. Poirer, A. Dascal, E. Fortin, M. Dionne, and M. Lorange. 2007. A portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of _C. difficile_-associated disease in Quebec. Clin. Infect. Dis. 44238-244. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 3411645-1651. [DOI] [PubMed] [Google Scholar]
- 12.Joseph, R., D. Demeyer, D. Vanrenterghem, R. van den Berg, E. Kuijper, and M. Delmee. 2005. First isolation of Clostridium difficile PCR ribotype 027, toxinotype III in Belgium. Euro. Surveill. 10E051020.4. www.eurosurveillance.org/ew/2005/051020.asp#4. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson, S., A. Lindberg, L. G. Burman, E. Norin, and T. Åkerlund. 2000. Toxins, butyric acid and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 685881-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson, S., L. G. Burman, and T. Åkerlund. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 1451683-1693. [DOI] [PubMed] [Google Scholar]
- 15.Kuijper, E. J., B. Coignard, P. Tull, ESCMID Study Group for Clostridium difficile, EU Member States, and the European Centre for Disease Prevention and Control. 2006. Emergence of _Clostridium difficile_-associated disease in North America and Europe. Clin. Microbiol. Infect. 6(Suppl.)S2-S18. [DOI] [PubMed] [Google Scholar]
- 16.Kuijper, E. J., J. T. van Dissel, and M. H. Wilcox. 2007. Clostridium difficile: changing epidemiology and new treatment options. Curr. Opin. Infect. Dis. 20376-383. [DOI] [PubMed] [Google Scholar]
- 17.Kuijper, E. J., R. J. van den Berg, S. Debast, C. E. Visser, D. Veenendaal, A. Troelstra, T. van der Kooi, S. van den Hof, and D. W. Notermans. 2006. Clostridium difficile ribotype 027, toxinotype III, the Netherlands. Emerg. Infect. Dis. 12827-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of _Clostridium difficile_-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 19.MacCannell, D. R., T. J. Louie, D. B. Gregson, M. Laverdiere, A. C. Labbe, F. Laing, and S. Henwick. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 442147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mani, N., and B. Dupuy. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. USA 985844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matamouros, S., P. England, and B. Dupuy. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 641274-1288. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 23.Norén, T., T. Åkerlund, E. Bäck, L. Sjöberg, I. Persson, I. Alriksson, and L. G. Burman. 2004. Molecular epidemiology of hospital-associated and community-acquired Clostridium difficile infection in a Swedish county. J. Clin. Microbiol. 423635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norén, T., T. Åkerlund, M. Wullt, L. G. Burman, and M. Unemo. 2007. Mutations in fusA associated with posttherapy fusidic acid resistance in Clostridium difficile. Antimicrob. Agents Chemother. 511840-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pépin, J., L. Valiquette, M. E. Alary, P. Villemure, A. Pelletier, K. Forget, K. Pepin, and D. Chouinard. 2004. _Clostridium difficile_-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pépin, J., L. Valiquette, and B. Cossette. 2005. Mortality attributable to nosocomial _Clostridium difficile_-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 1731037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pépin, J., N. Saheb, M. A. Coulombe, M. E. Alary, M. P. Corriveau, S. Authier, M. LeBlanc, G. Rivard, M. Bettez, V. Primeau, M. Nquyen, C. E. Jacob, and L. Lanthier. 2005. Emergence of fluoroquinolones as the predominant risk factor for _Clostridium difficile_-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 411254-1260. [DOI] [PubMed] [Google Scholar]
- 28.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 403470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svenungsson, B., A. Lagergren, E. Ekwall, B. Evengård, K. O. Hedlund, A. Kärnell, S. Löfdahl, L. Svensson, and A. Weintraub. 2000. Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin. Infect. Dis. 30770-778. [DOI] [PubMed] [Google Scholar]
- 30.Svenungsson, B., L. G. Burman, K. Jalakas-Pörnull, Å. Lagergren, J. Struwe, and T. Åkerlund. 2003. Epidemiology and molecular characterization of Clostridium difficile strains from patients with diarrhea: low disease incidence and evidence of limited cross-infection in a Swedish teaching hospital. J. Clin. Microbiol. 414031-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tachon, M., C. Cattoen, K. Blanckaert, I. Poujol, A. Carbonne, F. Barbut, J. C. Petit, and B. Coignard. 2006. First cluster of C. difficile toxinotype III, PCR-ribotype 027 associated disease in France: preliminary report. Euro. Surveill. 11E060504.1. www.eurosurveillance.org/ew/2006/060504.asp#1. [DOI] [PubMed] [Google Scholar]
- 32.Valiquette, L., B. Cossette, M. P. Garant, H. Diab, and J. Pepin. 2007. Impact of a reduction in the use of high-risk antibiotics on the course of an epidemic of _Clostridium difficile_-associated disease caused by the hypervirulent NAP1/027 strain. Clin. Infect. Dis. 45(Suppl.)S112-S121. [DOI] [PubMed] [Google Scholar]
- 33.van Dijck, P., V. Avesani, and M. Delmée. 1996. Genotyping of outbreak-related and sporadic isolates of Clostridium difficile belonging to serogroup C. J. Clin. Microbiol. 343049-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 3661079-1084. [DOI] [PubMed] [Google Scholar]
- 35.Wullt, M., and I. Odenholt. 2004. A double-blind randomized controlled trial of fusidic acid and metronidazole for treatment of an initial episode of _Clostridium difficile_-associated diarrhoea. J. Antimicrob. Chemother. 54211-216. [DOI] [PubMed] [Google Scholar]
- 36.Wullt, M., M. L. Hagslätt, and I. Odenholt. 2003. Lactobacillus plantarum 299v for the treatment of recurrent _Clostridium difficile_-associated diarrhoea: a double-blind, placebo-controlled trial. Scand. J. Infect. Dis. 35365-367. [DOI] [PubMed] [Google Scholar]
- 37.Wullt, M., L. G. Burman, M. H. Laurell, and T. Åkerlund. 2003. Comparison of AP-PCR typing and PCR-ribotyping for estimation of nosocomial transmission of Clostridium difficile. J. Hosp. Infect. 55124-130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]