Genome-Scale Metabolic Network Analysis of the Opportunistic Pathogen Pseudomonas aeruginosa PAO1 (original) (raw)
Abstract
Pseudomonas aeruginosa is a major life-threatening opportunistic pathogen that commonly infects immunocompromised patients. This bacterium owes its success as a pathogen largely to its metabolic versatility and flexibility. A thorough understanding of P. aeruginosa's metabolism is thus pivotal for the design of effective intervention strategies. Here we aim to provide, through systems analysis, a basis for the characterization of the genome-scale properties of this pathogen's versatile metabolic network. To this end, we reconstructed a genome-scale metabolic network of Pseudomonas aeruginosa PAO1. This reconstruction accounts for 1,056 genes (19% of the genome), 1,030 proteins, and 883 reactions. Flux balance analysis was used to identify key features of P. aeruginosa metabolism, such as growth yield, under defined conditions and with defined knowledge gaps within the network. BIOLOG substrate oxidation data were used in model expansion, and a genome-scale transposon knockout set was compared against in silico knockout predictions to validate the model. Ultimately, this genome-scale model provides a basic modeling framework with which to explore the metabolism of P. aeruginosa in the context of its environmental and genetic constraints, thereby contributing to a more thorough understanding of the genotype-phenotype relationships in this resourceful and dangerous pathogen.
With sequenced genomes now routinely being made available to the public, detailed annotations and various publicly available genomic resources have enabled the formation of genome-scale models of metabolism for a wide variety of organisms (12, 21, 26, 42, 63, 65). A wealth of data from well-controlled experiments, coupled with advancements in methods for computational network analysis, have allowed these models to aid interrogation of metabolic behavior. In addition, an iterative process to model development—cycles of in silico model predictions, experimental (i.e., wet lab) validation, and subsequent model refinement—has enabled the development of methods that have contributed to biological discovery, such as in determination of likely drug targets in Mycobacterium tuberculosis (3, 26), metabolic engineering of cells for production of valuable compounds (5, 32, 34), and development of novel frameworks for contextualizing high-throughput “-omics” data sets (15, 24, 64).
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium that is capable of surviving in a broad range of natural environments, although it is mostly known for its role as an opportunistic pathogen (40, 60, 72). While P. aeruginosa is generally found in aerobic environments, it is able to thrive anoxically and, notably, to denitrify (58). It was also recently shown that P. aeruginosa can form biofilms under microaerobic (i.e., very low oxygen) conditions similar to those found in the lungs of cystic fibrosis (CF) patients (1). These features strongly contribute to the notable success of P. aeruginosa in chronically infecting the lungs of CF patients, nearly all of whom have lifelong P. aeruginosa infections starting at an early age (49, 70). P. aeruginosa is also a serious pathogen in nosocomial infections and various acute infections in immunocompromised patients, such as severe burns and urinary tract infections (39, 49, 68). Part of the reason for this remarkable ecological success is thought to be the considerable metabolic versatility and flexibility of P. aeruginosa (62), which renders the study of the metabolism of this life-threatening microbe crucial to the understanding of its pathogenicity and opportunistic nature.
We present a genome-scale metabolic reconstruction of P. aeruginosa PAO1 (hereafter referred to as PAO1), called in silico strain iMO1056 following an often-used naming convention (48). This model accounts for 1,056 genes encoding 1,030 proteins that catalyze 883 reactions (Fig. 1a). Gene-protein-reaction (GPR) associations are accounted for in the reconstruction, as well as the stoichiometry and thermodynamically derived directionality of all included reactions. This reconstruction process led to the reannotation of several open reading frames (ORFs). The model was tested against high-throughput substrate utilization experiments (90% matched the usage of common substrates) and two published sets of genome-wide knockout data (85% accuracy of essentiality predictions). Several additional predictions were made regarding PAO1 physiology and virulence, and these may be tested subsequently. This genome-scale reconstruction and subsequent constraint-based modeling enable integration of high-throughput data to generate novel, testable hypotheses that will assist in exploring the physiology of PAO1 and assessing its relevance for pathology.
FIG. 1.
Reconstruction statistics. (a) Statistics on the P. aeruginosa PAO1 genome and the iMO1056 reconstruction. *, see reference 62; **, reaction confidences were based on the scale for protein name confidences set out at www.pseudomonas.com. This scale is described in Materials and Methods. The confidence value for the PAO1 genome is from a version (19 June 2007) of the annotation at www.pseudomonas.com. (b) Numbers of genes participating in different metabolic processes in the iMO1056 metabolic reconstruction.
MATERIALS AND METHODS
Network reconstruction.
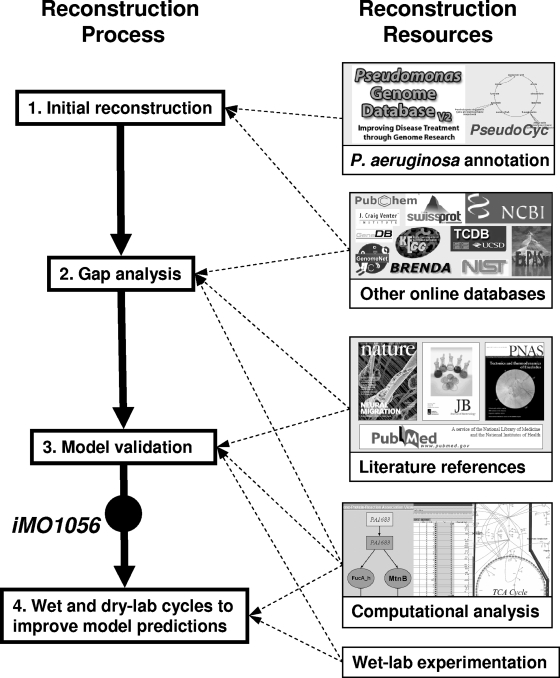
The reconstruction process is outlined schematically in Fig. 2, and details are described elsewhere (47). The initial draft of iMO1056 was built from the annotated PAO1 genome available at the Pseudomonas Genome Database (PseudoCAP; www.pseudomonas.com), supplemented by literature mining and BLAST searches (69). Biological information databases such as EXPASY, KEGG, and TCDB were used to link annotated genes to proteins and proteins to reactions (19, 28, 51), and confidence values were added to reactions based on the reported confidence values from the PseudoCAP annotation. These GPR associations link genetic data to reactions in the model and allow the model to predict effects of genetic perturbations on metabolic phenotype. GPR associations are defined through Boolean logic, where, for example, two protein subunits that combine as a complex to perform one enzymatic function would have an “and” relationship with relation to that reaction, whereas two isozymes that are each independently capable of catalyzing a reaction would have an “or” relationship.
FIG. 2.
Schematic of the network reconstruction process for iMO1056. Resources used for the reconstruction are shown on the right, and the reconstruction process is shown on the left. (1) The P. aeruginosa annotation was used along with online resources to generate an initial reconstruction of metabolism. (2) Next, computational analysis was used in conjunction with extensive literature mining and online database searches to identify and fill gaps in metabolism. (3) The model was then validated with data sets from the literature and experimental data generated as a part of this study. (4) The current model version, iMO1056, is now ready for the next step, in which the model will continually be improved through cycles of computational hypothesis generation followed by experimental validation.
Gaps in metabolic pathways necessary for cell growth and production of major virulence factors were filled through careful literature mining and homology and context analysis with BLAST at the Pseudomonas Genome Database and NCBI websites and other online resources (53). Genes whose functions were inferred via these sources were assigned reaction identification confidence values based on the guidelines of the Pseudomonas Genome Database genome annotation effort for protein name confidence values. This concordance ensures that confidence values for key findings in our study are consistent with confidence values for gene functions in the currently published annotation (69). The PAO1 biomass reaction (defined as a relative weight of metabolites necessary to synthesize biomass, as described previously) (66) was assumed to be similar to the published Escherichia coli biomass reaction, albeit with some minor variations to account for PAO1-specific data, as described in the supplemental material. Energy efficiency of the electron transport chain in PAO1 was approximated by assuming that P. aeruginosa grown aerobically would have a similar P/O ratio to published values for the model prokaryote Escherichia coli (41).
Confidence classes for reactions were assigned based on the PseudoCAP protein name confidence rating system (69), which rates the confidences of gene-protein associations in the PAO1 annotation. We extended the system to rating confidences for reaction associations. There are four levels in this rating system, as follows: class 1, which accounts for genes whose functions have been demonstrated experimentally with PAO1; class 2, which accounts for genes whose functions were inferred via homology with an experimentally characterized protein in another organism; class 3, which represents gene-protein associations based on the presence of conserved amino acid motifs; and class 4, which accounts for genes with similarity to other genes of unknown function. We adhered to this rating system for genes whose functions were newly annotated in this study (see Table 2, “proposed confidence” column), and for genes which were not newly annotated, we assigned reaction confidences equivalent to the protein name confidences in the annotation.
TABLE 2.
Proposed annotation refinements
| Gene locus | Current annotation (www.pseudomonas.com) | Proposed functional reannotation | ||||||
|---|---|---|---|---|---|---|---|---|
| Current protein identification | Confidence class (1 = high, 4 = low) | New reaction assignment | Proposed confidence class | Protein name | Protein ID | Evidence | Reference | |
| PA0143 | Nonspecific ribonucleoside hydrolase | 1 | Purine nucleosidase (adenosine), purine nucleosidase (inosine) | 1 | Nuh | EC 3.2.2.1 | Specific function outlined in reference; nonspecific ribonucleoside hydrolase annotation ID (with class I confidence) implies that there are more reactions catalyzed by this enzyme | 23 |
| PA0215, PA0216 | Probable transporter | 3 | Malonate transport | 2 | MadL, MadM | TC-2.A.70.1.1 | Biolog data show growth on malonate; annotation assigns PA0215 and PA0216 as having 71% and 81% homology with MadL and MadM, respectively, from Malonomonas rubra | 54 |
| PA0313, PA0314 | Probable permease of ABC transporter, probable binding protein component of ABC transporter | 3 | l-Cysteine transport via ABC system | 3 | YecS, FliY | ? | Reference gives strong physiological evidence and BLAST E values (<10e−45) | 30 |
| PA0761, PA1004 | l-Aspartate oxidase | 2 | l-Aspartate oxidase (aerobic/anaerobic) | 2 | NadB, NadA | EC 1.4.3.16? | Reaction not utilizing O2 is necessary to produce spermidine under anaerobic conditions, as shown by gap analysis and explained in references | 10, 17 |
| PA1514 | Conserved hypothetical protein | 4 | Ureidoglycolate hydrolase | 3 | YbbT | EC 3.5.3.19 | Biochemical evidence for pathway; gene locus identified from annotation as 44% similar to ureidoglycolate hydrolase (DAL3) from Saccharomyces cerevisiae; listed in NCBI database as ureidoglycolate hydrolase | 23, 37 |
| PA1684 | Probable oxidase | 3 | 1,2-Dihydroxy-3-keto-5-methylthiopentene dioxygenase | 2 | MtnD | EC 1.13.11.54, EC 1.13.11.53 | Biochemical evidence for pathway; EC 1.13.11.54 occurs when Fe2+ binds the enzyme, EC 1.13.11.53 occurs when Ni2+ binds the enzyme | 19, 57 |
| PA1818 | Probable Orn/Arg/Lys decarboxylase | 3 | Orn/Arg/Lys decarboxylase | 2 | LdcC | EC 4.1.1.18 | Modeling evidence, since PAO1 grows on lysine; also physiological evidence from references; BLAST search of lysine decarboxylases from Burkholderia cenocepacia HI2424 vs P. aeruginosa PAO1 genome gave an E value of 0; Orn and Arg functions are also in reconstruction | 18, 44, 45 |
| PA2080 | Hypothetical protein | 4 | Kynureninase | 1 | KynU | EC 3.7.1.3 | Genetic evidence, using kynU knockout strain and analyzing PQS production; in ExPASy, this reaction is one of several in the EC numbers; listed as kynureninase in NCBI database | 14 |
| PA2366 | Conserved hypothetical protein | 4 | Uricase | 2 | PuuD | EC 1.7.3.3 | Genetic evidence; BLAST search of uricases of Pseudomonas aeruginosa Ps-x vs P. aeruginosa PAO1 genome gave an E score of 0 | 23, 50 |
| PA2385 | PvdQ | 1 | 3-Oxo-C12-homoserine lactone acylase, C4-homoserine lactone acylase | 1 | PvdQ | ? | Reaction described in reference | 23 |
| PA2579 | Hypothetical protein | 4 | l-Tryptophan:oxygen 2,3-oxidoreductase (decyclizing) | 1 | KynA | EC 1.13.11.11 | Genetic evidence | 14 |
| PA3004 | Probable nucleoside phosphorylase | 3 | 5′-Methylthioadenosine phosphorylase | 1 | MtnP | EC 2.4.2.28 | Detailed sequence analysis and genetic evidence provided in reference; listed as 5′-methylthioadenosine phosphorylase in NCBI database | 57 |
| PA3169 | Probable initiation factor 2 subunit | 3 | 5-Methylthioribose-1-phosphate isomerase | 2 | MtnA | EC 5.3.1.23 | Gene association from reference; biochemical evidence for pathway; listed in NCBI database as 5-methylthioribulose-1-phosphate isomerase | 57 |
| PA4425 | Probable phosphoheptose isomerase | 3 | Sedoheptulose 7-phosphate isomerase | 2 | GmhA | ? | Modeling evidence for sedoheptulose 7-phosphate isomerase; BLAST search of PA4425 against the NCBI database showed an E value of 2e−59 vs sedoheptulose 7-phosphate isomerase of Ralstonia eutropha H16 | 31 |
| PA4457 | Conserved hypothetical protein | 4 | Arabinose-5-phosphate isomerase | 2 | KdsD | EC 5.3.1.13 | BLAST search of PA4457 vs NCBI database gave an E value of 3e−130 for arabinose 5-phosphate isomerase of Pseudomonas fluorescens Pf-5; modeling evidence, since necessary for KDO synthesis | |
| PA5175 | CysQ protein | 2 | 3′,5′-bisphosphate nucleotidase | 2 | CysQ | EC 3.1.3.7 | BLAST search of PA5175 identified it by strong homology (E value of 10e−113) with Pseudomonas mendocina Ymp 3′(2′),5′-biphosphate nucleotidase; modeling evidence for this reaction |
Formulation of S matrix.
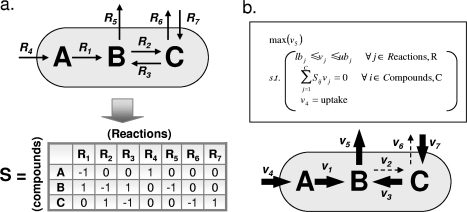
In order to perform flux balance analysis (FBA), a network must first be represented in the form of a stoichiometric matrix (S matrix), as shown in Fig. 3a. By convention, rows of the S matrix represent metabolites and columns represent reactions. Reaction substrates have negative coefficients in the S matrix, while products have positive coefficients. The S matrix for a network wholly describes the stoichiometry of reactions in the network. The S matrix also accounts for transport reactions across the cell membrane, represented as reactions interconverting intracellular and extracellular compounds, often coupled with hydrolysis of ATP (in the case of ATP-binding cassette transporters) or exploitation of a chemical gradient (e.g., H+ symport or antiport). Figure 3a, for example, illustrates four transport reactions, namely, R4, R5, R6, and R7.
FIG. 3.
FBA. (a) The conversion of a metabolic system into an S matrix is shown for a simple prototype. Columns represent reactions, and rows represent compounds. Reaction stoichiometries are represented with negative coefficients for substrates and positive coefficients for products of a given reaction. (b) (Top) Standard form of an FBA problem. Flux through the objective reaction (_v_5) is maximized subject to thermodynamic, stoichiometric, and uptake constraints, as follows: reaction fluxes (vj) must lie between lower and upper bounds (lb and ub), metabolite concentrations are fixed over time (S · v = 0), and the flux of a limiting carbon source (_v_4) is set to some uptake value. (Bottom) Flux values calculated by FBA for the prototype network are represented by arrows.
FBA.
FBA is a computational method that calculates reaction fluxes, their distribution within a metabolic network, and on this basis, growth yields (Fig. 3) (33). In order to obtain flux predictions, the metabolic network is assumed to be at steady state, meaning that concentrations of intracellular metabolites do not vary during the course of a simulation. This steady-state assumption, which makes FBA results directly comparable to data from cells at a fixed growth rate (i.e., in exponential phase), is necessary since kinetic parameters are not accounted for in iMO1056.
In many FBA applications, a linear biomass reaction is defined and assumed to be the objective toward which the flux distribution of a network is optimized. The biomass reaction represents a weighted ratio of components forming the dry weight of a cell, as well as hydrolysis of ATP to account for the energy needs involved in growth and cellular maintenance (66). Optimization for biomass is justified by the assumption that bacteria have been optimized evolutionarily for growth, and experimental studies have verified the rationality of this assumption (16, 55). This optimization step is necessary due to the underdetermined nature of metabolic networks, as a range of flux profiles are possible for a given network. FBA optimization is constrained by the stoichiometry of the network and by upper and lower bounds on reactions, as set by thermodynamic and substrate uptake constraints. In addition, conditional constraints, such as gene knockouts and alternative gene regulation, can be placed on the system to simulate experimental conditions (22, 59).
To perform FBA, the flux of the biomass reaction is maximized given the steady-state condition, as follows: S · v = 0, where S is the S matrix representing the network (Fig. 3a) and v is the vector of all fluxes in the network (Fig. 3b, top panel). Ultimately, FBA yields a predicted optimal growth yield and a flux distribution through the system that will support this optimal growth (Fig. 3b, bottom panel). See the supplemental material for a discussion of constraints used in FBA simulations in this study.
Biolog experiments.
PAO1 was tested for the ability to utilize various carbon sources, using Biolog GN2 microplates (Biolog Inc., Hayward, CA). All procedures were performed as indicated by the manufacturer. Bacteria were grown overnight at 28°C on a Biolog universal growth agar plate. Samples were swabbed from the surface of the plate and suspended in GN inoculating fluid. Each well of the microplate was inoculated with 150 μl of bacterial suspension, and the plate was incubated at 28°C for 24 h. Subsequently, the microplate was read by a microplate reader, and the results were analyzed with MicroLog3 4.20 software.
The assay performed with the Biolog plate is a measurement of the reduction of tetrazolium dye after 24 h, indicating whether P. aeruginosa has been respiring actively during that period. Active respiration in minimal medium indicates utilization of the sole carbon source provided.
Technical computing methods.
FBA optimizations were performed on a Dell computer with 2 GB of RAM and a 1.8-GHz Intel Centrino processor linked to a dedicated database server with Simpheny software (Genomatica, Inc.). Transposon knockout data were handled in Excel (Microsoft Corp.), and statistical analysis of transposon data was performed in Matlab R2006a (Mathworks, Inc.).
RESULTS
Metabolic reconstruction of P. aeruginosa PAO1.
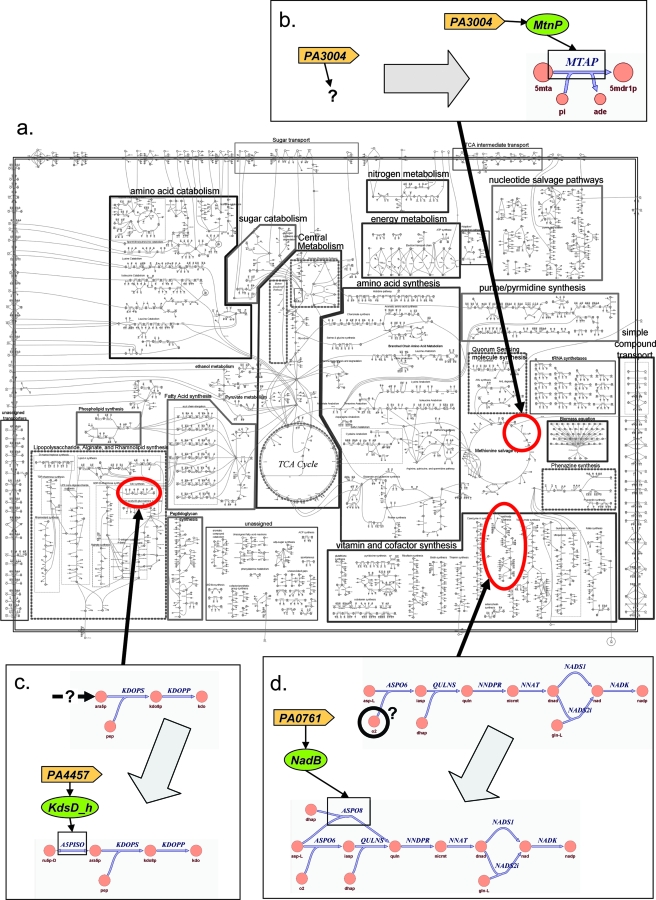
We generated a constraint-based, genome-scale representation of PAO1's metabolism. The reconstruction accounts for 1,056 genes involved in 883 metabolic reactions (Fig. 1a), including anabolic pathways necessary for the synthesis of all major cellular biomass components from basic metabolic precursors (see Fig. 4a for a complete network map). As described in Materials and Methods, the reconstruction process included an initial model-building stage based heavily on public databases and literature, a gap-filling stage in which missing steps in essential metabolic pathways were refined, and finally, an extension and validation stage that included expansion of the model by comparison with Biolog substrate utilization data, comparison of in silico predictions with physiological data, and validation against two experimentally determined sets of likely essential genes.
FIG. 4.
Reannotation processes. (a) Full network map of iMO1056. Metabolic processes are grouped together, and an effort was made to link pathways to their major carbon donors and sinks in central metabolism, although many metabolites occupy two or more locations on the map. All internal boxes are meant to highlight metabolic processes rather than to denote cellular localizations. The double line surrounding the map represents the cell barrier. (b, c, and d) Several types of reannotation that went into iMO1056. Large gray arrows denote the reannotation process: initial annotation statuses are shown before the arrows, and reannotated gene functions are shown after the arrows. The location of each newly annotated gene is highlighted on the map. (b) PA3004 was functionally identified by literature mining. (c) A gap in the KDO synthesis pathway was identified, and PA4457 was found by a BLAST search to fulfill the missing function. (d) NAD initially could not be synthesized anaerobically in iMO1056, since O2 was required for NAD synthesis. However, literature mining revealed a putative alternate stoichiometry for the l-aspartate oxidase reaction (ASPO8) that does not require oxygen.
Aside from accounting for all major pathways necessary for growth of P. aeruginosa, an effort was made to reconstruct some of the major pathways associated with virulence for this bacterium. The metabolism of P. aeruginosa is involved in a number of virulence processes, such as quorum sensing (11, 23), expression of lipopolysaccharide and rhamnolipids (61), and notoriously, the switch from the nonmucoid to mucoid form and associated production of the exopolysaccharide alginate (46). Table 1 lists the virulence-associated pathways accounted for in iMO1056 and some relevant features of these pathways.
TABLE 1.
Virulence factors in P. aeruginosaa
| Virulence factor | No. of reactions | Proteins involved | Major metabolic precursors |
|---|---|---|---|
| Rhamnolipids | 5 | RhlA, RhlB, RhlC, PhaC1, PhaC2 | dtdpddm, acyl-ACP |
| Alginate | 6 | Alg44, Alg8, AlgE, AlgF, AlgG, AlgI, AlgJ, AlgK, AlgL, AlgX | GDP-d-mannose, acetyl-ACP |
| Phenazines | 7 | PhzA1, PhzA2, PhzB1, PhzB2, PhzC1, PhzC2, PhzD1, PhzD2, PhzE1, PhzE2, PhzF1, PhzF2, PhzG1, PhzG2, PhzH, PhzM, PhzS | Chorismate |
| PQS | 5 | KynA, KynB, KynU, PqsA, PqsB, PqsC, PqsD, PqsH | Tryptophan, 3-oxodecanoyl-ACP |
| AHL | 5 | Las , PvdQ, Rhl | SAM, acy -ACP |
| Lipopolysaccharide components | |||
| Lipid A | 12 | HtrB, HtrB2, Kds, KdsA, KdsB, KdtA, LpxA, LpxB, LpxC, LpxD, LpxK | ara5p, pep, uacgam, acyl-ACP |
| Core oligosaccharide | 14 | HldD, RfaD, HldE, GmhB, GmnA, RmlD, WaaC, WaaF, WaaG, WaaP, WapP | s7p, UDP-glucose, dtdpddm, l-alanine |
| A band O antigen | 4 | WbpL, WbpX, WbpY, WbpZ | uacgam, GDP-d-rhamnose |
| B band O antigen | 10 | WbpA, WbpB, WbpC, WbpD, WbpE, WbpH, WbpI, WbpJ, WbpK, WbpL, WbpM | uacgam, acetyl-ACP |
| Other | 2 | WzX, WzY, WzZ | Assembles O antigen and core |
Systematic refinement of genome annotation.
A major value of a manual model-building effort is the careful revision of the current genome annotation based on literature evidence encountered during the model-building process, BLAST searches, and gap closures. Annotation refinements represent (i) knowledge that was in the literature but was overlooked in the original annotation and (ii) new hypotheses that came directly from the model-building process. Specifically, the identification of network gaps (instances where a metabolite can be consumed but not produced or vice versa) using FBA optimization pinpointed reactions that are necessary for a pathway to function but that are peripheral to the pathway and thus were overlooked during historical pathway characterization. Table 2 provides a list of recommended annotation refinements for PAO1 that arose from the reconstruction process. In some cases, such as the annotation refinements for the nuh gene (Table 2), the function of the gene was not changed, but rather, specific reactions which are important for certain cellular processes were assigned to already annotated genes.
The first level of annotation refinement is the assignment of a specific function to a previously uncharacterized gene based on literature evidence or a BLAST search. An example of this in iMO1056 is the gene mtnP (PA3004), which was annotated in PseudoCAP as a “probable nucleoside phosphorylase” but was subsequently found in the literature to have the specific function of a 5′-methylthioadenosine phosphorylase (Fig. 4b) (57). The mtnP gene was first identified (57) through detailed sequence analysis, and then its function was confirmed (57) by knocking out mtnP in a Δ_metZ_ strain of P. aeruginosa and observing a loss of growth on minimal medium supplemented with methionine, homocysteine, or methylthioadenosine (57). mtnP is necessary for the function of the methionine salvage pathway in P. aeruginosa and was previously characterized in the literature but not in the P. aeruginosa annotation, and its inclusion in iMO1056 is an example of how the manual reconstruction process can benefit annotation efforts in a way that would not be immediately obvious from automatic model building. In this case, the reconstruction process led us to investigate the methionine salvage cycle and to uncover a reaction that was necessary for the pathway to function but which would have been difficult to pinpoint without a rational model-building approach.
The next level of gene annotation refinement is the assignment of a new function based on gap analysis. This type of annotation refinement occurs when a gap in a pathway renders model growth infeasible, and the missing reaction is subsequently identified by literature mining or a homology search. For example, the previously uncharacterized conserved hypothetical protein PA4457 was identified as an arabinose-5-phosphate isomerase gene (kdsD), which was lacking in the original PAO1 annotation (Fig. 4c). Gap analysis revealed a need for the production of d-arabinose-5-phosphate in the network to enable synthesis of 3-deoxy-d-manno-2-octulosonate (KDO). KDO links the backbone of the lipid A moiety to the core oligosaccharide of lipopolysaccharide and is essential for growth of PAO1 (20). A series of BLAST searches in the nonredundant NCBI database revealed that PA4457 (thus far annotated as a gene coding for a hypothetical protein) had 79% identity over a length of 326 amino acids to the protein KdsD carried by Pseudomonas fluorescens Pf-5 (score, 501; E value, 3 × 10−130). KdsD is an arabinose-5-phosphate isomerase, the enzyme that catalyzes the interconversion of d-arabinose 5-phosphate and d-ribulose 5-phosphate (38). This reannotation and its subsequent incorporation into the model enabled iMO1056 to grow in silico.
Gap analysis also enables the reconciliation of conflicting knowledge in the literature. An earlier version of the PAO1 reconstruction was unable to produce biomass in the absence of oxygen. It was discovered through gap analysis that the inability of iMO1056 to grow without oxygen was due to the oxygen consumption of the enzyme l-aspartate oxidase (NadB), which catalyzes the first step in the production of NAD. Literature mining revealed that while NadB has a strict requirement for O2 when assayed in vitro, it has been suggested that another electron acceptor might suffice in the absence of elemental oxygen in vivo (17). In the network reconstruction, we included an alternate reaction stoichiometry for NadB that had been suggested previously (17), and we thus eliminated the oxygen requirement for growth (Fig. 4d). Despite this literature analysis, however, it is noteworthy that knocking out nadB is not lethal for P. aeruginosa in vivo (10). This result suggests that another pathway for NAD biosynthesis likely exists. More detailed biochemical study is necessary to determine if there is another anaerobic path for NAD synthesis in PAO1 and whether NadB can truly function in the absence of oxygen.
Gap analysis was also used to refine the stoichiometry and directionality of several reactions that were incorrect or vague in online databases. For instance, the reaction catalyzed by the electron transport enzyme NADPH-quinone oxidoreductase (PA4975; EC 1.6.5.5) was found upon analysis of free ATP production to be necessarily irreversible, despite its being listed as reversible in the EXPASY database (19). Such network refinements would be difficult to systematize without a genome-scale model that accounts for the interdependency of reactions and metabolites.
Growth predictions.
The in silico biomass yield of iMO1056 on glucose minimal medium under steady-state (continuous culture) conditions was calculated using FBA. In silico constraints used for minimal medium conditions are outlined in the supplemental material. The maximum specific growth rate was determined to be 1.048 h−1, with a specific glucose uptake rate of 10 mmol Glc · g dry weight−1 · h−1, giving an in silico biomass yield of 0.1048 g dry weight · (mmol Glc)−1. This value for in silico biomass yield falls within the range of experimentally determined values for biomass yield for P. aeruginosa under similar conditions, which were reported previously (67) as 0.094, 0.085, and 0.118 g dry weight · (mmol Glc)−1 at temperatures of 30°C, 38°C, and 41°C, respectively. This value for in silico biomass yield is also within 20% of the maximum biomass yield determined experimentally for P. aeruginosa ATCC 9027 under aerobic growth conditions in another study [0.088 g dry weight · (mmol Glc)−1] (7).
P. aeruginosa is renowned for its ability to survive by denitrification in anaerobic environments (13, 58). The pathway for denitrification of nitrate was included in the reconstruction, and iMO1056 is able to grow anaerobically, with an in silico biomass yield of 0.0846 g dry weight · (mmol Glc)−1, when nitrate is provided under glucose limitation. This in silico biomass yield is within 25% of an experimental value for maximum biomass yield reported previously for P. aeruginosa ATCC 9027 under anaerobic growth conditions [0.067 g dry weight · (mmol Glc)−1] (7). The ratio of maximum anaerobic yield to maximum aerobic yield reported previously (7) is 0.76, which differs from the ratio obtained in silico (0.81) by only 6%, indicating a good match between in silico versus in vivo (experimental) ratios of anaerobic to aerobic yield.
Phenotyping data.
A Biolog study was performed for P. aeruginosa by cultivating PAO1 in minimal medium supplemented with various carbon sources (as described in Materials and Methods). Of the 95 carbon sources tested, 30 could be compared directly to in silico growth of iMO1056 (by assuming that substrate utilization indicates growth, which is usually but not always true). Disagreements between the Biolog data and the in silico growth simulations were used as hypotheses for refining the network reconstruction. The remaining 65 carbon sources included 15 that were present as intracellular metabolites in iMO1056 but for which there were no transporters assigned and 50 that are not currently accounted for in the iMO1056 reconstruction. Of these, 14 showed a growth phenotype in the Biolog data, indicating that incorporation of these compounds into the model might be a future step in iMO1056 model expansion.
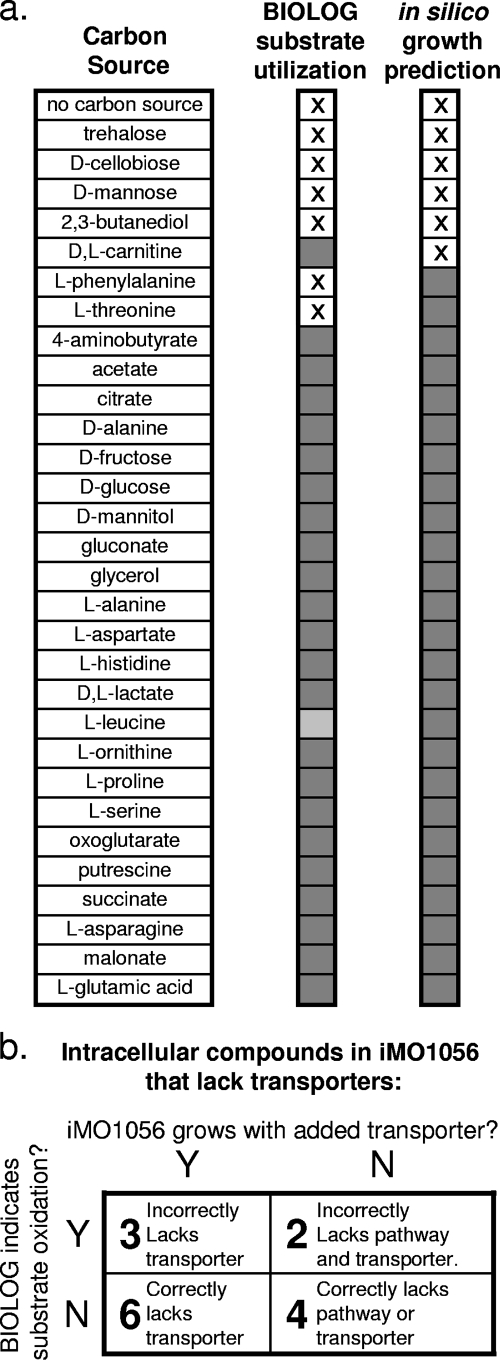
Computational experiments were performed to determine the growth of iMO1056 on the 30 Biolog carbon sources that were directly testable with iMO1056 (Fig. 5a). The in silico growth experiments were performed in Simpheny by using FBA, as described in Materials and Methods. Disagreements between Biolog substrate utilization data and in silico growth simulations were investigated through gap analysis and literature mining, and these discrepancies were rectified where possible. In one case (l-leucine), the Biolog data were unclear about the growth of PAO1, so that data point in Fig. 5a is shaded lighter than those for the other carbon sources but is still considered to show a growth phenotype. After completion of pathways for substrates which initially conflicted with the Biolog data set, PAO1 viability matched iMO1056 viability under 27 of 30 conditions (Fig. 5a). This 90% match indicates that the core metabolism and various catabolic pathways of PAO1 were sufficiently reconstructed in iMO1056 for the model to properly predict catabolism of a variety of common substrates, including many amino acids and several common sugars.
FIG. 5.
Biolog validation. The results of a Biolog study of P. aeruginosa viability on different carbon sources in minimal medium were compared with in silico viability predictions for iMO1056 obtained via FBA. (a) Thirty-one carbon sources tested in Biolog were directly comparable with in silico predictions. The results of this comparison are shown here. An “X” indicates no growth, while shaded boxes in the Biolog column indicate that growth occurred. Lightly shaded boxes in the Biolog column indicate weak results that are close to the sensor threshold for growth and were included as a growth phenotype for analyses. (b) Fifteen carbon sources tested in Biolog are intracellular compounds in iMO1056 but did not sustain in silico growth since they lacked appropriate transporters. The number of carbon sources from this group belonging to each permutation of Biolog growth (yes or no) and in silico growth (yes or no) is represented in the table, as well as a brief description of what each combination of Biolog growth and in silico growth indicates about iMO1056 regarding the set of carbon sources.
The 15 Biolog carbon sources that were intracellular metabolites but did not have transporters in iMO1056 gave in silico no-growth phenotypes due to the inability of iMO1056 to uptake these compounds from the environment. We evaluated the cause of these in silico phenotypes by adding a temporary transport reaction to iMO1056 for each carbon source in turn and assessing the new transporter-augmented models for in silico growth. Figure 5b indicates the results of this study, where numbers in the table represent the numbers of carbon compounds out of 15 total that fall into each of four possible permutations (growth versus nongrowth and in silico versus Biolog data). The 10 carbon sources on which PAO1 did not grow in the Biolog study (and thus matched the in silico results for iMO1056) were grouped into the “correctly lacks transporter” and “correctly lacks either transporter or pathway” categories, depending on whether the transporter-augmented model for that carbon source was able to grow (Fig. 5b). The five carbon sources on which PAO1 did grow in the Biolog study (which thus contradicted the in silico results for iMO1056) were grouped into the similar categories “incorrectly lacks transporter” and “incorrectly lacks pathway and transporter,” indicating the likely cause of the mismatch between Biolog data and in silico simulation. These categorizations will help to focus future annotation refinement efforts. The full list of Biolog results can be found in the supplemental material.
Gene essentiality prediction and validation.
A list of genes predicted to be essential for in silico growth of iMO1056 was compiled and compared with a set of in vivo essential genes (i.e., genes that are reported to be required for growth) from the literature. The set of in vivo essential genes encompasses genes deemed essential in both of two independent genome-scale transposon mutant studies of PAO1 (25, 35). This overlapping data set was chosen because it has a higher confidence value for gene essentiality than the candidate essentials from either transposon study individually, since the inclusion of two transposon studies increases coverage of the PAO1 genome and thus reduces the number of genes erroneously labeled essential. The set of in vivo essentials includes all genes in the PAO1 genome that were not knocked out in either of the two transposon studies. In silico gene essentiality predictions for iMO1056 were calculated in the Simpheny platform via FBA as previously described (27).
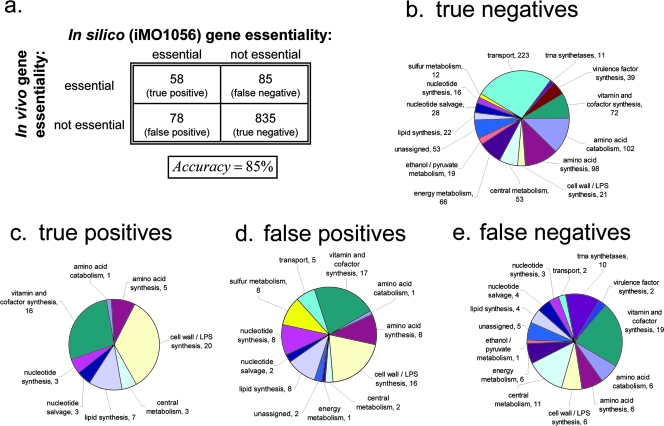
Genes in the in vivo essential set but not in the iMO1056 reconstruction were assumed to be involved either in nonmetabolic functions or in metabolic functions peripheral to central metabolism and were thus not included in our essentiality analysis. The total accuracy of essentiality predictions was 85% (Fig. 6a). This accuracy is comparable to the 94% accuracy achieved in a recent reconstruction of Bacillus subtilis, which also used a genome-scale knockout set for model validation (42). Of genes in the iMO1056 reconstruction, in silico essentiality predictions matched in vivo essentiality for 41% of in vivo essential genes and 91% of in vivo nonessential genes (Fig. 6a). Although the total accuracy of predictions is significant, these discrepancies (particularly in matching in vivo essentiality) deserve further discussion (as follows) and can serve as a collection of hypotheses that can subsequently be tested.
FIG. 6.
Gene essentiality comparison. FBA-derived (in silico) iMO1056 essentiality predictions were compared with an experimentally generated (in vivo) candidate essential gene set for P. aeruginosa. (a) Genes in iMO1056 were broken into the following four groups: true-positive (essential in vivo and in silico), true-negative (nonessential in vivo and in silico), false-positive (nonessential in vivo but essential in silico), and false-negative (essential in vivo but nonessential in silico) groups. The specific pathways represented in true-negative (b), true-positive (c), false-positive (d), and false-negative (e) groups are indicated. Numbers on pie charts indicate numbers of genes belonging to each group.
First, it is informative to note the functions of genes that mismatched between the in silico and in vivo essentiality predictions. Figure 6b, c, d, and e show the functional distribution of genes in iMO1056 that are nonessential in vivo and in silico (true-negative results), essential in vivo and in silico (true-positive results), essential in silico but not in vivo (false-positive results), and essential in vivo but not in silico (false-negative results), respectively. A simple analysis of these figures can explain much of the discrepancy. For instance, among the false-negative set, 10 genes encode tRNA synthetases. These synthetases catalyze the formation of aminoacyl-tRNAs in preparation for protein synthesis. These reactions were included in iMO1056 for the sake of completeness, but the tRNAs represent gaps in the model and so are not truly involved in any part of model function (although the tRNAs are gaps in the sense that they cannot participate in metabolic pathways in the iMO1056 model, and they were included in the reconstruction because they represent genes with known enzymatic functions). Considering these 10 reactions to be nonmetabolic and removing them from iMO1056 would improve the match between the in silico essential gene set and the set of in vivo essential genes in iMO1056 from 41% to 44%.
Vitamin and cofactor synthesis genes make up the largest number of genes in both the false-positive and false-negative categories (Fig. 6d and e), and they make up the second largest number of genes in the true-positive category as well (Fig. 6c). The high discrepancy between in silico and in vivo predictions of essentiality for vitamin and cofactor synthesis genes might be due in part to uncertainty about the vitamin and cofactor content of the LB medium on which the transposon mutants were grown. Since only trace amounts of vitamins are required for cell survival, it is possible that some essential vitamins were present at low concentrations in the LB medium but were not accounted for in our in silico LB medium. This is especially relevant considering previous analyses of yeast extract composition, which showed a high variance in vitamin concentration between extracts from different manufacturers (42; see the supplemental material). Furthermore, since the iMO1056 biomass reaction was based on that of E. coli, it is possible that nutritional differences between the two bacteria contribute to errant predictions about PAO1 vitamin and cofactor essentiality. It is therefore likely that growth experiments on better-defined media will be particularly helpful in elucidating the true essentiality of genes in vitamin and cofactor synthesis pathways.
Second, it is important that the in vivo gene essentiality predictions from the transposon mutant sets are not completely accurate in assessing gene essentiality. An example of a gene that was predicted to be essential in vivo despite in actuality being a nonessential gene is PA0555, encoding the enzyme fructose-1,6-biphosphate aldolase (FBPA). This gene was classified as essential in vivo because transposon mutants with a disrupted FBPA gene were not identified in either PAO1 transposon study, but previous literature shows that the FBPA knockout is not lethal for PAO1 (2). FBPA was correctly predicted to be nonessential in the in silico data set.
The in vivo PAO1 transposon studies used in this analysis discuss the possibility of errors in their candidate essential gene sets, and it was suggested in one of the studies that the number of candidate essential genes in that study (678 total) may overestimate the true number of essential genes by 40 to 50% (25). This overestimate prediction was based on an analysis of the number of ORFs that would likely escape transposon mutagenesis, assuming that all base pairs have equal susceptibility to transposon insertion, including essential genes. Due to its higher coverage of the PAO1 genome, the set of overlapping essentials from the two PAO1 studies should have a higher confidence than that for candidate essentials from either study alone, but the combined transposon set yields an in vivo candidate essential set that is reduced in size by only 34 genes, which does not nearly approximate the 40 to 50% discrepancy suggested (25). Hence, some of the mismatches between the in vivo essentials and the in silico essentials are likely due to overestimation of gene essentiality in the in vivo studies, which would translate into false-negative results in the in silico predictions of essentiality.
Additionally, in one of the transposon studies used in our analysis (35), transposons were reported to likely disrupt genes downstream within an operon, but in the other study (25), an outward-facing neomycin phosphotransferase promoter was inserted as part of the transposon in an attempt to reduce the disruption of downstream genes. It is somewhat unclear from the study, however, how successful the neomycin phosphotransferase promoter actually was in preventing disruption of downstream genes. We therefore undertook an analysis of whether genes downstream of transposon insertions are generally disrupted in vivo, a phenomenon that would cause some nonessential genes to be classified as essential in vivo due to transposons in those genes disrupting downstream essential genes. In order to investigate this phenomenon, we performed a statistical analysis of the number of in vivo essential genes <1,000 bp downstream of genes classified as giving false-negative results. Consistent with the reports for the two transposon studies, we determined that the false-negative results were significantly more likely to have essential genes downstream of them than a set of random genes from the in vivo study that reported polar disruption of downstream genes (indicating possible disruption of downstream essential genes by transposons) (35) but that false-negative results were not more likely to have essential genes downstream of them than a set of random genes from the in vivo study with the neomycin phosphotransferase promoter (indicating a lack of disruption of downstream essential genes by transposons) (25). The result for the in vivo study reporting polar disruption of downstream genes (35) might have been due to the study's much lower coverage of the genome than that of the other study (23% versus 88% of ORFs disrupted). Regardless, this analysis suggests that in a genome-scale transposon study, an outward-facing neomycin phosphotransferase promoter is effective in preventing disruption of downstream genes (see the supplemental material for more details).
DISCUSSION
We present iMO1056, a genome-scale reconstruction of P. aeruginosa PAO1 that accounts for 1,056 genes encoding 1,030 proteins that are involved in 883 reactions. Specifically reconstructed pathways included those necessary for growth and for production of common virulence factors, including alginate, rhamnolipids, phenazines, and quorum-sensing molecules. iMO1056 was validated with experimental growth rate data from literature, Biolog viability data on multiple carbon sources, and a genome-scale gene essentiality set derived from two independent transposon mutant studies of P. aeruginosa. Model predictions matched well with experimental data in many cases.
Building a genome-scale reconstruction of PAO1 offered insights that would have been difficult to obtain through any other means, including evidence for annotation refinements, the ability to quantitatively predict growth yields and secretion rates under various conditions, and a potential way to probe metabolic stresses incurred by the production of various virulence factors that may be explored in future studies. During the model-building process, we were forced to approach each metabolic pathway quantitatively and comprehensively, so previous gaps in knowledge were highlighted and investigated in a rational manner. This process of gap analysis involves coupling in silico growth simulations with bioinformatic searches and literature mining to fill holes in otherwise known pathways and offers a unique tool for identifying areas of metabolism that require further elucidation. Through gap analysis of P. aeruginosa metabolism, we were able to refine the annotation of a number of genes with respect to their function, directionality, or stoichiometry. Some of these changes are highlighted in Table 2. These annotation refinements represent many crucial metabolic functions of P. aeruginosa, and without a systematic network-level approach to guide our analysis, it would have been difficult to highlight these weak points in current knowledge.
As in any genome-scale annotation effort, the network reconstruction process for P. aeruginosa will require continued work in the future. Mismatches between in vivo and in silico essential genes have highlighted metabolic regions of uncertainty in current knowledge, and these discrepancies still require more work to be explained adequately. In addition, several pathways in the model remain incomplete due to a lack of available knowledge. These gaps include synthesis of cofactors (e.g., cobalamin and thiamine) and, notably, the entire fatty acid β-oxidation pathway, which exists in but has not been characterized extensively for pseudomonads, to our knowledge (52). The Biolog data also indicated some pathways that should be investigated further in the future, such as substrates that PAO1 utilized but which are not in iMO1056 or for which iMO1056 has no membrane transporter. Furthermore, some pathways were not included in the model despite having been studied in the literature, simply because they were peripheral to the main processes of interest in this study (growth and expression of virulence traits). Thus, for instance, pathways for degrading aromatic compounds and for handling xenobiotics were generally not included in the model, and these offer areas for model expansion in the future. Specific validation involving controlled growth experiments with P. aeruginosa will also be informative, and coupling wet lab and in silico experiments will lend greater insight into both specific functions and global properties of P. aeruginosa metabolism.
Additionally, a genome-scale reconstruction of P. aeruginosa metabolism enables the interrogation of several otherwise difficult research questions. For instance, it would be informative to compare the P. aeruginosa metabolic network with metabolic networks of other related but nonpathogenic species. This comparison would allow probing of properties such as pathway redundancy and growth burden of key virulence pathways and would offer insight into how these system-level properties might affect pathogenicity. Such a genome-scale network analysis between a pathogen and a nonpathogen has never been done, to our knowledge, and could provide significant insight into the mechanisms for disease and possible therapeutic targets. Another question of great interest involves the enumeration of selective pressures on P. aeruginosa in the CF lung environment. For example, P. aeruginosa samples obtained from sputum during the life of a CF patient have shown gene mutations and significant alterations in gene expression over time (4, 60). Notably, convergent evolution toward a loss of virulence factors by P. aeruginosa strains taken from multiple CF patients has been demonstrated, suggesting that virulence traits might be selected against once a stable infection has been achieved (60). The effect of the loss of the lasR gene, a master regulator of hundreds of virulence-related genes through the quorum-sensing system in P. aeruginosa, has also been evaluated (8, 56). It was shown that loss of the lasR gene conferred a growth advantage both to strains extracted from the lungs of CF patients and from strains that emerged by selection on rich medium, indicating that optimization of yield could act as a metabolic objective even in the CF lung (8). This result is counterintuitive, as much of the success of P. aeruginosa in chronic lung infections seems to lie in its ability to adopt a slow-growth phenotype, which would suggest that optimization of yield is perhaps not a strong selective pressure in that environment (6). Regardless of whether slow-growing mutants of P. aeruginosa in the CF lung environment are actually optimized for yield, these studies indicate that metabolic selective pressure might be a factor in the evolution of chronic CF strains. With the integration of simple regulatory rules into iMO1056, it will be possible to model different hypotheses about selective pressures in the lung and to analyze the causes for these selective processes.
In addition to the pathogenicity analysis and evolutionary studies outlined above, iMO1056 can serve as a valuable tool in interpreting and informing genome-scale transcriptomic studies of PAO1. Microarray analysis has attracted sizeable interest in the P. aeruginosa community and was recently used to determine genome-level expression changes under various stresses and conditions (13, 36, 71). Enabling a survey of system-level traits of an organism in a relatively unbiased way, microarrays are a crucial element in postgenomic analyses of cell phenotype. While gene expression data cannot be linked directly with metabolic fluxes, past studies have used gene expression data to indicate the regulation of whole pathways in metabolism, thus indicating global phenotypes (9, 12). Furthermore, metabolic reconstructions have been used to predict which genes in a network are likely to be coregulated, and overlaying expression data on a metabolic reconstruction can inform interpretation of microarrays within the context of a genome-scale model (9, 43).
P. aeruginosa is an organism of much interest for its various roles as an opportunistic pathogen (60). From chronic lifelong infections of the lungs of CF patients to acute, highly deadly infections of severe wounds in burn victims, the robustness and environmental diversity of P. aeruginosa are testament to its remarkable natural metabolic agility. We chose to focus our reconstruction effort on P. aeruginosa PAO1 since it is the most studied P. aeruginosa strain and is also the best-characterized strain, but with minor modifications to iMO1056 the model can be tailored to describe similar strains, such as the more virulent P. aeruginosa PA14 (62, 69). A genome-scale metabolic model represents a potentially enormous tool for rational drug design and prediction of cell phenotypes and, in conjunction with regulatory information, can serve in modeling disease processes and engineering therapeutic responses. For P. aeruginosa, a bacterium whose metabolic diversity is a major determinant of virulence, a metabolic network reconstruction will serve as an essential component in a multifaceted and effective response to disease.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Joanna Goldberg, Kenneth N. Timmis, Arvind Chavali, Erwin Gianchandani, Corinne Locke, and Rhiannon Franck for their thoughtful comments and contributions to this study. We also thank the reviewers for thoughtful comments that led to interesting new analyses and insights.
We acknowledge funding from the Whitaker Foundation, the National Science Foundation (CAREER grant 0643548), the National Institutes of Health (NIH Biotechnology training grant [M.A.O.]), the Kluyver Centre for Genomics (The Netherlands), the European Union (Project PROBACTYS; no. 21904), and ERA-NET SysMO (BMBF).
Footnotes
▿
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Alvarez-Ortega, C., and C. S. Harwood. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, P. C., R. I. Vanags, A. M. Chakrabarty, and P. K. Maitra. 1985. Fructose 1,6-bisphosphate aldolase activity is essential for synthesis of alginate from glucose by Pseudomonas aeruginosa. J. Bacteriol. 161458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beste, D. J., T. Hooper, G. Stewart, B. Bonde, C. Avignone-Rossa, M. E. Bushell, P. Wheeler, S. Klamt, A. M. Kierzek, and J. McFadden. 2007. GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol. 8R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bragonzi, A., D. Worlitzsch, G. B. Pier, P. Timpert, M. Ulrich, M. Hentzer, J. B. Andersen, M. Givskov, M. Conese, and G. Doring. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bro, C., B. Regenberg, J. Forster, and J. Nielsen. 2006. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 8102-111. [DOI] [PubMed] [Google Scholar]
- 6.Burns, J. L., L. Saiman, S. Whittier, D. Larone, J. Krzewinski, Z. Liu, S. A. Marshall, and R. N. Jones. 2000. Comparison of agar diffusion methodologies for antimicrobial susceptibility testing of Pseudomonas aeruginosa isolates from cystic fibrosis patients. J. Clin. Microbiol. 381818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., Q. Xia, and L. K. Ju. 2006. Competition between oxygen and nitrate respirations in continuous culture of Pseudomonas aeruginosa performing aerobic denitrification. Biotechnol. Bioeng. 931069-1078. [DOI] [PubMed] [Google Scholar]
- 8.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Deziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David, H., G. Hofmann, A. P. Oliveira, H. Jarmer, and J. Nielsen. 2006. Metabolic network driven analysis of genome-wide transcription data from Aspergillus nidulans. Genome Biol. 7R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVries, C. A., D. J. Hassett, J. L. Flynn, and D. E. Ohman. 1995. Genetic linkage in Pseudomonas aeruginosa of algT and nadB: mutation in nadB does not affect NAD biosynthesis or alginate production. Gene 15663-67. [DOI] [PubMed] [Google Scholar]
- 11.Deziel, E., F. Lepine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 1011339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duarte, N. C., S. A. Becker, N. Jamshidi, I. Thiele, M. L. Mo, T. D. Vo, R. Srivas, and B. O. Palsson. 2007. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA 1041777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschbach, M., K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 1864596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrow, J. M., III, and E. C. Pesci. 2007. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J. Bacteriol. 1893425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feala, J. D., L. Coquin, A. D. McCulloch, and G. Paternostro. 2007. Flexibility in energy metabolism supports hypoxia tolerance in Drosophila flight muscle: metabolomic and computational systems analysis. Mol. Syst. Biol. 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong, S. S., and B. O. Palsson. 2004. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat. Genet. 361056-1058. [DOI] [PubMed] [Google Scholar]
- 17.Foster, J. W., and A. G. Moat. 1980. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol. Rev. 4483-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fothergill, J. C., and J. R. Guest. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J. Gen. Microbiol. 99139-155. [DOI] [PubMed] [Google Scholar]
- 19.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 313784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman, R. C., C. C. Doran, S. K. Kadam, and J. O. Capobianco. 1988. Lipid A precursor from Pseudomonas aeruginosa is completely acylated prior to addition of 3-deoxy-d-manno-octulosonate. J. Biol. Chem. 2635217-5223. [PubMed] [Google Scholar]
- 21.Heinemann, M., A. Kummel, R. Ruinatscha, and S. Panke. 2005. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol. Bioeng. 92850-864. [DOI] [PubMed] [Google Scholar]
- 22.Herrgard, M. J., B. S. Lee, V. Portnoy, and B. O. Palsson. 2006. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 16627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heurlier, K., V. Denervaud, and D. Haas. 2006. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 29693-102. [DOI] [PubMed] [Google Scholar]
- 24.Ideker, T., V. Thorsson, J. A. Ranish, R. Christmas, J. Buhler, J. K. Eng, R. Bumgarner, D. R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292929-934. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10014339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamshidi, N., and B. O. Palsson. 2007. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst. Biol. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce, A. R., J. L. Reed, A. White, R. Edwards, A. Osterman, T. Baba, H. Mori, S. A. Lesely, B. O. Palsson, and S. Agarwalla. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 1888259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karsten, W. E., P. A. Tipton, and P. F. Cook. 2002. Tartrate dehydrogenase catalyzes the stepwise oxidative decarboxylation of d-malate with both NAD and thio-NAD. Biochemistry 4112193-12199. [DOI] [PubMed] [Google Scholar]
- 30.Kay, W. W., and A. F. Gronlund. 1969. Amino acid transport in Pseudomonas aeruginosa. J. Bacteriol. 97273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kneidinger, B., C. Marolda, M. Graninger, A. Zamyatina, F. McArthur, P. Kosma, M. A. Valvano, and P. Messner. 2002. Biosynthesis pathway of ADP-l-glycero-beta-d-manno-heptose in Escherichia coli. J. Bacteriol. 184363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koffas, M., and G. Stephanopoulos. 2005. Strain improvement by metabolic engineering: lysine production as a case study for systems biology. Curr. Opin. Biotechnol. 16361-366. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. M., E. P. Gianchandani, and J. A. Papin. 2006. Flux balance analysis in the era of metabolomics. Brief. Bioinform. 7140-150. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. Y., D. Y. Lee, and T. Y. Kim. 2005. Systems biotechnology for strain improvement. Trends Biotechnol. 23349-358. [DOI] [PubMed] [Google Scholar]
- 35.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. Brinkman, and R. E. Hancock. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lizewski, S. E., J. R. Schurr, D. W. Jackson, A. Frisk, A. J. Carterson, and M. J. Schurr. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 1865672-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto, H., S. Ohta, R. Kobayashi, and Y. Terawaki. 1978. Chromosomal location of genes participating in the degradation of purines in Pseudomonas aeruginosa. Mol. Gen. Genet. 167165-176. [DOI] [PubMed] [Google Scholar]
- 38.Meredith, T. C., and R. W. Woodard. 2003. Escherichia coli YrbH is a d-arabinose 5-phosphate isomerase. J. Biol. Chem. 27832771-32777. [DOI] [PubMed] [Google Scholar]
- 39.Mousa, H. A. 2005. Burn and scald injuries. East Mediterr. Health J. 111099-1109. [PubMed] [Google Scholar]
- 40.Nguyen, D., and P. K. Singh. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. USA 1038305-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noguchi, Y., Y. Nakai, N. Shimba, H. Toyosaki, Y. Kawahara, S. Sugimoto, and E. Suzuki. 2004. The energetic conversion competence of Escherichia coli during aerobic respiration studied by 31P NMR using a circulating fermentation system. J. Biochem. (Tokyo) 136509-515. [DOI] [PubMed] [Google Scholar]
- 42.Oh, Y. K., B. O. Palsson, S. M. Park, C. H. Schilling, and R. Mahadevan. 2007. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 28228791-28799. [DOI] [PubMed] [Google Scholar]
- 43.Papin, J. A., J. L. Reed, and B. O. Palsson. 2004. Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem. Sci. 29641-647. [DOI] [PubMed] [Google Scholar]
- 44.Perron, K., O. Caille, C. Rossier, C. Van Delden, J. L. Dumas, and T. Kohler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2798761-8768. [DOI] [PubMed] [Google Scholar]
- 45.Rahman, M., and P. H. Clarke. 1980. Genes and enzymes of lysine catabolism in Pseudomonas aeruginosa. J. Gen. Microbiol. 116357-369. [DOI] [PubMed] [Google Scholar]
- 46.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56309-322. [DOI] [PubMed] [Google Scholar]
- 47.Reed, J. L., I. Famili, I. Thiele, and B. O. Palsson. 2006. Towards multidimensional genome annotation. Nat. Rev. Genet. 7130-141. [DOI] [PubMed] [Google Scholar]
- 48.Reed, J. L., T. D. Vo, C. H. Schilling, and B. O. Palsson. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 1711209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saeed, H. M., Y. R. Abdel-Fattah, M. M. Berekaa, Y. M. Gohar, and M. A. Elbaz. 2004. Identification, cloning and expression of Pseudomonas aeruginosa Ps-x putative urate oxidase gene in Escherichia coli. Pol. J. Microbiol. 53227-236. [PubMed] [Google Scholar]
- 51.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato, S., M. Hayashi, S. Imamura, Y. Ozeki, and A. Kawaguchi. 1992. Primary structures of the genes, faoA and faoB, from Pseudomonas fragi B-0771 which encode the two subunits of the HDT multienzyme complex involved in fatty acid beta-oxidation. J. Biochem. (Tokyo) 1118-15. [DOI] [PubMed] [Google Scholar]
- 53.Schaffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge, Y. I. Wolf, E. V. Koonin, and S. F. Altschul. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 292994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaffitzel, C., M. Berg, P. Dimroth, and K. M. Pos. 1998. Identification of an Na+-dependent malonate transporter of Malonomonas rubra and its dependence on two separate genes. J. Bacteriol. 1802689-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuetz, R., L. Kuepfer, and U. Sauer. 2007. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 10115833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekowska, A., V. Denervaud, H. Ashida, K. Michoud, D. Haas, A. Yokota, and A. Danchin. 2004. Bacterial variations on the methionine salvage pathway. BMC Microbiol. 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma, V., C. E. Noriega, and J. J. Rowe. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shlomi, T., Y. Eisenberg, R. Sharan, and E. Ruppin. 2007. A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol. Syst. Biol. 3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soberon-Chavez, G., F. Lepine, and E. Deziel. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68718-725. [DOI] [PubMed] [Google Scholar]
- 62.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406959-964. [DOI] [PubMed] [Google Scholar]
- 63.Sun, J., X. Lu, U. Rinas, and A. P. Zeng. 2007. Metabolic peculiarities of Aspergillus niger disclosed by comparative metabolic genomics. Genome Biol. 8R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teusink, B., F. H. van Enckevort, C. Francke, A. Wiersma, A. Wegkamp, E. J. Smid, and R. J. Siezen. 2005. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 717253-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiele, I., T. D. Vo, N. D. Price, and B. O. Palsson. 2005. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J. Bacteriol. 1875818-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varma, A., and B. O. Palsson. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 603724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verstraete, W., and J. P. Voets. 1978. Evaluation of yield and maintenance coefficients, expressed in carbon units, for Pseudomonas fluorescens and P. aeruginosa. Z. Allg. Mikrobiol. 18135-141. [DOI] [PubMed] [Google Scholar]
- 68.Wagenlehner, F. M., and K. G. Naber. 2006. Current challenges in the treatment of complicated urinary tract infections and prostatitis. Clin. Microbiol. Infect. 12(Suppl. 3)67-80. [DOI] [PubMed] [Google Scholar]
- 69.Winsor, G. L., R. Lo, S. J. Sui, K. S. Ung, S. Huang, D. Cheng, W. K. Ching, R. E. Hancock, and F. S. Brinkman. 2005. Pseudomonas aeruginosa genome database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33D338-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, Z., and C. D. Lu. 2007. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 1893945-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, J., H. Li, J. Wang, Z. Dong, S. Mian, and F. S. Yu. 2004. Role of EGFR transactivation in preventing apoptosis in Pseudomonas aeruginosa-infected human corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 452569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]