AGO1 Homeostasis Entails Coexpression of MIR168 and AGO1 and Preferential Stabilization of miR168 by AGO1 (original) (raw)
. Author manuscript; available in PMC: 2008 Apr 18.
Summary
Arabidopsis ARGONAUTE1 (AGO1) encodes the RNA slicer enzyme of the microRNA (miRNA) pathway and is regulated by miR168-programmed, AGO1-catalyzed mRNA cleavage. Here, we describe two additional regulatory processes required for AGO1 homeostasis: transcriptional coregulation of MIR168 and AGO1 genes, and posttranscriptional stabilization of miR168 by AGO1. Disrupting any of these regulatory processes by using mutations or transgenes disturbs a proper functioning of the miRNA pathway. In contrast, minor perturbation leads to fine-tuned posttranscriptional adjustment of miR168 and AGO1 levels, thereby maintaining a proper balance of other miRNAs, which, together with AGO1, control the mRNA levels of miRNA targets. We suggest that miR168 stabilization occurs at the level of silencing-complex assembly and that modulating the efficiency of assembling miRNA-programmed silencing complexes will also be important in other contexts.
Introduction
MicroRNAs (miRNAs) are 21 nt endogenous RNAs that can regulate gene expression posttranscriptionally (Bartel, 2004). They are processed from stem-loop structures of longer primary transcripts through the action of RNase III enzymes. In animals, the RNase III Drosha makes a first cut that produces pre-miRNAs, and the RNase III Dicer liberates miRNA:miRNA* duplexes from pre-miRNAs (Kim, 2005). In plants, the RNase III DICER-LIKE 1 (DCL1) makes both cuts (Park et al., 2002; Reinhart et al., 2002; Kurihara and Watanabe, 2004). After processing, the miRNA strand of the miRNA:miRNA* duplex is loaded into an Argonaute (AGO) protein, which has a single-stranded RNA binding PAZ domain and an RNa-seH-like PIWI domain, which catalyzes either mRNA cleavage or translation repression (Tomari and Zamore, 2005). In plants, miRNAs are loaded into AGO1, which acts as an RNA slicer, similar to human AGO2 (Baumberger and Baulcombe, 2005; Qi et al., 2005).
Although several actors in the miRNA pathway have been genetically or biochemically characterized, little is known about how they are regulated to maintain the homeostasis of the miRNA pathway. Two miRNAs, miR168 and miR162, direct the cleavage of AGO1 and DCL1 mRNAs, respectively, indicating that these miRNAs themselves feedback regulate the activity of the miRNA pathway (Xie et al., 2003; Vaucheret et al., 2004). We previously reported that ago1 null alleles exhibit reduced levels of miRNAs and increased levels of the corresponding target mRNAs (Vaucheret et al., 2004). In addition, plants that express a miR168-resistant AGO1 mRNA (4m-AGO1) exhibit developmental defects, indicating that the role of AGO1 in the miRNA pathway and its regulation by miR168 are crucial for plant development (Vaucheret et al., 2004). Here, we describe two additional layers of regulation: the transcriptional coregulation of AGO1 and MIR168 genes and the stabilization of miR168 by AGO1. Our results indicate that posttranscriptional AGO1-mediated stabilization of miR168 works in concert with miR168-programmed, AGO1-catalyzed cleavage of AGO1 mRNA to maintain AGO1 homeostasis, proper miRNA pathway functioning, and normal plant development.
Results and Discussion
miR168 Overaccumulates in 4m-AGO1 Plants
We previously reported that expressing a miR168-resistant AGO1 gene (4m-AGO1) in Arabidopsis results in a series of developmental defects, suggesting that the excess AGO1 protein interferes with the functioning of the miRNA pathway (Vaucheret et al., 2004). Analysis of miR168 accumulation in 4m-AGO1 plants exhibiting either a mild or severe 4m-AGO1 phenotype (Figure 1A) revealed that miR168 overaccumulated 3- to 5-fold (Figure 1C). The degree of miR168 overaccumulation (Figure 1C) directly correlated with both the severity of the phenotype (Figure 1A) and the level of AGO1 mRNA overaccumulation (Figure 1B). The perturbation of miR168 was specific to 4m-AGO1 expression and not simply due to the expression of a miRNA-resistant mRNA, because plants expressing miRNA-resistant CUC1 or ARF17 (5m-CUC1 or 5m-ARF17), (Mallory et al., 2004a, 2005), accumulated miR168 at wild-type levels (see Figure S1 in the Supplemental Data available with this article online). With the exception of miR159 and miR165/166, which accumulated at levels 2- to 3-fold higher in 4m-AGO1 plants than in wild-type plants (Figure 1C), the other nine miRNAs analyzed accumulated at levels similar to those of wild-type plants, indicating that miR168 accumulation was the most sensitive to 4m-AGO1 expression.
Figure 1. miR168 Accumulation Is Increased in 4m-AGO1 Transformants and Is Unchanged in Hypomorphic dcl1 Mutants.
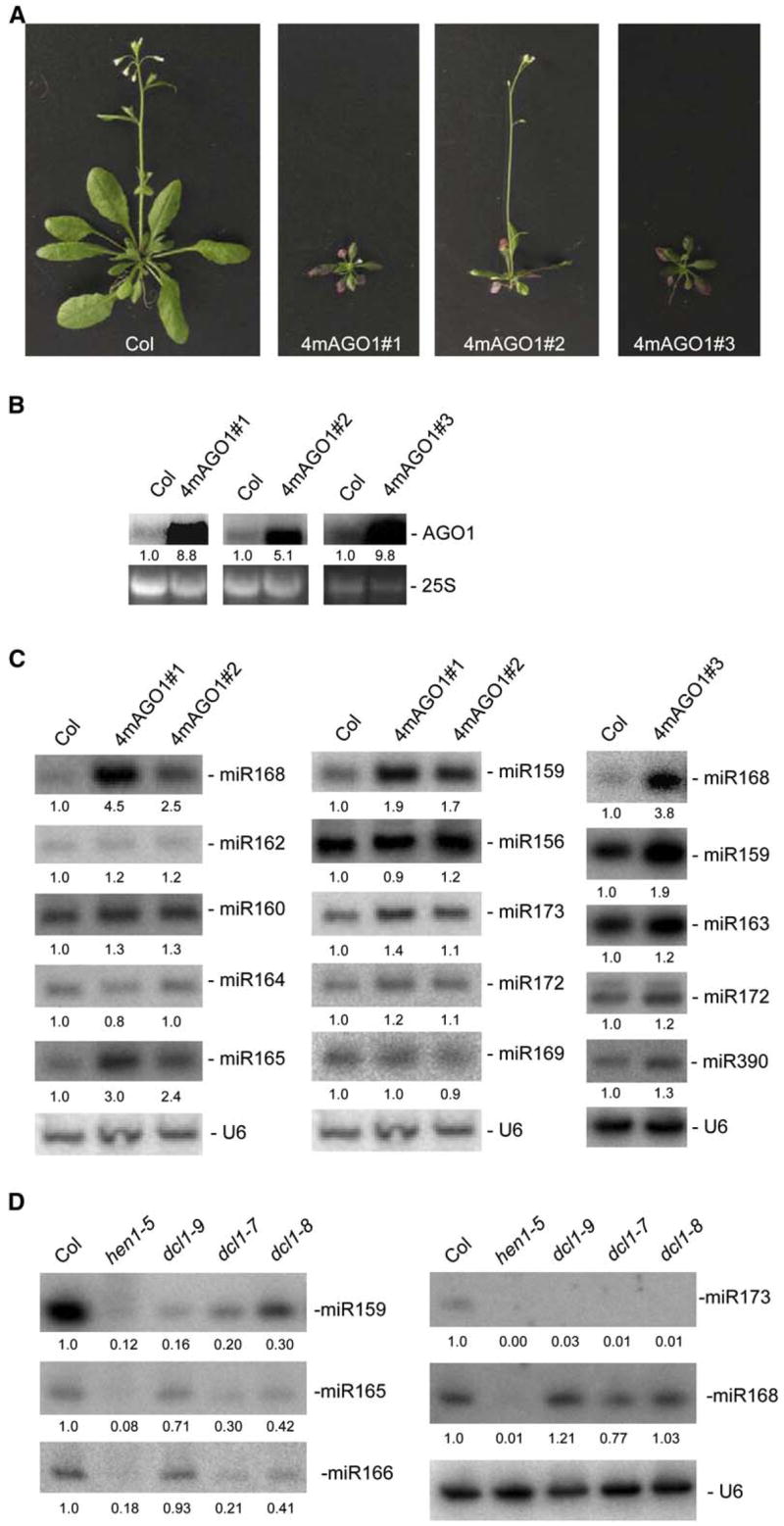
(A) Phenotypes of 35-day-old mild and severe 4m-AGO1 transformants compared with a 35-day-old wild-type Col plant.
(B) AGO1 mRNA accumulation in wild-type Col and 4m-AGO1 transformants was determined by RNA gel blot analysis using 10 μg of RNA. Ethidium bromide staining of 25S rRNA is shown as a loading control. Normalized values of mRNAs to 25S rRNA are indicated, with mRNA levels in Col control plants set at 1.0.
(C) miRNA accumulation in wild-type Col and 4m-AGO1 transformants was determined by RNA gel blot analysis using 10 μg of the same RNA in (B). Blots were successively hybridized to different probes complementary to miRNAs and a probe complementary to U6 as a loading control. Normalized values of miRNA to U6 RNA are indicated, with miRNA levels in Col set at 1.0.
(D) miRNA accumulation in wild-type Col and hen1 and dcl1 mutants was determined by RNA gel blot analysis using 10 μg of RNA. A single blot was successively hybridized to different probes complementary to miRNAs and a probe complementary to U6 as a loading control. Normalized values of miRNA to U6 RNA are indicated, with miRNA levels in Col set at 1.0.
MicroRNAs that Are the Most Sensitive to Changes in AGO1 Levels Are the Least Sensitive to Changes in DCL1 Activity
Null dcl1 mutants are embryo lethal, but three viable hypomorphic dcl1 mutants have been recovered during developmental screens (Schauer et al., 2002). Previous analyses revealed that miRNA accumulation is strongly reduced in these mutants, with the exception of miR166 (Park et al., 2002; Reinhart et al., 2002; Kasschau et al., 2003; Mlotshwa et al., 2005; Kurihara et al., 2006). However, the effect of dcl1 mutations on miR168 accumulation has never been reported. We found that miR168 accumulated at similar levels in wild-type plants and all three dcl1 mutants (Figure 1D). In addition, miR159 and miR165/166 accumulation was reduced in dcl1 mutants but remained detectable (Figure 1D). This contrasts sharply with other miRNAs, such as miR173, which were below detectable levels in all three mutants (Figure 1D and data not shown). These results point to a particular behavior of miR168 and, to a lesser extent, of miR159 and miR165/166 and indicate that the miRNAs that are the least sensitive to impairment in DCL1 activity are the most sensitive to changes in AGO1 levels (Figure 1C).
miR168a Stem-Loop Intermediate, miR168a*, and Processed Loop Overaccumulate in 35Sx2-MIR168a Plants but Not in 4m-AGO1 Plants
Several mechanisms could account for the increased level of miR168 in 4m-AGO1 plants, including increased transcription of MIR168 genes, increased processing of MIR168 transcripts, or increased stability of miR168. To determine the predominant mechanism for miR168 overaccumulation, we compared the levels of MIR168 transcript processing intermediates and mature products in wild-type plants, 4m-AGO1 plants, and plants expressing the MIR168a transcript under the control of a strong, broadly expressed promoter.
To generate MIR168a-overexpressing plants, we cloned a 416 nt fragment of the MIR168a gene corresponding to EST H77158 downstream of the Cauliflower mosaic virus 35S promoter with a double enhancer (Figure 2A) and introduced this construct into wild-type Col plants. RNA blot analysis revealed an overaccumulation of miR168 in several lines (Figure 2B). In addition to mature miR168, other MIR168a-processing products and intermediates overaccumulated in 35Sx2-MIR168a transformants (Figures 2B and 2C). These RNAs included miR168a* (the 21 nt RNA from the other arm of the stem loop), the 64 nt loop that connects miR168 and miR168*, and the 106 nt stem-loop intermediate. The detection of the 106 nt stem-loop intermediate (Figure 2B) indicated that at least some primary MIR168a transcript was cut at the stem first, similar to the Drosha-mediated cut of animal pri-miRNAs (Kim, 2005). Whether the cut that liberates the 64 nt loop always occurs after the cut at the stem, similar to the Dicer-mediated cut of animal pre-miRNAs (Kim, 2005), or whether a first cut can occur at the loop followed by a second cut at the stem, is unknown. A 2-fold increase in the accumulation of miR168, miR168*, MIR168a loop, and MIR168a stem-loop intermediate also was observed in the T-DNA insertion line SALK 113514 (Figure 2C). In this line, an inverted T-DNA repeat is inserted 60 bp upstream of the 5′ end of the MIR168a EST H77185 (Figure 2A). One T-DNA copy is intact, while the other is truncated at the left border at position +41 relative to the transcription start of the 35S promoter, resulting in a transcriptional fusion between the 35S promoter carried by the T-DNA and the MIR168a transcribed region. Because this line could be considered as an activation-tagged line, we refer to it as mir168a-1d. The increased accumulation of mature miR168 and other MIR168a-processing products and intermediates in line mir168a-1d that resulted from substituting the native MIR168a promoter by the 35S promoter in the genomic context of the MIR168a gene confirmed that the increased accumulation of mature miR168 and other MIR168a-processing products and intermediates in 35Sx2-MIR168a transformants was not due to some aberrancy of the 35S-MIR168a construct.
Figure 2. The Stem-Loop Intermediate and the Processed Loop Overaccumulate in 35Sx2-MIR168 Plants and the SALK 113514 Line but Not in 4m-AGO1 Plants.
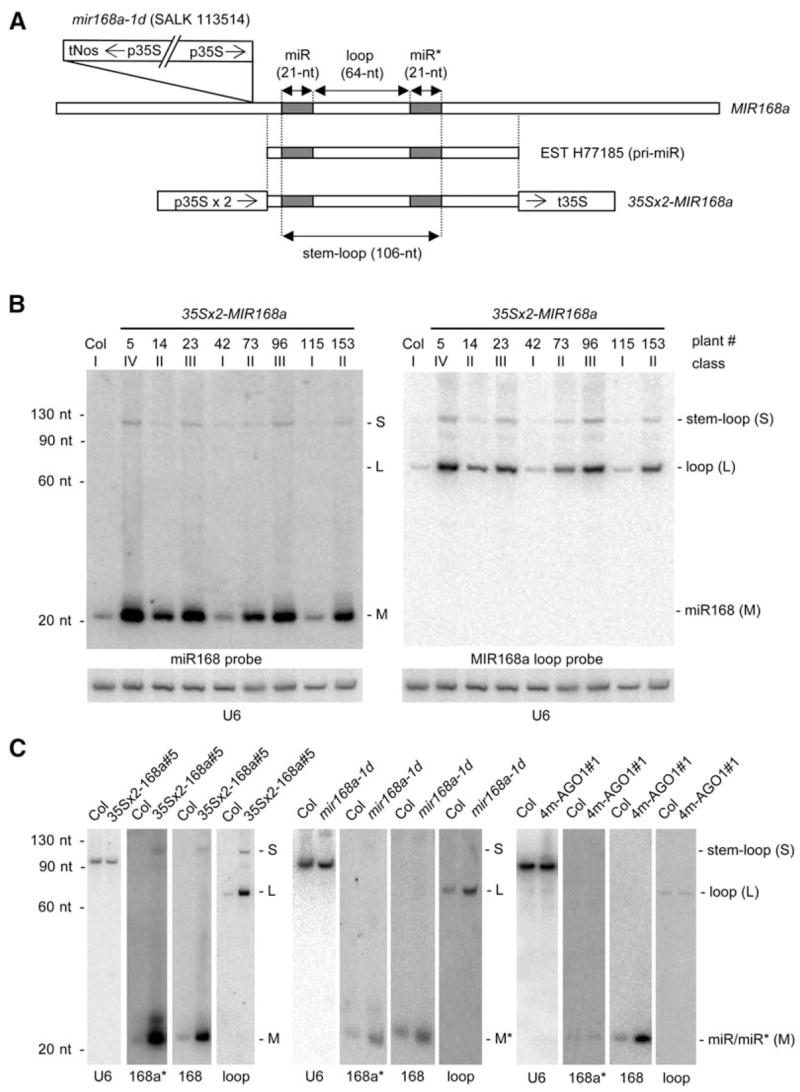
(A) Structures of the T-DNA in the mir168a-1d mutant (SALK 113514) relative to the MIR168a locus, the MIR168a EST H77185, and the 35Sx2-MIR168a transgene. In mir168a-1d, the insertion of a truncated, inverted T-DNA repeat substitutes the native MIR168a promoter with the 35S promoter. The 35Sx2-MIR168a construct was generated by inserting the MIR168a region corresponding to EST H77185 between the 35S promoter with a double enhancer (35Sx2) and the 35S terminator. The 106 nt stem-loop intermediate corresponds to the RNA product expected after a Drosha-like cut at the stem. The 64 nt loop corresponds to the RNA product expected after a Dicer-like cut at the loop. The positions of the mature miR168 and miR168a* are indicated by gray boxes.
(B) Mature miR168, processed loop, and stem-loop intermediate accumulation in Col and 35x2-MIR168a transformants. RNA gel blot analysis of 10 μg of RNA with a probe complementary to miR168 (left), a probe complementary to the processed loop (right), and U6 as loading control (below). Note that the increase in processed loop and stem-loop intermediate correlates with the increase in mature miRNA.
(C) Stem-loop, miR168, miR168*, and loop accumulation in Col, 4m-AGO1#1, mir168a-1d, and 35-MIR168a#5. RNA gel blot analysis of 10 μg of RNA with miR168, miR168a*, and miR168a-loop probes reveals that only mature miR168 overaccumulates in 4m-AGO1 plants, whereas the 106 nt stem-loop intermediate, mature miRNA and miRNA*, and the 64 nt processed loop overaccumulate in mir168a-1d and 35-MIR168a#5 plants. The positions of the stem-loop intermediate (S), processed loop (L), and miR168 and miR168* (M) are noted at the right. The probe is indicated below each blot. Hybridizations with a probe complementary to U6 are shown as a loading control.
In contrast to mir168a-1d and 35Sx2-MIR168a plants, 4m-AGO1 plants did not have altered stem-loop intermediate, processed loop, or miR168* accumulation (Figure 2C), suggesting that neither the transcription of MIR168 genes nor the processing of MIR168 transcripts is increased in 4m-AGO1 plants. Having ruled out substantial changes in transcription or processing, we concluded that the increase in miR168 in 4m-AGO1 plants was due to increased stability of the mature miR168. That is, increased AGO1 protein (a reasonably inferred consequence of increased AGO1 mRNA observed in 4m-AGO1 plants) results in increased stability of the mature miR168. This model implies that AGO1 is normally limiting for the stabilization of miR168 in wild-type plants. Note that we disfavor a model in which miR168 stabilization would result from unproductive use of miR168 by RISC due to 4m-AGO1 mRNA being resistant to miR168-directed cleavage because plants expressing other miRNA-resistant mRNAs, such as 5m-CUC1 or 5m-ARF17 (Mallory et al., 2004a; Mallory et al., 2005), accumulate the corresponding miRNAs, miR164 and miR160, at wild-type levels (Figure S1).
Overexpression of miR168 Impacts the Accumulation of miR165/166 and miR159
35Sx2-MIR168a transformants exhibited qualitatively consistent phenotypes (Figure 3A). The most altered plants exhibited serrated and adaxialized leaves (Figures 3A and 3B), flowered late compared with wild-type plants (Figure 3A), and had altered floral phyllotaxy (Figure 3C), reminiscent of the phenotypes of hypomorphic ago1 alleles (Morel et al., 2002; Kidner and Martienssen, 2004). RNA blot analysis of miR168 accumulation revealed a tight correlation between the severity of the phenotype and the level of miR168 overaccumulation (Figure 3D). AGO1 mRNA accumulation was reduced ~5- to 10-fold in the 35Sx2-MIR168a line with the most severe phenotype (Figure 3E).
Figure 3. miR168 Overaccumulation in 35Sx2-MIR168a Transformants Phenocopies Hypomorphic ago1 Mutants and Results in Downregulation of Other Small RNAs.
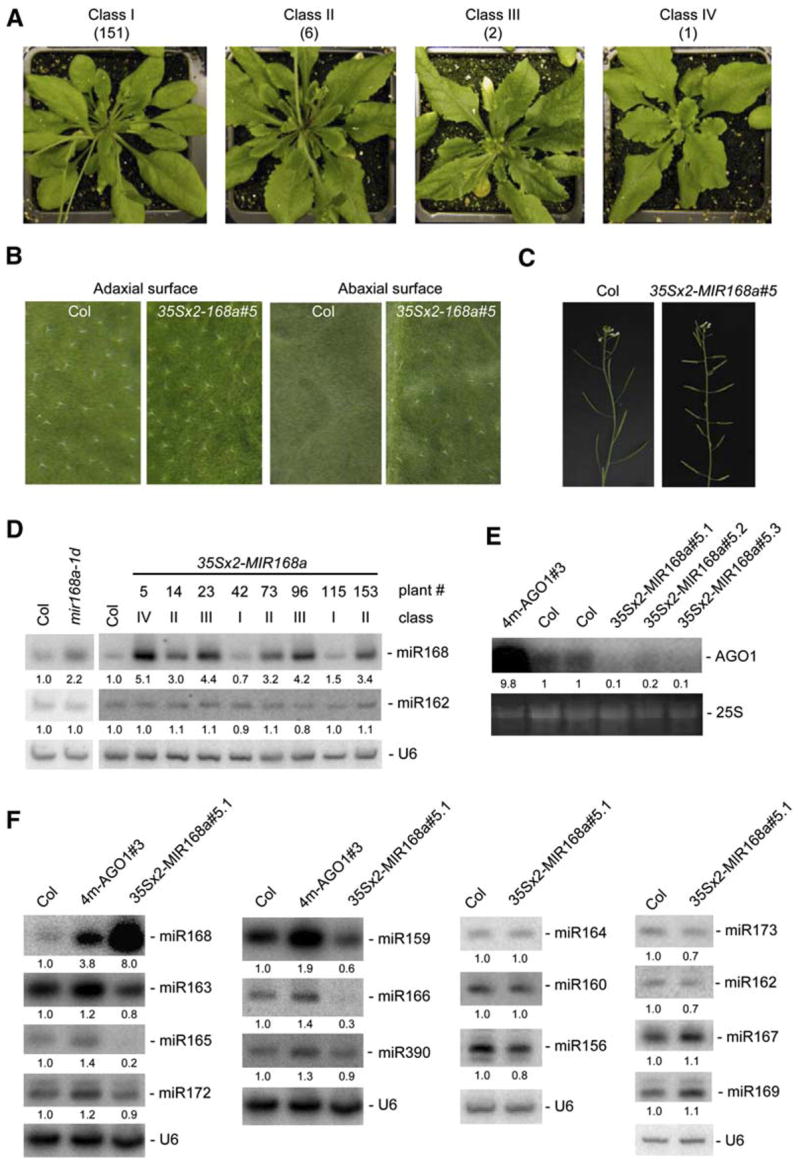
(A) Phenotypes of representative 35Sx2-MIR168 transformants. Among 160 transformants, 151 could not be distinguished from wild-type Col (class I), whereas nine exhibited leaf serration. Six of these nine transformants flowered like Col (class II), two flowered 5–7 days later (class III), and one flowered 12 days later (class IV) than wild-type Col.
(B) Leaf abaxial(lower)andadaxial(upper)surfaces of wild-type Col and 35Sx2-MIR168a transformants. Trichomes are abundant on the adaxial surface but not the abaxial surface of wild-type Col leaves. In contrast, trichomes are abundant on both the adaxial and abaxial surfaces of 35Sx2-MIR168a#5 leaves, indicative of leaf adaxialization.
(C) Altered floral phyllotaxy of 35Sx2-MIR168a transformants. The class IV, 35Sx2-MIR168a#5 plant has siliques that radiate out from the inflorescence stem at altered angles and positions relative to wild-type Col.
(D) miR168 accumulation in 35Sx2-MIR168a transformants and the SALK 113514 line was determined by RNA gel blot analysis using 10 μg of RNA. The phenotypic class and the plant number are indicated above each lane. Blots were successively hybridized to a probe complementary to miR168 and probes complementary to miR162 and U6 as loading controls. Normalized values of miR168 and miR162 to U6 RNA are indicated, with miRNA levels in Col set at 1.0. The structure of the T-DNA in the SALK 113514 line relative to the MIR168a locus is shown in Figure 2A.
(E) AGO1 mRNA accumulation in the 4m-AGO1#3 plant, two independent wild-type plants (Col), and three sibling progenies of the 35Sx2-MIR168a#5 transformant was determined by RNA gel blot analysis using 10 μg of RNA. Ethidium bromide staining of 25S rRNA is shown as a loading control. Normalized values of mRNAs to 25S rRNA are indicated, with mRNA levels in Col control plants set at 1.0.
(F) miRNA and tasiRNA accumulation in wild-type Col, 4m-AGO1#3, and 35Sx2-MIR168a#5 plants was determined by RNA gel blot analysis using 10 μg of RNA. Blots were successively hybridized to different probes complementary to small RNAs and a probe complementary to U6 as a loading control. Normalized values of miRNA to U6 RNA are indicated, with miRNA levels in Col set at 1.0.
The phenotype of 35Sx2-MIR168a transformants likely was due to changes in AGO1 and other miRNA levels. Indeed, the accumulation of miR159 and miR165/166, which was increased in 4m-AGO1 plants, although to a lesser extent than miR168 (Figure 1C), was symmetrically decreased in class IV 35Sx2-MIR168a transformants (Figure 3F). The perturbations of miR159 and miR165/166 accumulation in 35Sx3-MIR168a plants likely are related directly to changes in AGO1 levels and do not result from a general effect of 35S-driven MIR gene expression, because 35S:MIR164b plants, which overaccumulated miR164 (Mallory et al., 2004a), accumulated miR168, miR159, and miR165/166 at wild-type levels (Figure S2). These results confirm that miR159 and miR165/166 are sensitive to perturbations in AGO1 levels, although not as sensitive as miR168.
35Sx2-MIR168a transformants showed leaf adaxialization (Figure 3C), reminiscent of hypomorphic ago1 mutants (Kidner and Martienssen, 2004) and gain-of-function phb, phv, or rev mutants that are insensitive to miR165/166-directed repression, owing to mutations in miR165/166 complementary sites (McConnell et al., 2001; Emery et al., 2003; Tang et al., 2003; Mallory et al., 2004b). This observation suggests that the decrease in miR165/166 accumulation could contribute to the leaf adaxialization in 35Sx2-MIR168a transformants. In addition to decreased miR165/166 accumulation, the decreased AGO1 mRNA levels in 35Sx2-MIR168a transformants (Figure 3E) also may contribute to the adaxialized phenotype, because AGO1 is required for proper regulation of miRNA targets, including PHB and PHV (Kidner and Martienssen, 2004; Vaucheret et al., 2004).
A Posttranscriptional Autoregulatory Loop Model for the Homeostasis of AGO1 Levels
We propose that AGO1 and miR168 participate in a posttranscriptional autoregulatory loop consisting of miR168-guided AGO1-catalyzed cleavage of AGO1 mRNA and AGO1-mediated stabilization of miR168, which allows the maintenance of AGO1 homeostasis and the proper functioning of the miRNA pathway. The observation that miR168, the miRNA that regulates AGO1, is the miRNA most sensitive to altered AGO1 levels (Figure 1C) fits nicely within this scheme, in that minor perturbations in the level of AGO1 protein would induce changes in miR168 activity that in turn bring AGO1 levels back to proper levels before the functions of the other miRNAs are affected. To explain the unusual sensitivity of miR168 to AGO1 levels, we propose that the loading of miR168 duplexes onto AGO1 is very inefficient. As a result, a high proportion of miR168 duplexes produced by DCL1 are not loaded onto AGO1 and are instead degraded. Because of this inefficiency, AGO1 is limiting for formation of the miR168-silencing complex. More AGO1 leads to more loading of miR168; less AGO1 leads to less loading of this miRNA. In contrast, the other miRNAs are loaded more efficiently, such that most are already maximally loaded in the presence of normal AGO1 levels. More AGO1 cannot increase the loading of these miRNAs, and a modest decrease in AGO1 also has little impact.
This model implies that a perturbation in the level of miR168, AGO1 mRNA, or AGO1 protein would induce compensatory adjustments to bring AGO1 levels back to equilibrium. We tested this hypothesis by expressing a wild-type AGO1 mRNA or an AGO1 mRNA perfectly complementary to miR168 (0m-AGO1) in an ago1 null allele (Supplemental Results and Figure S3). AGO1 mRNA accumulated at an ~2-fold lower level in ago1-1::0m-AGO1 plants compared with ago1-1::WT-AGO1 plants, consistent with the ~2-fold increased cleavage rate of 0m-AGO1 RNA observed in vitro in wheat germ extract. miR168 accumulated at an ~2-fold lower level in ago1-1::0m-AGO1 plants compared with ago1-1::WT-AGO1 plants, whereas accumulation of the processed MIR168a loop was similar in the two plants. The decrease in the accumulation of miR168 likely was due to reduced AGO1 mRNA (and AGO1 protein) levels because of the increased cleavage efficiency of 0m-AGO1 mRNA. In this scenario, the new equilibrium between miR168 and AGO1 levels maintains miRNA homeostasis and proper RISC functioning, consistent with the absence of developmental defects in ago1-1::0m-AGO1 plants, supporting our posttranscriptional autoregulatory loop model.
Irreversible disruption of this loop in 35S-MIR168 or 4m-AGO1 plants prevents the maintenance of the equilibrium between AGO1 and miR168 levels. For example, 4m-AGO1 plants have higher levels of AGO1 protein, which can bind a higher amount of miR168 produced by DCL1, thus protecting more miR168 from degradation. However, this elevated level of miR168 cannot downregulate the level of AGO1 mRNA and AGO1 protein because 4m-AGO1 plants express an AGO1 mRNA that is resistant to miR168-guided cleavage. With the exception of miR165/166 and, to a lesser extent, miR159, which seem to have limited loading efficiency, similar to miR168, levels of other miRNAs are affected marginally in 4m-AGO1 plants because they are already maximally loaded onto AGO1 in wild-type cells. 35Sx2-MIR168a and mir168a-1d plants, which both overaccumulate miR168 and miR168* due to increased transcription, likely have much higher levels of miR168 duplexes. The association of more miR168 duplexes with AGO1 would lead to increased miR168 accumulation, decreased AGO1 accumulation, and reduced accumulation of miR159 and miR165/166, which are the miR-NAs that, apart from miR168, are the most sensitive to decreased AGO1 levels in ago1 mutants (Vaucheret et al., 2004) and elevated AGO1 levels in 4m-AGO1 plants (Figures 1C and 3F).
Our model provides a plausible explanation as to why the miRNAs that were most sensitive to changes in AGO1 levels appeared to also be least sensitive to decreases in DCL1 activity. We suggest that the processing of all miRNAs, including miR168, is substantially compromised in the hypomorphic dcl1 mutants. As a consequence, there are far fewer miRNAs to be loaded into AGO1, and the amount of free AGO1 greatly increases. Because AGO1 is limiting for loading of miR168, the increase in free AGO1 offsets the lower amount of processed miR168; although there is much less processed miR168 in the dcl1 hypomorphs, a higher fraction of the miR168 that is produced is loaded into AGO1. The same would apply for miR159 and miR165/166 but to a lesser extent because these miRNAs are less sensitive to AGO1 levels. The other miRNAs, which are already maximally loaded in the presence of normal AGO1 levels, do not benefit from increased free AGO1; their levels drop in the dcl1 hypomorphs without a counteracting effect of free AGO1.
AGO1 and MIR168 Genes Have a Common Expression Pattern
The posttranscriptional regulatory loop proposed above implies that miR168 might be present in all cells expressing AGO1. To test this hypothesis, we compared the patterns of a GUS reporter transgene expression in plants when driven by the AGO1 versus MIR168a promoters. GUS expression in both pAGO1-GUS and p168a-GUS plants was highest in the shoot and root apical meristems, but, in transformants that showed the strongest GUS expression in the meristem, GUS expression also was detectable in vascular tissues of leaves and roots (Figure 4A and data not shown).
Figure 4. AGO1 and MIR168a Promoters Exhibit Similar Transcriptional Regulation, and Expression of the AGO1 cDNA under the Control of the MIR168a Promoter Rescues the Null ago1-1 Allele.

(A) Histochemical staining of representative transformants carrying the GUS reporter transgene expressed under the control of the AGO1 (pAGO1-GUS) or MIR168a promoter (p168a-GUS).
(B) Phenotype of a wild-type plant (Col), a null ago1 allele (ago1-1), and a representative ago1-1 transformant carrying the pMIR168a-AGO1 construct (ago1-1::p168a-AGO1).
AGO1 Expressed under the Control of the MIR168a Promoter Rescues ago1 Mutants
Our expression analysis of AGO1 and MIR168a promoters suggests that AGO1 and MIR168 genes are coregulated transcriptionally to ensure proper functioning of the miRNA pathway. However, other regions of these genes may have regulatory roles that could differentiate the expression of their final products. We tested further the coexpression hypothesis by analyzing transgenic ago1 mutants that expressed a wild-type copy of the AGO1 mRNA under the control of the MIR168a promoter. To this end, a full-length AGO1 cDNA was cloned downstream of the MIR168a promoter, and the resulting construct (p168a-AGO1) was introduced into the null ago1-1 allele. A construct consisting of the AGO1 cDNA with no promoter (noPro-AGO1) was used as a negative control. Only the p168a-AGO1 construct restored development and fertility (Figure 4B) and accumulation of ta-siRNAs (Figure S4), which depend on miRNA-guided AGO1 slicer activity for their production (Vazquez et al., 2004; Allen et al., 2005). These results indicate that miR168 is expressed everywhere that AGO1 is needed and that coexpressing AGO1 and miR168 in the same cell types is compatible with proper plant development.
Concluding Remarks
Some miRNAs have been proposed to clear regulatory transcripts from previous developmental stages, thereby facilitating a more rapid and robust transition to a new expression program (Rhoades et al., 2002). Supporting this idea, in situ hybridizations have shown that the accumulation patterns of miR165 and miR172 are complementary to those of their respective targets, PHB and AP2 (Chen, 2004; Kidner and Martienssen, 2004). Also supporting this idea, expression of plant miRNAs inversely correlates with expression of their targets (Axtell and Bartel, 2005). Here we show an example of a different paradigm: a miRNA (miR168) and its target mRNA (AGO1) that are transcriptionally coregulated in time and space. Furthermore, this target regulates the level of the miRNA in addition to being regulated by it. This fine-tuned adjustment of AGO1 and miR168 levels corresponds to a posttranscriptional autoregulatory loop that maintains a proper balance of the miRNA pool and AGO1 levels, which together control the mRNA levels of all miRNA targets. We suggest that modulating the efficiency of assembling miRNA-programmed silencing complexes will also be important in other contexts, as will the regulation of miRNAs by their own targets.
Experimental Procedures
Plant Material
The ago1-1 mutant has been described before (Bohmert et al., 1998). The Salk Institute Genomic Analysis Laboratory generated the sequence-indexed T-DNA insertion line SALK_113514 (Alonso et al., 2003), and the Arabidopsis Biological Resource Center at Ohio State University supplied seeds.
Molecular Cloning, Plant Transformation, and In Vitro Cleavage Assay
The 4m-AGO1 and 4m-MIR168a constructs deriving from the AGO1 and MIR168a genes, respectively, have been described before (Vaucheret et al., 2004).
The 35Sx2-MIR168a construct was made as follows: the MIR168a region corresponding to EST H77185 was amplified as a 415 nt fragment (position 56590–57005 on BAC T5K18) using a forward primer that introduces a HindIII site (5′-GCGCGCAAGCTTCTCTCTTCTCT TTCTTCATATCCC-3′) and a reverse primer that introduces an EcoRI site (5′-GCGCGCGAATTCAACATTTGGGCAAACAAAAGGAGAC-3′) and cloned between the HindIII and EcoRI sites in the pLBR19 vector. The p35S-MIR168a-t35S fragment was excised as a KpnI-XbaI and cloned between the KpnI and XbaI sites in the pBin+ binary vector.
The 0m-AGO1 cDNA and 0m-AGO1 genomic construct were made as follows: mutations were introduced in the miR168 complementary site of the AGO1 cDNA (Bohmert et al., 1998) or of a Pac1-Kpn1 fragment corresponding to the 5′ half of the AGO1 gene (Vaucheret et al., 2004) using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) using the following primer pair: 5′-CCGCA GAGACAATCAGTTCCCGACCTGCACCAAGCGACCTCACCTACTT ATCAAGCGG-3′ and 5′-CCGCTTGATAAGTAGGTGAGGTCGCTTG GTGCAGGTCGGGAACTGATTGTCTCTGCGG-3′. The wild-type and mutagenized DNAs were sequenced to ensure that no other mutations had been introduced. The mutagenized Pac1-Kpn1 fragment was transferred from pKS+ into the pBin+ vector containing the 3′ half of the gene to reconstitute a complete AGO1 gene (Vaucheret et al., 2004).
The construct pAGO1-GUS was a translational fusion with 2000 nt of the AGO1 promoter and 678 nt of the AGO1 pre-mRNA, which included the 5′ UTR intron. The AGO1 fragment was excised from the wild-type AGO1 gene as a Xho-PpuM1 fragment, blunt ended at the PpuM1 sites, and cloned between the Sal1 and SmaI sites in the pBI101.2 binary vector.
The construct p168a-GUS was a transcriptional fusion with a 1339 bp fragment of the MIR168a promoter (position −1358 to −19 relative to the beginning of the hairpin); positions were chosen so as to exclude an ATG located at position −15 that could compromise the use of the GUS reporter initiation codon and to keep the entire promoter region previously used to express the compensatory miRNA 4m-miR168 from a 4m-MIR168a transgene, which rescued the 4m-AGO1 phenotype (Vaucheret et al., 2004). The MIR168a promoter was amplified from the 4m-MIR168a construct in pKS+ using a universal forward primer and a reverse primer that introduces a BamHI site (5′-GGATCCCTTTAGACAGGGATATGAAGAAAGA-3′), blunt ended, cut with BamHI, and cloned between the BamHI and bluntended HindIII sites in the pBI101.2 binary vector.
The p168a-AGO1 construct was made as follows: the MIR168a promoter was excised from the p168a-GUS construct and cloned between the SmaI and BamHI sites in the pBin+ binary vector. Then the AGO1 cDNA was excised from pZL1 (Bohmert et al., 1998) and cloned between the Sal1 and HindIII sites in the pBin+ binary vector.
The 4m-AGO1, 0m-AGO1, pAGO1-GUS, p168a-GUS, p168a-AGO1, and 35S-MIR168a constructs were transferred to Agrobacterium tumefaciens by triparental mating, and Arabidopsis plants were transformed using the floral dip method (Bechtold and Pelletier, 1998; Clough and Bent, 1998). Collected seeds were surface sterilized and plated on a medium supplemented with 50 μg/ml kanamycin for selecting transformants. Plants were grown under cool white light in long days (16 hr of light, 8 hr of dark) at 23ºC.
AGO1 and 0m-AGO1 RNAs containing the miR168 complementary site were generated by linearizing the pZL1 vector containing the AGO1 or 0m-AGO1 cDNA (Bohmert et al., 1998) with SacI and transcribing with T7 RNA polymerase. Wheat germ lysate preparation, cap labeling, and in vitro cleavage assays were performed as described (Mallory et al., 2004b).
RNA Analysis
Total RNA was isolated from rosette leaves as described (Vaucheret et al., 2004). RNA gel blot analysis of low molecular weight RNAs was carried out on denaturing 15% polyacrylamide gels followed by blotting to a nylon membrane (Genescreen Plus, PerkinElmer Inc.) as described (Vaucheret et al., 2004). Blots were hybridized with gamma-ATP 32P end-labeled oligonucleotides. Blots were rehybridized with an end-labeled oligonucleotide probe complementary to U6. For mRNA gel blot analysis, RNA was separated on denaturing 1% agarose gels and transferred to nylon membrane (Genescreen Plus, PerkinElmer Inc.). Blots were hybridized with an alpha-UTP 32P-labeled RNA probe complementary to AGO1 mRNA in ULTRAhyb buffer at 68ºC (Ambion). AGO1 RNA probes were generated by PCR using primers 5′-CTACAGGGATGGAGTCAGTGAGGGAC-3′, 5′-GGCCGTAATACGACTCACTATAGGCTCCCACTAGCCATTGAGCCACTG-3′ followed by T7-mediated in vitro transcription.
Supplementary Material
Acknowledgments
We thank G. Tang and P. Zamore for wheat germ extracts, B. Bartel and D. Dugas for 35S:MIR164b RNA, and V. Gasciolli for homozygous seeds of line mir168a-1d. The Salk Institute Genomic Analysis Laboratory generated the sequence-indexed T-DNA insertion lines, and the Arabidopsis Biological Resource Center at Ohio State University supplied seeds. We also thank members of the Vaucheret and Bartel labs for stimulating discussions. This work was partly supported by a grant from the European Commission (Riboreg program) to H.V. and by NIH grants (to D.P.B. and A.C.M.). D.P.B. is an HHMI investigator.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. micro-RNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGO-NAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APE-TALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DB, Bartel B. Micro-RNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004a;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5 ′ region. EMBO J. 2004b;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mlotshwa S, Schauer SE, Smith TH, Mallory AC, Herr JM, Jr, Roth B, Merchant DS, Ray A, Bowman LH, Vance VB. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell. 2005;17:2873–2885. doi: 10.1105/tpc.105.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in micro-RNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M, Reinhart B, Lim L, Burge C, Bartel B, Bartel D. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.