Global regulatory logic for specification of an embryonic cell lineage (original) (raw)
Abstract
Explanation of a process of development must ultimately be couched in the terms of the genomic regulatory code. Specification of an embryonic cell lineage is driven by a network of interactions among genes encoding transcription factors. Here, we present the gene regulatory network (GRN) that directs the specification of the skeletogenic micromere lineage of the sea urchin embryo. The GRN now includes all regulatory genes expressed in this lineage up to late blastula stage, as identified in a genomewide survey. The architecture of the GRN was established by a large-scale perturbation analysis in which the expression of each gene in the GRN was cut off by use of morpholinos, and the effects on all other genes were measured quantitatively. Several _cis_-regulatory analyses provided additional evidence. The explanatory power of the GRN suffices to provide a causal explanation for all observable developmental functions of the micromere lineage during the specification period. These functions are: (i) initial acquisition of identity through transcriptional interpretation of localized maternal cues; (ii) activation of specific regulatory genes by use of a double negative gate; (iii) dynamic stabilization of the regulatory state by activation of a feedback subcircuit; (iv) exclusion of alternative regulatory states; (v) presentation of a signal required by the micromeres themselves and of two different signals required for development of adjacent endomesodermal lineages; and (vi) lineage-specific activation of batteries of skeletogenic genes. The GRN precisely predicts gene expression responses and provides a coherent explanation of the biology of specification.
Keywords: gene regulatory networks, network subcircuits, sea urchin embryo, skeletogenic micromeres
Developmental gene regulatory networks (GRNs) are models that explain the causal sequence of combinatorial interactions among genes encoding transcription and signaling factors. The architecture of a GRN gives the map of functional interactions among these genes and provides a direct guide to the regulatory logic of developmental control (1). Many regulatory genes are required to program the specification and differentiation of a given embryonic cell lineage or the progressive organization of any given part of an animal embryo. When mature, a GRN should indicate the causal _cis_-regulatory transactions at the relevant modular control elements of all of the genes in the network. The architecture of a GRN model can thus be experimentally authenticated by direct _cis_-regulatory analysis (2). GRNs explain developmental phenomenology at the system level, by reference to its source, the genomic control apparatus. It follows that, in principle, a GRN should explicitly show why all aspects of a developmental process occur the way they do.
Here we test this claim. For several years we have been assembling and authenticating at the _cis_-regulatory level a GRN for endomesoderm specification in the sea urchin embryo, from earliest cleavage to just before gastrulation (1–3). The endomesoderm comprises the endodermal cell types of the vegetal plate that give rise to the gut of the embryo; the vegetal plate mesoderm, which differentiates into pigment cells and several other cell types of the late embryo; and the skeletogenic mesenchyme. In modern sea urchins the skeletogenic mesenchyme stems from a specific lineage deriving from four fifth cleavage founder cells, known as the “large micromeres.” This article is focused on that portion of the overall GRN that refers to the specification and differentiation of the skeletogenic micromere lineage, as it is this domain of the overall GRN that contains the most nearly complete population of relevant regulatory genes.
In the course of the Strongylocentrotus purpuratus genome project (4), all gene models that included DNA recognition domains, i.e., which predicted genes encoding transcription factors, were studied experimentally (5–9). It was determined by quantitative PCR (QPCR) whether the gene is expressed in embryogenesis. The spatial domains of all regulatory genes expressed at possibly significant levels in the period up to late-gastrula (48 h in S. purpuratus) were then investigated by whole-mount in situ hybridization (WMISH). Every known regulatory gene expressed specifically in the skeletogenic micromere lineage has now been incorporated into the portion of the GRN that pertains to this lineage. Thus, if the GRN indeed states the roles of all of the regulatory players involved in skeletogenic micromere lineage specification, then it should be capable of providing us with a qualitative explanation for all of the functions these cells execute during this period of development.
The specification of an embryonic cell lineage is traditionally defined as the process by which it achieves its developmental identity. In mechanistic terms specification is the acquisition of a given regulatory state (2), where regulatory state is the sum of the activities of the transcription factors expressed in the cell nuclei. Therefore, at root the process of specification depends on the regulatory activation (and repression) of genes encoding transcription factors, which is why a GRN may provide a direct explanation of a specification event at the genomic sequence level. However, specification is not a one-step process. For a cell lineage arising very early in embryogenesis, the initial function that must be executed is transcriptional interpretation of whatever regulatory cues are spatially inherited in the portion of the egg cytoplasm, which is incorporated by the lineage founder cells; somehow these cues must be transduced into regulatory gene expression. Then this initial state, which is always transient, has to be expanded and stabilized. Signaling genes must be activated, for no embryonic cell lineage develops silently with respect to its neighbors, and in the case of the skeletogenic micromere lineage, the signals it emits early in development are crucial to the development of these neighboring lineages. And as the phase of specification ends, the regulatory drivers of the differentiation gene batteries that the lineage will express must be brought into play, and these downstream genes activated.
In the following, we first recapitulate the developmental features of the skeletogenic micromere lineage in the process of specification, as it is these features that require causal explanation, and then, one by one, address the underlying genomic control functions. This study does not include the cytoarchitectural organization of the egg, which is set up before the zygotic genomic apparatus begins to operate, nor processes that take place from gastrulation on, after the end of the period to which our GRN refers.
Results
The Skeletogenic Micromere Lineage.
The external or descriptive biology of the skeletogenic micromere lineage, and of skeletogenesis in the sea urchin embryo, have been the subject of many detailed reviews (10–12). Here, we touch only on the salient aspects of the pregastrular specification of this lineage. The micromere lineage arises as follows: the first two cleavages are vertical and orthogonal, the third horizontal and equatorial, but the fourth, which is also horizontal, is unequal in the vegetal (lower) half. The polar micromeres collectively contain ≈8% of the egg volume, their sister cells (macromeres) contain ≈42%, and the animal cells (mesomeres) contain the remaining 50%. The unequal position of the cleavage plane producing the micromeres depends on structural features of the egg cortex in the lower quadrant of the egg (13). The fifth cleavage is also unequal, producing four “small micromeres” and the four large micromeres (Fig. 1A). The small micromeres play no further role in embryonic development. They are quiescent, pluripotential cells, the descendants of which replicate extensively only after embryogenesis is complete, when they contribute to multiple components of the adult body plan (11). From early on their regulatory state is different from that of their sister cells, the large micromeres (Fig. 1B). In S. purpuratus the skeletogenic large micromeres divide twice more while residing in the vegetal plate, and then after few hours, before invagination of the gut, they individually ingress into the blastocoel. They divide once more, and after gastrulation all 32 of the micromere descendants participate in skeletogenesis. They express a distinct suite of genes throughout and carry out no embryological functions other than skeletogenesis. The GRN pertains to the time from the appearance of the fourth-cleavage micromeres to their ingression.
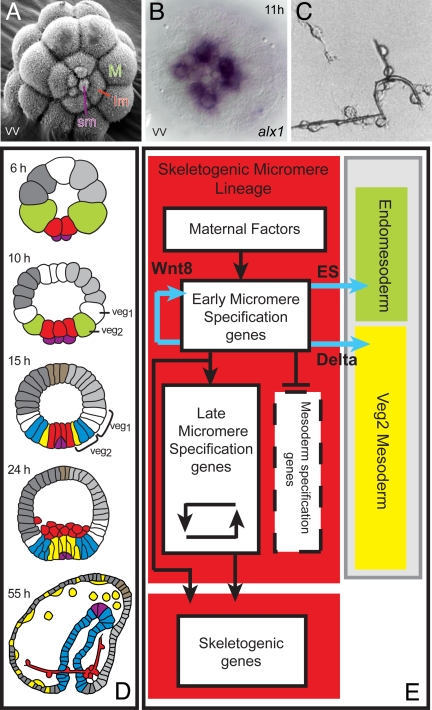
Fig. 1.
Regulatory specificity and specification functions of the skeletogenic micromere lineage. (A) SEM image of fifth-cleavage embryo viewed from vegetal pole (vv), displaying small micromeres (sm), large micromeres (lm), and macromeres (M) (photograph by J. B. Morrill and L. Marcus, 2005). The embryos in this and all following figures are ≈70 μm in diameter. (B) Distinct regulatory state in large and small micromeres: expression of alx1 at midcleavage stage in large micromeres only. (C) Synthesis of calcite biomineral skeletal rods in vitro by descendants of isolated micromeres cultured in sea water with 2% horse serum. [Reproduced with permission from ref. 48 (Copyright 1991, Japanese Society of Developmental Biologists).] (D) Territorial components of the sea urchin embryo in lateral view: green, macromeres; red, skeletogenic micromere lineage; purple, small micromeres; yellow, nonskeletogenic mesoderm; blue, gut endoderm; brown, apical neurogenic territory; dark gray, aboral ectoderm; light gray, oral ectoderm. Stages are: 6 h, fifth cleavage; 10 h, seventh cleavage; 15 h, early blastula; 24 h, mesenchyme blastula showing skeletogenic micromere lineage ingressed; 55 h, late gastrula with forming skeleton. (E) Process diagram (4), summarizing specification functions. Color coding of background is as in D; signal ligands produced by skeletogenic micromere lineage cells are in blue, transcriptional regulatory functions are in black.
Two classical experiments foreshadow our analysis. In 1936 Hörstadius (10) showed that if micromeres are transplanted to the top of the embryo they induce the adjacent presumptive ectodermal cells to instead form endomesoderm, including a second gut. This experiment, which has been repeated with molecular markers (14), means that micromeres execute signaling functions that have inductive effects on adjacent cells. We now know that between fourth and ninth cleavage (5–12 h after fertilization) the micromeres in fact express three different intercellular signaling ligands: (i) Wnt8, which enhances nuclearization of β-catenin in recipient cells, including themselves (15–17); (ii) a still undefined “early signal (ES),” which is received by adjacent cells in the fourth- to sixth-cleavage interval and is required by them for the normal process of endomesoderm specification (18); and (iii) from seventh cleavage, the Notch ligand Delta. Reception of this signal causes the ring of cells then immediately adjacent to the micromere lineage to assume mesodermal fate (19, 20).
A second prescient experiment was done by Okazaki (21), who showed that if fourth-cleavage micromeres are isolated and cultured, they proceed to divide the proper set number of times and then produce biomineral skeletal rods in vitro (Fig. 1C). The skeletogenic micromeres thus contain whatever regulatory inputs are required to cause their autonomous specification. The genes encoding the biomineralization and cell function proteins that are exclusively expressed in the skeletogenic micromere lineage began to be identified in the 1980s (for complete repertoire, see ref. 22). Using them as developmental probes, it became apparent that many of these genes are turned on even while the cells of the skeletogenic lineage reside within the vegetal plate, before ingression and any overt skeletogenesis. Combined with Okazaki's experiment, these results indicated that the autonomous capabilities of the skeletogenic micromere lineage extend all of the way down to the activation of batteries of differentiation genes.
In Fig. 1D the disposition of the skeletogenic micromere lineage with respect to the remainder of the embryo can be seen, which is relevant in particular to the targets of the signals expressed by the micromere lineage. In Fig. 1E is what we term a “process diagram” (1) for the specification of this lineage. It enables us to begin to think mechanistically about the organization of the GRN directing specification. The autonomy of specification requires the existence of maternal factors of some kind that can affect transcriptional expression, and each of the other white boxes in the process diagram should contain a population of control genes that execute the functions indicated by the blue arrows (signal expression), the black arrows (direct transcriptional activation), and the barred stem (transcriptional repression) in Fig. 1E.
Solving the Network.
By focusing on regulatory genes expressed specifically in the emerging micromere lineage we focus on the immediate generators of the lineage regulatory state. If we can include all of these, then all functions dependent on the regulatory state as it changes should become accessible. Two concerns are that in the genomewide analysis that formed the background to this work we might have missed some key regulatory players, if these are expressed at very low levels; and that by studying only specifically expressed genes, we are excluding ubiquitously expressed factors that may nonetheless contribute to the process. However, the threshold of significance set for transcript level in the global analyses of regulatory gene expression cited above were conservative (≈150 molecules per embryo), and if even these small amounts were localized to the micromeres they should be visualized by WMISH. Therefore, we believe that the only specifically expressed micromere lineage regulators that are likely to be missing from our study would be ones that encode transcription factors with yet uncharacterized DNA binding domains, an increasingly rare class. Ubiquitously expressed factors indeed contribute to the specific quantitative performance of individual _cis_-regulatory modules, as shown by studies that approach the function of every known transcription factor target site (e.g., ref. 23). But those factors that are zygotically expressed in a ubiquitous manner are not likely to cause lineage-specific decisions to occur. Indeed one such factor is included in the micromere lineage GRN (hnf6) and its ancillary “booster” function is illustrative.
Two inputs at the very beginning of the process of micromere lineage specification provided an experimental lever useful for distinguishing genes that genuinely participate in this lineage. All such genes, for reasons that will become apparent in the next section, (i) must be shut down by interference with nuclearization of β-catenin [by injection of a truncated dominant negative form of _cadherin_ mRNA, Δ-_cadherin_ (24)]; and (ii) must be ectopically activated by injection of mRNA encoding the Pmar1 repressor (25). Both methods were used in screens for regulatory genes that contribute to micromere lineage specification. A complete list of these regulatory genes is in supporting information (SI) Fig. S1.
The temporal sequence of gene activations and the forms of the mRNA accumulation time courses contain important information, and each was measured by QPCR at high resolution (Fig. S2). As a general guide, in S. purpuratus embryos at 15°C the step time between activation of a regulatory gene and activation of its target gene is on the order of 2–3 h (26). The most significant source of insight into GRN architecture came from perturbation of regulatory gene expression by injection into the egg of gene-specific morpholino antisense oligonucleotides (morpholinos). In sea urchin embryos this method provides a specific and effective means of knocking down gene expression that is particularly suitable for large-scale, systemwide perturbation studies (3). The effects of perturbations of gene expression were systematically measured by QPCR assessment of alterations in levels of transcripts of all other genes in the GRN, at various developmental times, and when warranted, by WMISH (perturbation data are listed in Fig. S3). Several _cis_-regulatory studies on important genes in the GRN also contributed at key nodes, as discussed in the following. Morphological phenotypes are in our experience poor guides to the roles of regulatory genes: they show up late and are interpretable only ex post facto, after the architecture of the GRN is known.
The Genomic Program for Initial Specification of the Micromere Lineage.
There are at least three molecular features of regulatory significance that are particular to the fourth-cleavage micromeres as soon as they are born. The GRN shows that these spatially anisotropic features are used as inputs to set up the initial transcriptional regulatory state of the micromeres. During oogenesis the docking protein Disheveled (Dsh) is localized at the vegetal cortex of the oocyte (ref. 13 and Fig. S4) and it has been shown that this causes β-catenin nuclearization in the prospective endomesoderm founder cells (micromeres and macromeres). Dsh regionally prevents the degradation of cytoplasmic β-catenin (27), which transits to the nucleus, forming an active complex with the Tcf transcription factor. The β-catenin transcriptional cofactor is first localized in the nuclei of micromeres, immediately at the fourth-cleavage stage, constituting the initial unique character state of these cells. Concomitantly, the maternal transcription factor SoxB1 enters the nuclei of all early cleavage blastomeres except the micromeres (ref. 28 and Fig. S4). The relevance of this, a second micromere-specific character state, is that SoxB1 is believed to act as an antagonist of the transcriptional cofactor function of β-catenin. A third micromere-specific anisotropy is the precocious nuclearization in fourth-cleavage micromeres of another maternal transcription factor, Otx (ref. 29 and Fig. S4).
The GRN subcircuit shown in Fig. 2A explains how these transient, anisotropic character states are transduced into a lineage-specific regulatory state. The first genes in the sequence are the pmar1 genes (there is a cluster of several very similar pmar1 genes). The pmar1 genes are activated by the β-catenin/Tcf transcription complex plus Otx, i.e., by two of the micromere-specific inputs just enumerated (25, 30). Expression of pmar1 in the micromere lineage (Fig. 2B) is specific and transient, detectable initially right at late fourth cleavage, and gone 12–15 h later. Furthermore, it was shown in the following way that although many genes in the endomesoderm respond to β-catenin/Tcf, the pmar1 genes alone are responsible for the first step of micromere specification and indeed are sufficient to cause it. Blocking β-catenin nuclearization by injection of Δ-cadherin mRNA blocks micromere specification (Fig. 2C and refs. 16 and 31). Thus if the normal micromeres are replaced by transplanted Δ-cadherin-expressing micromeres from another embryo, no skeletogenic micromere lineage forms. But skeletogenic micromere lineage specification is entirely rescued if exogenous pmar1 mRNA and Δ-cadherin mRNA are present in the transplanted micromeres (30). This transplantation experiment can even be done successfully with cells from the part of the embryo normally fated to become ectoderm providing they contain pmar1 mRNA. Furthermore, if pmar1 mRNA is made to be present in all cells at the same levels as normally in the micromeres, the whole embryo turns into skeletogenic mesenchyme (ref. 25 and Fig. 2E). So pmar1 is necessary and sufficient; no other β-catenin/Tcf target need be involved.
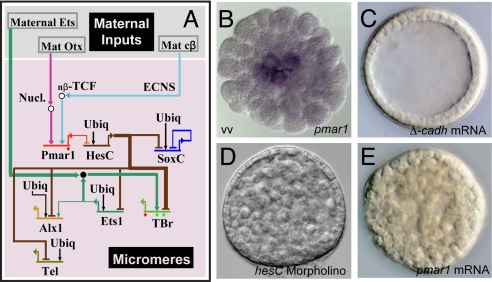
Fig. 2.
Initial regulatory state and circuitry of the double negative gate. (A) Portion of GRN indicating initial inputs to the pmar1 gene, the double negative gate, and the target regulatory genes. The GRN in this and succeeding figures is represented in BioTapestry software (49). Thick lines indicate inputs validated by _cis_-regulatory analysis. ECNS, early cytoplasmic nuclear localization system. (B) WMISH display of pmar1 transcripts in micromeres, midcleavage, (vv, vegetal view). (C) Effect of blocking β-catenin nuclearization by injection of Δ-cadherin mRNA; all endomesodermal specification is blocked, including that of the micromere lineage, as shown earlier by others (16, 31). (D) Transformation of gastrula-stage embryo into solid ball of mesenchyme by ectopic expression of pmar1 mRNA. (E) Transformation of gastrula-stage embryo into solid ball of mesenchyme by use of anti-hesC morpholino.
The factor encoded by the pmar1 genes is a transcriptional repressor (25), and the second element in the subcircuit is the immediate target gene of this repressor. This gene encodes a second repressor, HesC (32). HesC is expressed zygotically early in cleavage, in all cells of the embryo except the micromeres, where Pmar1 prevents its expression. The primary regulatory genes of the skeletogenic micromere specification GRN are subject to direct HesC repression. The mechanism that unlocks the initial micromere regulatory state is therefore a double negative gate (28).
The five primary target regulatory genes of the double negative gate are indicated in Fig. 2A. They are four essential regulators of downstream micromere lineage function: alx1 (29), ets1 (9, 33), tbr (34, 35), and tel (9), plus SoxC, which in this lineage plays no role as it turns itself off shortly after activation. The expression of all these genes relies on ubiquitous activator inputs, but they are expressed specifically in micromeres and silent everywhere else because they are subject to direct HesC repression. Therefore, if either hesC expression is blocked or pmar1 is globally expressed, the target genes of the double negative gate are activated all over the embryo (25, 32). The consequence is to turn the whole embryo into mesenchymal cells expressing genes normally activated in the skeletogenic micromere lineage (Fig. 2 D and E). The key feature of a double negative gate is that it achieves its end in two ways: it promotes expression where it works and actively prevents it everywhere else. The GRN subcircuit shown in Fig. 2A takes the embryo from the initial anisotropies of the newly born micromeres, to the expression of a new, lineage-specific regulatory state.
The Genomic Program for Progression and Stabilization of the Regulatory State.
It is now a predictable feature of GRNs for developmental specification processes that the initial transcriptional functions by which a regulatory state is initiated are transient, and that the next portion of the regulatory apparatus to be deployed will include a dynamic state stabilization device (2). By this is meant a transcriptional feedback subcircuit that functions to lock the specification in a regulatory embrace, so to speak, in that its components drive one another's expression. This feature is present in exactly the expected place in the GRN architecture, as shown in Fig. 3A. Here, we see that immediately downstream of the initial four regulatory genes activated by the double negative gate (Fig. 2A), three additional genes are activated: one encoding the Ets family factor Erg (9), and the other two, hex and tgif, encoding homeobox factors (5). These genes are engaged in interlocking positive double feedback loops (erg with hex; and hex with tgif). Interference with expression of any of these genes severely depresses transcript levels of the others (Fig. S3). Thus they dynamically stabilize all aspects of the regulatory state downstream of themselves. In addition to tgif, the outputs of hex go to erg, to differentiation genes as we see below, and to foxo, a late activated regulatory gene the functions of which occur beyond the time frame of this GRN. In addition to hex, the outputs of tgif go back to the key regulatory gene alx1. This gene is also a cross-regulatory target of the ets1 gene, among the initial double negative gate genes, and ets1 expression may lock itself on by autoactivation. The tbr and alx1 genes also provide inputs into the additional skeletogenic differentiation genes dri and foxb. The proper function of most of these regulatory genes is essential for expression of skeletogenic differentiation genes, and interference with their expression produces a skeleton-minus phenotype, as illustrated in Fig. 3 B–G for tbr, hex, tgif, ets1, and alx1. Additional morphological effects are also observed for some genes, for easily understood reasons: ets1 also is later expressed in nonskeletogenic mesoderm, and both it and alx1 target genes are required for ingression of the skeletogenic cells (ref. 36 and Fig. 3 F and G). This is not the case for tbr, hex, or tgif: interference with their expression results in embryos with a full complement of ingressed cells of the micromere lineage but these cells are unable to create the biomineral skeleton. (Fig. 3 C–E).
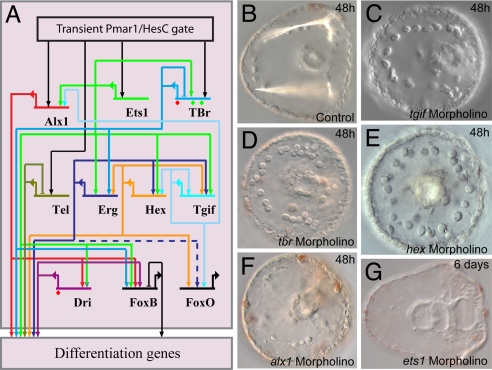
Fig. 3.
State stabilization circuitry. (A) Portion of GRN displaying feedback circuitry and linkages among the regulatory genes that comprise the climax regulatory state of the skeletogenic micromere lineage during its specification period. Dashed line into foxo indicates the input from erg, which although supported by QPCR data, could be indirect, via other parallel linkages shown in the GRN. (B) Control late gastrula embryo displaying birefringent skeletal rods. (C–G) Failure of skeletogenesis when expression of the indicated regulatory genes is blocked [as also shown earlier for _tbr_ (35) and alx1 (29)].
The Genomic Program for Expression of Intercellular Signals.
As indicated in Fig. 1, the micromere lineage expresses three distinct signals, all essential for development. The GRN provides a direct explanation for why each of these is expressed (Fig. 4A).
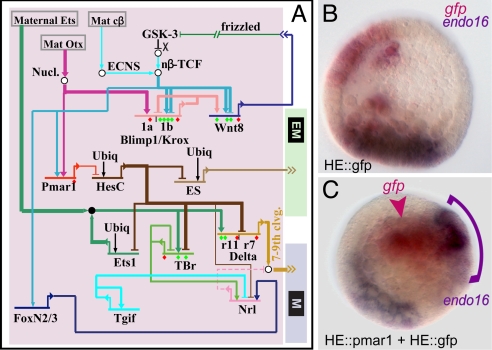
Fig. 4.
Control of signaling functions by the double negative gate. (A) Portion of the GRN displaying regulatory circuitry by which expression of signals (Wnt8, ES, Delta) is controlled in the micromere lineage. (B and C) Demonstration using double WMISH that the ES signal is a target of the double negative gate. (B) Control blastula stage embryo in which endogenous endo16 expression is displayed in purple in the vegetal plate and GFP mRNA produced by a construct under control of a hatching enzyme (HE) _cis_-regulatory module is shown in red, in the ectoderm. (C) Embryo bearing two _cis_-regulatory constructs under HE _cis_-regulatory control, one expressing pmar1, and the other expressing GFP, which marks the location of the ectopic pmar1 expression. The constructs are concatenated together in the egg and are incorporated together into the same cells. Ectopic endo16 transcript (purple arch) can be seen adjacent to the cells expressing gfp and pmar1 mRNA (red arrowhead), evidently in response to the ectopic production of the ES. [Reproduced with permission from ref. 30 (Copyright 2003, Elsevier).]
The first signal is Wnt8. By late fourth-cleavage stage the micromeres begin to transcribe the wnt8 gene, and both the network perturbation analysis and a direct _cis_-regulatory study (15, 37) demonstrate that the inputs required for its expression in the micromere lineage are Blimp1 and β-catenin/Tcf. To initiate wnt8 expression, the micromeres initially rely on the maternal β-catenin localization system, which is activated precociously in these cells, as discussed above. However, β-catenin/Tcf is itself a product of the signal transduction system driven by reception of the Wnt8 signal. Therefore, an intercellular feedback circuit is set up, as each _wnt8_-expressing cell also causes the adjacent recipient cells to drive more β-catenin into its nucleus and further express wnt8; in S. purpuratus for ≈7 h all cells of the micromere lineage both send and receive this signal. Then its expression in the micromere lineage is shut down. As discussed in detail in ref. 15, this is because the Blimp1 input into the wnt8 gene disappears due to blimp1 autorepression. The blimp1 gene plays no other essential early role in the skeletogenic micromere lineage other than to provide input into the wnt8 gene.
The second signal is the yet undefined ES. The existence of this signal is revealed by the ability of micromeres when transplanted to induce a second endomesodermal axis, and by the failure of normal endomesodermal development when micromeres are removed, providing that operation is done before sixth cleavage (18). As shown in Fig. 4A, the gene encoding the ES is under control of the double negative gate. This linkage is demonstrated by the experiment reproduced in Fig. 4 B and C (30), which shows that localized ectopic expression of pmar1 causes adjacent cells to produce endo16 mRNA, which is an endomesoderm-specific marker, in consequence of having received an ectopic ES input. Furthermore, transplanted micromeres or mesomeres bearing Δ-cadherin plus pmar1 mRNA, as above, can also induce a second gut (30). Therefore, production of the ES is also controlled by the double negative gate.
Expression of the third signal, the Notch ligand Delta, is also controlled by the double negative gate. As predicted by the circuitry in Fig. 4A, the relevant _cis_-regulatory module of the delta gene responds to global pmar1 expression by driving ectopic reporter expression (2), and the gene behaves in the same way in embryos bearing hesC morpholino (32). The requirement of this circuitry is that the delta gene be turned on by a ubiquitous activator to account for its ubiquitous expression when the pmar1-hesC double negative gate is circumvented, as in these last experiments. This ubiquitous input turns out to be an Ets factor (unpublished data). Although zygotically expressed in the skeletogenic micromere lineage because it is one of the double negative gate targets, the ets1 gene is also maternally expressed, and WMISH shows that in cleavage-blastula-stage embryos the maternal transcript is present everywhere. The ancillary signaling gene neuralized (nrl), which may control the level of the Delta ligand (38), is also under control of the double negative gate. No regulatory apparatus further downstream of the double negative gate affects any of these signals.
To step back from these detailed aspects of mechanism for a moment, we have here three examples of direct genomic regulatory control of developmental signaling functions, two of which (wnt8 and delta) have been confirmed by mutational _cis_-regulatory demonstration (for review see ref. 2). These are essential functions: blockade of wnt8 expression and disruption of the wnt8 intercellular feedback loop plays havoc with endomesodermal specification, including that of the skeletogenic micromere lineage (2, 15); and blockade of the micromere lineage expression of delta or of the reception and transduction of this signal by the Notch pathway in the adjacent cells severely affects their mesodermal specification and abolishes pigment cell differentiation (19, 20, 39). In these examples we see that intercellular developmental signaling functions are directly controlled by means of the genomic _cis_-regulatory code, just as are the cell autonomous regulatory functions considered in the previous section.
The Genomic Program for Repression of Alternative Fate.
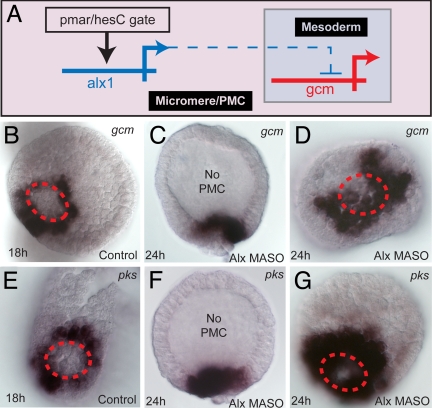
A further function of the skeletogenic micromere lineage is cryptic, evident only under experimental circumstances. This is the exclusion of the alternative fate that is actually assumed by the adjacent nonskeletogenic mesoderm. In the normal course of events the reception of the Notch signal causes transcriptional activation of the gcm regulatory gene in these cells, via the Su(H) _cis_-regulatory target sites of this gene (39). Downstream of this gene are differentiation genes that encode pigment synthesis pathway enzymes (39, 40), including polyketide synthase (Pks). In Fig. 5A we see that a role of the micromere lineage regulator alx1 is to repress gcm in that lineage. Thus if alx1 expression is blocked, expression of gcm and pks genes spreads across the vegetal plate, including the micromere domain, rather than being confined to the surrounding nonskeletogenic mesoderm (Fig. 5 B–G). These cells are thereupon transfated to nonskeletogenic mesoderm fate, and they produce extranumerary pigment cells (Fig. S5). Conversely, if alx1 mRNA is introduced into the egg, gcm expression is dramatically reduced (Fig. S5). This internal constraint may be essential, for the reason that the micromere lineage cells might otherwise respond as do their neighbors to the Delta ligand that each is expressing. This “exclusion function” is typical of embryonic specification systems (41). In the micromere lineage the alx1 exclusion function contributes to the autonomy of the specification process.
Fig. 5.
Regulatory exclusion of mesodermal fate. (A) GRN subcircuit showing repression of gcm transcription by alx1 in micromere lineage (dashed line indicates this is not known to be a direct interaction). (B–D) WMISH demonstration of derepression of gcm in embryos in which alx1 expression is blocked: control (B); alx1 morpholino, lateral view (C); vegetal view comparable to B (D). (E–G) Same for the pks gene, a pigment cell differentiation gene downstream of gcm. Red dashed circle delimits the skeletogenic lineage territory in the vegetal plate.
The Genomic Program for Activation of Differentiation Gene Batteries.
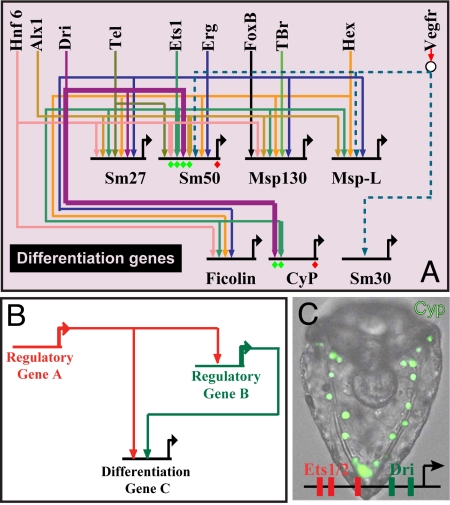
We arrive now at the climax of the specification process, the activation of the sets of genes (22) that actually constitute the skeletal biomineral and cause the cells to execute the many cell biology functions required for skeletal deposition. Although not all of these genes are activated before ingression, many are (Fig. S6). As shown in Fig. 6A, these differentiation genes require as drivers products of all of the now familiar components of the skeletogenic regulatory state (alx1, ets1, tbr, tel, erg, hex, foxb, dri) and in addition a factor that is at this stage ubiquitously present, Hnf6 (42). The maternal tbr and ets1 messages are also ubiquitously expressed, but by a few hours into cleavage only the zygotic skeletogenic lineage transcripts are present. We have seen explicitly in Figs. 2 and 3 the regulatory linkages that cause each of these genes (except hnf6) to be transcribed in the micromere lineage. The GRN includes only a sample of the many differentiation genes. However, at this level of the GRN, the nature of the circuit architecture changes dramatically. Almost all of the linkages to the differentiation genes are feeds forward (A > C; A > B; B > C): This motif is shown diagrammatically in Fig. 6, and in Table S1 the identities of the A and B drivers are indicated for each differentiation gene. The alx1 and erg genes play both roles, but the ets1 gene (9) is used in these gene batteries only as the A driver, and it is the most common of these. The nature of these subcircuits presuppose _cis_-regulatory modules in the differentiation genes that use the respective multiple positive inputs, and it has been verified by _cis_-regulatory studies of the sm50 gene (43) and the cyclophilin gene (ref. 44 and Fig. 6C). Both the A and B inputs contribute to the level of expression of the differentiation genes, as shown for three examples in Fig. S7 (43, 44), and the design ensures that these inputs are coordinated, even though the A and B genes themselves have nonidentical drivers.
Fig. 6.
Control of the differentiation gene batteries. (A) Portion of GRN displaying regulatory linkages to differentiation genes. sm27 and sm50 are biomineral matrix genes (27) expressed during the specification period. sm30 is a matrix gene expressed only after ingression, probably under control of signals from the ectoderm. Msp130, msp-L, ficolin, and cyclophilin (Cyp) are genes encoding cell biology functions of skeletogenesis (20). All genes shown on the top row encode transcription factors (compare Fig. 4) except for the receptor vegfR. (B) Canonical feed forward design of linkages to differentiation genes; compare Table S1. (C) GFP expression in skeletogenic mesenchyme, driven by a _cis_-regulatory construct from the cyclophilin differentiation gene. In the cases where there are multiple inputs shown of factors that might see the same target sites, such as Ets and Erg, and Tgif and Hex, and these genes are also interlocked in regulatory loops, the inputs shown could be redundant. [Reproduced with permission from ref. 44 (copyright 2005, Elsevier).]
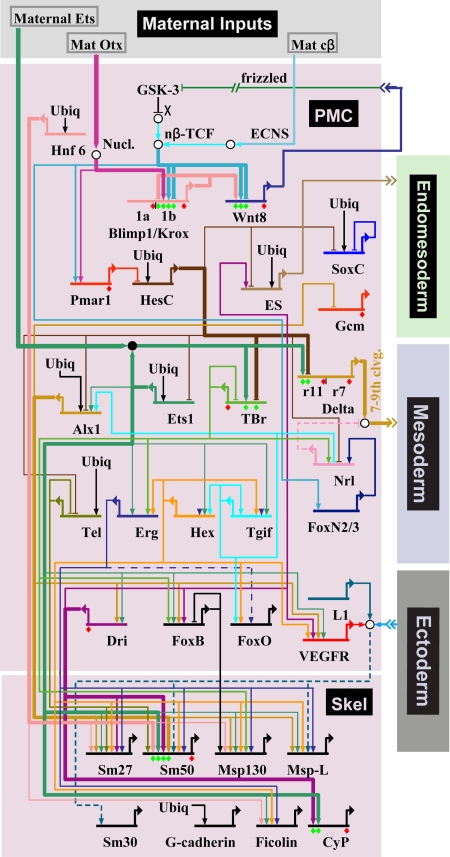
We have now traversed the specification and gene expression functions of the skeletogenic micromere lineage from its birth until its transformation to a differentiated state (Fig. 1). The entire GRN is shown in Fig. 7. A satisfying aspect is that we are aware of no unsolved problems in explaining why any of the regulatory genes go on in this lineage, although some ubiquitously present activators remain to be identified, as does the ES, and only a sample of the differentiation genes is yet included. Furthermore, there will emerge many interesting _cis_-regulatory aspects that will enrich our understanding of the genomic regulatory code as _cis_-regulatory analysis is further extended throughout the GRN.
Fig. 7.
Overall current GRN for specification of the skeletogenic micromere lineage.
Discussion
The regulatory relationships displayed in this article show how genomic control logic causes the skeletogenic micromere lineage to execute each of the zygotic functions in the specification process that the developmental biologist can identify. Thus we support the claim made at the outset that once it includes all or almost all specifically expressed regulatory genes, a GRN constitutes an explanation of why the events of development occur. The design principles of the skeletogenic micromere GRN are unlikely to be peculiar or unique, and because it may be the first GRN to achieve its level of sufficiency, it is useful to step back and ask what some of these principles are.
Modular Logic Executed by Modular Circuitry.
Although the overall GRN pictured in Fig. 7 looks at first sight to be a continuous tangle of interactions, the “sequential jobs” analysis we present here shows that each of the functions it carries out is controlled through a separate subcircuit, or module, of the GRN, and each subcircuit executes a distinct regulatory logic. The different modular components of a GRN evolve separately at diverse rates, a key to the process of body plan evolution (45, 46). Furthermore, and of future practical significance, comprehension of GRN modularity will provide a guide to re-engineering development. But the modularity of a GRN can be perceived only when it includes all or most of its essential components, and its modular structure can be regarded as one of the fundamental properties of GRNs that emerges only from system-level analysis.
The Individual Roles of Regulatory Genes.
At the periphery of the GRN the regulatory genes that drive differentiation gene expression play similar roles, just as specified in Table S1. Indeed they are forced into lockstep by the feed forward design. But in the internal portions of the GRN (Figs. 2–5) there is no such uniform design, and each regulatory gene is connected to others in a unique fashion. This is an important difference between the interior of a GRN and its periphery, where just as typified in Fig. 6A, the individual genes of differentiation gene batteries are linked in parallel circuits. It follows that experimental and computational approaches useful for gene battery analysis, which depend on coherence or similarities in mode of expression, are often not applicable to the solution of the internal portions of developmental GRNs.
Feedback Circuitry and Stabilization of Regulatory State.
Examples now abound in developmental GRNs where the developmental process begins with installation of a transient regulatory state that is then locked down by activation of a direct dynamic transcriptional feedback circuit (cases ranging from the postembryonic development of mammalian pancreas to Drosophila heart are reviewed in ref. 2). The tgif-hex and erg-hex loops in the skeletogenic micromere GRN are canonical examples. Outputs of these genes not only serve as drivers for one another but also feed multiple other nodes of the GRN, so that their activity ensures the persistence of surrounding activities. The surprise, when these features began to emerge from experimental GRN analysis, was that the lockdown of regulatory state is transcriptionally dynamic rather than passive, as so many examples of nontranscriptional state lockdowns mediated by chromatin level biochemistry were already known. However, most of the latter occur in contexts of terminal differentiation or later development in mammals and in Drosophila. It remains to be seen whether dynamic lockdown is a major feature only of embryogenesis, body plan formation, and early organogenesis, or conversely, whether chromatin level lockdowns are also set in train in early development. All that can be said is that the dynamic feedback lockdowns are essential and required in every one of the several cases where they have been observed in sea urchin embryo GRNs (for endomesoderm (2, 3), and there are additional examples in both oral and aboral ectoderm GRNs (unpublished data).
GRN Kinetics.
Although this is not a kinetics-centered analysis, the high-resolution time-course data in Fig. S2 permit a very simple conclusion with respect to the real-time pace of the developmental events controlled by the GRN: that the rate of developmental progress is probably controlled just by the average macromolecular synthesis and turnover kinetics for S. purpuratus embryos at 15°C. As noted above (26), the typical interval between successive regulatory gene activations in this system is 2–3 h, and indeed we see that the peak of pmar1 transcript accumulation in the skeletogenic micromeres is at 8 h; transcripts of the initial set of double negative gate targets peak at 10–12 h; transcripts of their targets peak at 14 h, and transcripts of their targets, the tertiary genes, peak at 16 h. It is not necessary to posit any particular temporal “coordination” device for this process.
Significance of GRNs.
In conclusion, it is our view that the spatial causes of developmental events after the earliest stages of dependence on egg cytoarchitecture are essentially all programmed in the genomic control system. Much of cell biology and biochemistry is engaged in the means by which development and differentiation materialize. The GRN controls not the downstream operation of these functions, but why they are deployed at a particular time and place. So it must be, because the program for development is an inherited feature of the genome. Informational logic is represented for any given event of zygotic development in the underlying GRN. In the earlier phases of the life cycle the main transactions are informational in significance, and that is why specification is the particular province of GRN analysis, and can only be understood in terms of a GRN.
Materials and Methods
All methods used in this work have been described: perturbation analysis and mRNA expression (27), microinjection and QPCR (30, 47), and in situ hybridization (8, 25). Morpholinos (Gene Tools) were injected into fertilized eggs at final concentrations of 200–400 μM. See Table S2 for morpholino sequences. Alx1 mRNA (36) was injected at 10–30 ng/μl.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Jina Jun for superb and essential technical assistance and Prof. Ellen Rothenberg and Dr. Andy Ransick for valuable comments. This research was supported by National Institutes of Health Grants HD-37105 and GM-61005, the Lucile P. Markey Charitable Trust, and the Camilla Chandler Frost Fellowship (to P.O.).
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 5951.
References
- 1.Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic; 2006. [Google Scholar]
- 3.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 4.Sodergren E, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 6.Howard-Ashby M, et al. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 8.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Horstadius S. The mechanics of sea urchin development, studied by operative methods. Biol Rev Cambridge Philos Soc. 1939;14:132–179. [Google Scholar]
- 11.Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: Summary and some proposed mechanisms. Development. 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 12.Wilt FH, Ettensohn CA. Morphogenesis and Biomineralization of the Sea Urchin Larval Endoskeleton. Weinheim, Germany: Wiley; 2007. [Google Scholar]
- 13.Croce J, et al. Maternal determinants in the vegetal cortex are required to activate the sea urchin endomesoderm gene regulatory network. Proc Natl Acad Sci USA. 2008 in press. [Google Scholar]
- 14.Ransick A, Davidson EH. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- 15.Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 16.Wikramanayake AH, Huang L, Klein WH. β-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci USA. 1998;95:9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wikramanayake AH, et al. Nuclear β-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 18.Ransick A, Davidson EH. Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development. 1995;121:3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- 19.Sherwood DR, McClay DR. Identification and localization of a sea urchin Notch homologue: Insights into vegetal plate regionalization and Notch receptor regulation. Development. 1997;124:3363–3374. doi: 10.1242/dev.124.17.3363. [DOI] [PubMed] [Google Scholar]
- 20.Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 21.Okazaki K. Spicule formation by isolated micromeres of the sea urchin embryo. Am Zool. 1975;15:567–581. [Google Scholar]
- 22.Livingston BT, et al. A genomewide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 23.Yuh CH, Bolouri H, Davidson EH. Cis-regulatory logic in the endo16 gene: Switching from a specification to a differentiation mode of control. Development. 2001;128:617–629. doi: 10.1242/dev.128.5.617. [DOI] [PubMed] [Google Scholar]
- 24.Miller JR, McClay DR. Characterization of the role of cadherin in regulating cell adhesion during sea urchin development. Dev Biol. 1997;192:323–339. doi: 10.1006/dbio.1997.8740. [DOI] [PubMed] [Google Scholar]
- 25.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 26.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitzel HE, et al. Differential stability of β-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131:2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- 28.Kenny AP, Kozlowski D, Oleksyn DW, Angerer LM, Angerer RC. SpSoxB1, a maternally encoded transcription factor asymmetrically distributed among early sea urchin blastomeres. Development. 1999;126:5473–5483. doi: 10.1242/dev.126.23.5473. [DOI] [PubMed] [Google Scholar]
- 29.Chuang CK, Wikramanayake AH, Mao CA, Li X, Klein WH. Transient appearance of Strongylocentrotus purpuratus Otx in micromere nuclei: Cytoplasmic retention of SpOtx possibly mediated through an α-actinin interaction. Dev Genet. 1996;19:231–237. doi: 10.1002/(SICI)1520-6408(1996)19:3<231::AID-DVG6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 30.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 31.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 32.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: HesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa D, et al. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech Dev. 1999;80:41–52. doi: 10.1016/s0925-4773(98)00192-0. [DOI] [PubMed] [Google Scholar]
- 34.Croce J, Lhomond G, Lozano JC, Gache C. ske-T, a T box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech Dev. 2001;107:159–162. doi: 10.1016/s0925-4773(01)00470-1. [DOI] [PubMed] [Google Scholar]
- 35.Fuchikami T, et al. T-brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development. 2002;129:5205–5216. doi: 10.1242/dev.129.22.5205. [DOI] [PubMed] [Google Scholar]
- 36.Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- 37.Minokawa T, Wikramanayake AH, Davidson EH. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 38.Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- 39.Ransick A, Davidson EH. cis-Regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 40.Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development. 2003;130:4587–4596. doi: 10.1242/dev.00647. [DOI] [PubMed] [Google Scholar]
- 41.Oliveri P, Davidson EH. Development: Built to run, not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- 42.Otim O, Amore G, Minokawa T, McClay DR, Davidson EH. SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Dev Biol. 2004;273:226–243. doi: 10.1016/j.ydbio.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 43.Makabe KW, Kirchhamer CV, Britten RJ, Davidson EH. Cis-regulatory control of the SM50 gene, an early marker of skeletogenic lineage specification in the sea urchin embryo. Development. 1995;121:1957–1970. doi: 10.1242/dev.121.7.1957. [DOI] [PubMed] [Google Scholar]
- 44.Amore G, Davidson EH. cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev Biol. 2006;293:555–564. doi: 10.1016/j.ydbio.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 45.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 46.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- 48.Kiyomoto M, Tsukahara J. Spicule formation-inducing substance in sea urchin embryo. Dev Growth Differ. 1991;33:443–450. doi: 10.1111/j.1440-169X.1991.00443.x. [DOI] [PubMed] [Google Scholar]
- 49.Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information