Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells (original) (raw)
Abstract
Increasing evidence suggests that HIV-1-specific cytotoxic T lymphocytes (CTLs) are a key host immune response to HIV-1 infection. Generation of CTL responses for prevention or therapy of HIV-1 infection has several intrinsic technical barriers such as antigen expression and presentation, the varying HLA restrictions between different individuals, and the potential for viral escape by sequence variation or surface molecule alteration on infected cells. A strategy to circumvent these limitations is the construction of a chimeric T cell receptor containing human CD4 or HIV-1-specific Ig sequences linked to the signaling domain of the T cell receptor ζ chain (universal T cell receptor). CD8+ CTLs transduced with this universal receptor can then bind and lyse infected cells that express surface HIV-1 gp120. We evaluated the ability of universal-receptor-bearing CD8+ cells from a seronegative donor to lyse acutely infected cells and inhibit HIV-1 replication in vitro. The kinetics of lysis and efficiency of inhibition were comparable to that of naturally occurring HIV-1-specific CTL clones isolated from infected individuals. Further study will be required to determine the utility of these cells as a therapeutic strategy in vivo.
Class I HLA-restricted CD8+ cytotoxic T lymphocytes (CTLs) are an important protective immune response in several viral infections and are probably a key element in HIV-1 infection. HIV-1-specific CTL activity has been identified in exposed but uninfected individuals (1–3) and has been correlated with the clearance of viremia in primary infection (4, 5). Infected individuals may have vigorous and broadly directed HIV-1-specific CTL activity of sufficient magnitude to detect without in vitro stimulation (6), and decline of this activity is associated with disease progression (7, 8). Several reports have suggested that those with long-term nonprogressing HIV-1 infection have more vigorous and broadly directed CTL responses than rapid progressors (9–11). Many investigators have verified the efficient inhibition of HIV-1 replication in vitro by CD8+ cells from infected individuals (12–17), and we have recently described the ability of HIV-1-specific CTL clones to mediate these effects (18). Thus, accumulating evidence supports the role of CTLs in controlling HIV-1 infection.
Attempts to generate protective or therapeutic immune responses with vaccines have been disappointing. Vaccination to generate cellular immune responses to HIV-1 infection has several intrinsic limitations. Initiation of a durable CTL response generally requires intracellular antigen expression and presentation. The class I HLA type of an individual limits the repertoire of HIV-1 epitopes that can be recognized, requiring any vaccine to present a large variety of epitopes to be generally useful. The epitopes expressed by a vaccine may vary within different strains of HIV-1, resulting in nonrecognition of certain isolates. HIV-1 may develop means of escape from CTL recognition such as the generation of unrecognized or antagonistic escape mutants, potential down-regulation of cell surface class I HLA, or alteration of viral antigen processing and presentation. Although a live attenuated vaccine has been shown to be protective in the rhesus macaque model (19), there is the potential hazard of an attenuated virus being pathogenic in certain individuals (20) or reverting to a more pathogenic phenotype in vivo.
To generate HIV-1-specific CTLs without such limitations, an in vitro gene therapeutic approach has been developed to produce CTLs bearing a chimeric T cell receptor (universal receptor, UR), as reported (21). Hybrid genes encoding HIV-1-specific URs were constructed by splicing the signaling domain of the T cell receptor ζ chain either to (i) the human CD4 molecule or (ii) the binding regions of an antibody specific for HIV-1 gp41. These genes were then inserted in a retroviral vector that could efficiently and stably transduce bulk human CD8 cells (22). Transduced cells (“UR-T cells”) therefore express a chimeric T cell receptor that triggers CTL activation and target cell lysis via the UR ζ domain upon binding to HIV-1 envelope on the surface of infected cells.
Such a strategy circumvents the difficulties of generating a cellular immune response in vivo and the necessity of accommodating different HLA restrictions; unlike native T cell receptors, class I HLA is not required for recognition by UR. Furthermore, because binding of HIV-1 gp120 to CD4 is a necessary step in the life cycle of all strains of HIV-1, the CD4-ζ UR should be broadly cross-reactive against all isolates and less prone to escape by sequence variation. Viral escape from CTLs by potential mechanisms such as surface class I down-regulation and altered viral antigen processing are also circumvented, again because binding does not depend on HLA expression or epitope presentation.
One potential drawback of such UR-T cells might be delayed recognition of infected cells. Epitope processing and presentation presumably occur early in the viral life cycle, as has been shown for other viruses such as vaccinia (39). Presentation of 10 or fewer epitope molecules may be sufficient for target cell lysis by CTLs (23–25). HIV-1-infected cells could, therefore, be susceptible to lysis before production of intact virions. A study of HIV-1-specific CTLs suggested that lysis can occur early in the viral replicative cycle (26). However, recognition by UR-T cells would require that infected cells express surface assembled viral envelope. Timing of cell lysis is obviously crucial in the control of viral replication, given the rapidity of the HIV-1 life cycle.
We tested the function of UR-T cell lines in a system previously developed to examine the efficiency and kinetics of infected cell killing by HIV-1-specific cytotoxic T lymphocytes (26). Acutely infected HIV-1-permissive CD4+ immortalized T cell lines were used as target cells in chromium release assays, and the efficiency and kinetics of recognition by UR-T cells was compared with that of HIV-1-specific CTL clones from infected individuals. We then evaluated the ability of UR-T cells to inhibit HIV-1 replication in vitro in coculture experiments using the same target cell lines, as well as primary monocytes and lymphocytes. As previously demonstrated for HIV-1-specific CTL clones (18), UR-bearing CD8+ T cells efficiently suppressed viral replication.
MATERIALS AND METHODS
Immortalized Target Cells.
The HIV-1-permissive cell lines T1 (HLA A2+, B14−) (27) and H9-B14 (H9 cells stably transfected with class I HLA B14 cDNA, HLA A2−, B14+) were maintained in RPMI 1640 medium (Sigma) supplemented with 20% heat-inactivated fetal calf serum (Sigma), 10 mM Hepes, 2 mM glutamine, penicillin (100 units/ml), and streptomycin (R20; 10 μg/ml). H9-B14 cells were generated as described (26). Near-confluent cells were split 1:2 in fresh medium the day before infection.
Primary Target Cells from HIV-1 Seronegative Donors.
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll gradient centrifugation. Primary monocyte cultures were established by adherence of 5 × 106 freshly isolated PBMCs per well in a polystyrene 24-well plate for 2 h at 37°C, followed by three washes and resuspension in R10. The remaining cells were >85% CD14+ and >99% CD3− by flow cytometric analysis (data not shown) and were infected 2 days after isolation. Primary CD4+ lymphocytes were generated from PBMCs by using a CD3:8-bispecific monoclonal antibody as described (28) without antiretroviral drugs and were >95% CD3+/CD4+ by flow cytometric analysis (data not shown). These cells were infected approximately 7 days after isolation/stimulation.
Naturally Occurring HIV-1-Specific CTL Clones.
From HIV-1-infected individuals, HIV-1-specific CTL clones were obtained by cloning of stimulated PBMCs at limiting dilution and characterized for specificity and HLA restriction as described (29, 30). The HLA A2-restricted CTL clone specific for an HIV-1 reverse transcriptase (RT) epitope (aa 77–85, SLYNTVATL) was 68A62 (designated 68/RT/A2). The HLA B14-restricted CTL clone specific for an HIV-1 Gag epitope (aa 298–306, DRFYKTLRA) was 15160 A49 (designated 15160/Gag/B14). Amino acids are numbered according to the HXB2 sequence. All CTL clones were maintained as described (26).
UR-T Cells.
As reported (21), bulk CD8+ cells from a HIV-1 seronegative individual were retrovirally transduced with UR constructs composed of the T cell receptor ζ chain and either (i) human CD4 (CD4-ζ) or (ii) a single chain antibody derived from a human anti-HIV-1 gp41 monoclonal antibody (SAb-ζ). The parent cell line T3 and the transduced cell lines T3F3 (expressing CD4-ζ) and T3F15 (expressing SAb-ζ) were maintained under the same conditions as the CTL clones above, with periodic restimulation using anti-CD3 antibody and irradiated feeder cells.
Synthetic Peptides.
Synthetic peptides corresponding to the HIV-1 epitopes RT aa 476–484 (ILKEPVHGV) and Gag aa 298–306 (DRFYKTLRA) were synthesized as free acids (model 432A peptide synthesizer, Applied Biosystems). Lyophilized peptides were reconstituted at 2 mg/ml in sterile distilled water with 10% dimethyl sulfoxide (Sigma) with or without 1 mM DTT (Sigma).
Virus Stocks.
The HIV-1 strain IIIB was taken from the supernatant fluid of freshly infected H9 cells. Viral titer (TCID50 units/ml) was determined by titration on C8166 cells as described (31). HIV-1 strain JR-CSF (32) was propagated and titered in phytohemagglutinin-stimulated PBMCs as described (33). First-passage primary isolate strains from subjects 115 (115v) and 18030 (18030v) were isolated by coculture of CD4+ cell lines from these individuals (generated as described above) with allogeneic phytohemagglutinin-stimulated PBMCs. All stocks were stored at −80°C until use.
Infection of Target Cells with HIV-1 for Lysis Assays.
T1 or H9-B14 cells were incubated with HIV-1 IIIB stock at the indicated multiplicity of infection (MOI) of 5 TCID50 units/cell for 4 h at 37°C, with intermittent agitation. The cells were then washed three times and resuspended at 5 × 105 cells per ml in R20. On the following 4 days, aliquots were taken for use as target cells in chromium release assays and for flow cytometric analysis of percentage of infected cells by intracellular HIV-1 p24 staining (clone KC57 labeled with fluorescein isothiocyanate, Coulter) as described (26). The remaining cells were centrifuged for harvesting of supernatant to assay TCID50 concentration by titration on C8166 cells as described (31) and p24 antigen concentration by ELISA (DuPont) and resuspended in fresh R20 at 5 × 105 cells per ml.
Chromium Release Assays.
Specific lysis was determined as described (26). Briefly, target cells consisted of infected or uninfected cells labeled with 100 μCi of Na2(51CrO4) (New England Nuclear; 1 Ci = 37 GBq) for 1 h, with or without peptide sensitization by adding peptide at 100 μg/ml during chromium labeling. Cytolytic activity was determined by adding 104 target cells to 5 × 104 CTL effector cells for 4-h incubations, followed by analysis of chromium release.
Infection of Target Cells with HIV-1 for Inhibition Assays.
T1, H9-B14, primary monocyte cultures, or primary CD4+ cells were incubated the indicated strain of HIV-1 at the described MOI for 4 h at 37°C, with intermittent agitation. The cells were then washed three times and resuspended in R10 with interleukin 2 (R10–50) at 50 units/ml for coculture with CTLs or UR-T cells.
Inhibition Assays.
T1 and H9-B14 cells acutely infected with HIV-1 IIIB at MOI 5 were cultured with HIV-1-specific CTL clones or with UR-T cells at a ratio of 1:1, 5 × 104 of each cell per well in duplicate in a 96-well round bottom plate, in 200 μl. Primary monocytes were infected with HIV-1 JR-CSF at MOI 10−2 and cocultured with UR-T cells at an effector/target ratio of 1:2, assuming a yield of 5 × 105 monocytes per well, in a 24-well plate in 2 ml. Primary CD4+ cells were infected with HIV-1 strains IIIB, JR-CSF, 115v, or 18030v at MOI 10−1 and added to a 24-well plate at 5 × 105 cells per well for coculture with UR-T cells at a ratio of 1:2 in 2 ml. All assays were performed in R10 with 50 units/ml IL-2. Half of the supernatant was harvested for quantitative HIV-1 p24 antigen capture ELISA (DuPont) and replenished with fresh medium at the indicated time points after infection.
RESULTS
Susceptibility to Lysis of Acutely Infected Cells by UR-T Cells and CTL Clones Is Similar.
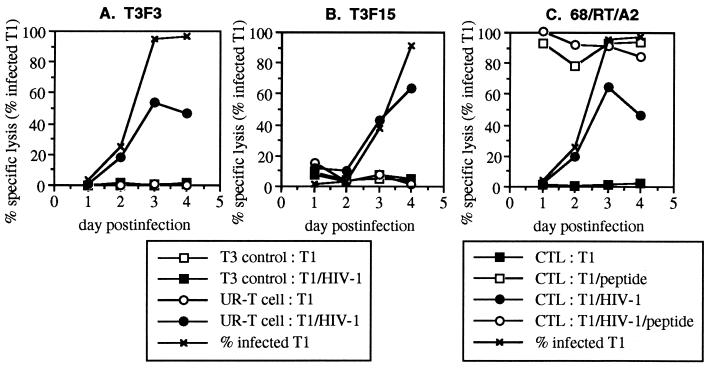
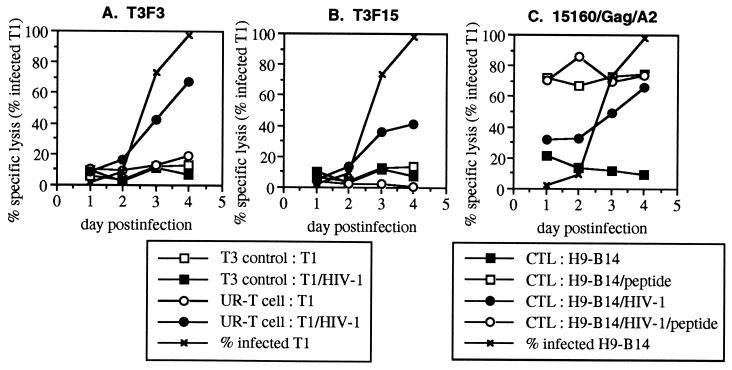
Several immortalized CD4+ cell lines were previously demonstrated to be suitable for acute synchronous infection with HIV-1 IIIB (18). Due to the availability of HLA B14- and A2-restricted CTLs specific for IIIB epitopes, H9-B14 (HLA A2− B14+) and T1 (HLA A2+ B14−) cell lines were chosen as target cells. We compared the ability of UR-T cells to that of naturally occurring HIV-1-specific CTL clones to lyse acutely infected T1 (Fig. 1) and H9-B14 (Fig. 2) cells in chromium release assays at daily time points after high-efficiency infection. T1 cells generally reached peak infection by day 3 after infection, with more than 95% of cells expressing intracellular p24 antigen by flow cytometric assessment (Fig. 1). Susceptibility to lysis by a naturally occurring CTL clone (68/RT/A2) closely paralleled that of intracellular p24 expression. Recognition of the infected T1 cells by the UR-T cell lines T3F3 and T3F15 also coincided with intracellular p24 expression. H9-B14 cell infection peaked on day 4 (Fig. 2), and again lysis by both a CTL clone (15160/Gag/B14) and UR-T cells (T3F3 and T3F15) followed kinetics similar to those of intracellular p24 expression. Recognition of acutely infected cells by both UR-T cell lines was, therefore, kinetically indistinguishable from that of naturally occurring CTL. Consistent with our previous report (26), the efficiency of lysis varied between CTL clones; the Gag-specific clone reached a peak level of lysis equivalent to that of cognate-peptide-sensitized controls, whereas lysis by the RT-specific clone plateaued below positive controls. There were no controls for maximal specific lysis by the UR-T cells; however, the absolute levels of lysis were intermediate to that of the Gag- and RT-specific CTLs, within the range of naturally occurring CTL clones.
Figure 1.
Lysis of acutely HIV-1-infected T1 cells by UR-T cells. T1 cells were acutely infected with HIV-1 IIIB and harvested daily for use as target cells in standard chromium release assays. Effector cells consisted of the UR-T cells T3F3 and T3F15, as well as a HLA A2-restricted CTL clone specific for RT (68/RT/A2). Controls included effector T3 cells (the parent nontransduced cell line of T3F3 and T3F15) and target infected or uninfected T1 cells with or without the addition of the cognate peptide for 68/RT/A2.
Figure 2.
Lysis of acutely HIV-1-infected H9-B14 cells by UR-T cells. H9-B14 cells were acutely infected with HIV-1 IIIB harvested daily for use as target cells in standard chromium release assays. Effector cells consisted of the UR-T cells T3F3 and T3F15 and a HLA B14-restricted CTL clone specific for Gag (15160/Gag/B14). The effector/target cell ratio was 5:1. Controls included effector T3 cells (the parent nontransduced cell line of T3F3 and T3F15) and target infected or uninfected H9-B14 cells with or without the addition of the cognate peptide for 15160/Gag/B14.
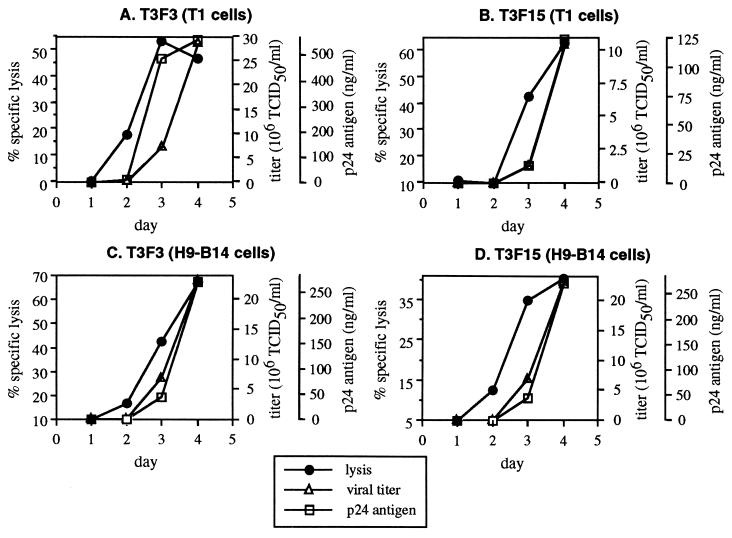
Susceptibility to Lysis by both UR-T Cells and CTL Precedes Significant Virion Production in Acutely Infected Cells.
The kinetics of lysis by UR-T cells and CTL clones were compared with the kinetics of virion production by acutely infected target cells. Daily HIV-1 production was determined for the acutely infected T1 and H9-B14 cultures from which cells were taken for the experiments depicted in Figs. 1 and 2. The supernatants from these cultures were quantitated for infectious viral titer and p24 antigen. Comparison of these parameters revealed a small but consistent lag of virion production behind lysis for both UR-T cells (Fig. 3) and CTL clones (data not shown; also ref. 26). This suggested that these effector cells were able to recognize infected cells early in the course of viral replication, as demonstrated for CTL clones (26). The ratios of infectious units produced to cells susceptible to lysis were also similar for UR-T cells and CTL clones (ref. 26 and data not shown).
Figure 3.
Temporal relationship of UR-T cell recognition and virion production in acutely infected T1 cells. Supernatants from the HIV-1 IIIB-infected T1 and H9-B14 target cells used in Figs. 1 and 2 were harvested daily for viral quantitation by p24 ELISA and viral titration. Viral production in the absence of CTLs is plotted against the curves for specific lysis for each UR-T cell as depicted in Figs. 1 and 2.
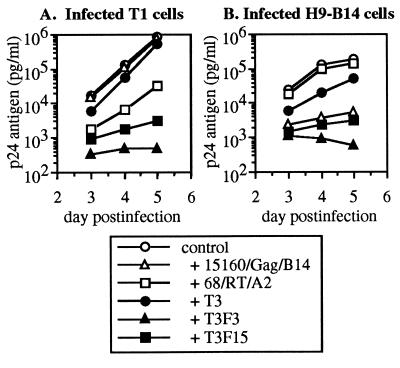
UR-T Cells Inhibit HIV-1 IIIB Replication in Immortalized CD4+ Cell Lines.
We next compared the inhibitory activity of UR-T cells with that of naturally occurring CTL clones on the same immortalized target cell lines. Cocultures of infected T1 (HLA A2+ and B14−) cells with the CTL clones revealed viral suppression by the A2-restricted clone and not by the B14-restricted clone (Fig. 4A). Conversely, infected H9-B14 cells (HLA B14+ and A2−) were inhibited by the B14-restricted clone and not the A2-restricted clone (Fig. 4B). In contrast, UR-T cells suppressed HIV-1 replication in both T1 and H9-B14 cells (Fig. 4 A and B). The efficiency of inhibition by T3F3 and T3F15 was superior to that of the Gag-specific (Fig. 4B) and RT-specific (Fig. 4A) CTL clones but less efficient than that of Env-specific HLA B14-restricted CTL clones (data not shown). UR-T cells therefore exerted antiviral activity comparable to that of naturally occurring CTL but without HLA restriction. Furthermore, the efficient suppressive activity of T3F3 (CD4-ζ UR containing) supports our observation that these cells themselves do not appear to be infectable (data not shown).
Figure 4.
Suppression of HIV-1 IIIB replication by UR-T cells in direct coculture with acutely infected T1 and H9-B14 cells. T1 (HLA A2+, B14−) and H9-B14 (B14+, A2−) cells were infected and cocultured 1:1 with UR-T cells, nontransduced T3 cells, and natural CTL clones 68/RT/A2 or 15160/Gag/B14. HIV-1 p24 antigen in the supernatant was quantitated on days 3–5 after infection.
UR-T Cells Inhibit Replication of Other HIV-1 Isolates in Primary Monocytes and Lymphocytes.
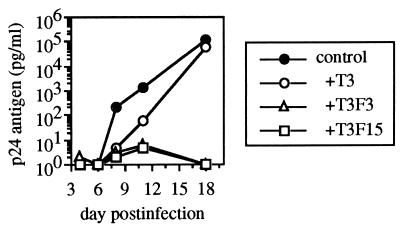
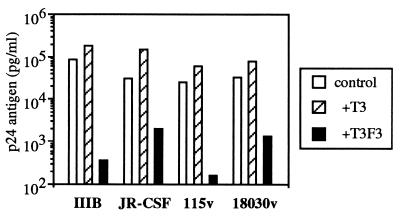
We also tested the ability of UR-T cells to inhibit the replication of other strains of HIV-1 in primary cells. Primary monocyte cultures from a seronegative individual were acutely infected with the monocytotropic (M-tropic) strain HIV-1 JR-CSF and cocultured with UR-T cells (Fig. 5). Viral replication was efficiently suppressed by the UR-transduced cell lines T3F3 and T3F15, whereas the nontransduced control parent cell line T3 was minimally inhibitory. The degree of inhibition was comparable to that seen with HIV-1-specific CTL clones on infected HLA-matched monocytes (data not shown). We further evaluated the ability of the UR-T cell line T3F3 to inhibit replication of diverse strains of HIV-1 in primary CD4+ lymphocytes from a seronegative donor (Fig. 6). T3F3 was inhibitory for the laboratory strains IIIB (T-lymphocytotropic) and JR-CSF (monocytotropic) and for first passage primary isolates from two infected individuals, 115v and 18030v, reducing p24 production by 100- to 1000-fold compared with the control cell line T3.
Figure 5.
Suppression of a monocytotropic HIV-1 strain in primary monocytes. Primary monocytes from a seronegative donor were acutely infected with the HIV-1 M-tropic strain JR-CSF and cocultured with UR-T cells or the control cell line T3. Supernatant was harvested at the indicated intervals for HIV-1 p24 quantitative ELISA.
Figure 6.
Suppression of diverse HIV-1 strains in primary lymphocytes. Primary lymphocytes from a seronegative donor were acutely infected with the laboratory strains IIIB (T-tropic) or JR-CSF (M-tropic), as well as the primary patient isolate strains 115v or 18030v, and cocultured with the UR-T cell line T3F3 or the control cell line T3. Day 7 p24 ELISA data is shown. Although the control CD8+ cell line T3 was variably stimulatory for p24 production in this and other experiments (data not shown) presumably due to alloreactive stimulation of the target cells, the UR-T cell line T3F3 was reproducibly inhibitory compared with T3.
DISCUSSION
Naturally occurring class I HLA-restricted HIV-1-specific CTLs have been shown to exert potent antiviral effects in an antigen-specific HLA-restricted fashion (18, 26). We have documented the ability of CTL to lyse acutely infected CD4+ cells and inhibit viral replication by up to 106-fold (sterilizing in vitro) and suggested that HIV-1-specific CTL mediate the antiviral activity of bulk CD8+ cells in infected individuals (18). Clinical correlations of CTL activity with clearance of viremia in primary infection (4, 5) and decline with disease progression (7, 8) provide further evidence that CTLs are an important immune response in HIV-1 infection. Augmentation of HIV-1-specific cellular immunity, particularly strategies that would circumvent the effects of viral variation on immune recognition (29, 34–37) and antagonism (38), therefore, might have clinical benefit.
In this study we evaluate the ability of CD8+ cells transduced with HIV-1-specific chimeric T cell receptors (URs) to lyse acutely infected cells and inhibit viral replication. We compare these abilities to that of naturally occurring HIV-1-specific CTL clones in vitro. Our findings suggest that these UR-T cells are comparable to naturally HIV-1-specific CTL clones in the kinetics and efficiency of infected cell lysis and in the ability to suppress viral replication. UR-T cells performed within the range of previously observed efficiencies for natural CTL clones of different specificities both in levels of cell lysis and degree of inhibition.
The comparable kinetics of HIV-1-infected cell lysis by UR-T cells and natural CTL clones is somewhat unexpected. Epitope processing and presentation by class I HLA molecules is an event that occurs early in the viral life cycle, as described in prior studies showing that virus-specific CTL lyse cells acutely infected with vaccinia (39) or HIV-1 (26) early in virion production. UR-T cells require presentation of intact envelope on the surface of infected cells for recognition, presumably a later event. We observe, however, that natural CTL clones and UR-T cells lyse acutely infected cells with similar kinetics. Evidence that fewer than 10 epitope–major histocompatibility complex complexes may be required to trigger lysis of a target cell by CTLs (23–25) indicates that natural CTLs are highly sensitive to presented antigen. Our data indicate that UR-T cells also are likely to function in the presence of small amounts of intact HIV-1 envelope on the cell surface. The similar kinetics of natural CTLs and UR-T cells indicates that viral envelope appears on infected cell surfaces with similar kinetics to processed antigen.
The ability of UR-T cells to kill infected cells early in virion production is also supported by our observation that they are potent inhibitors of viral replication. These cells were highly efficient at suppressing virus production in cells of various HLA types. This inhibition occurred at levels within the range observed for natural CTL clones of different specificities. The ability of cells from seronegative individuals transduced with HIV-1-specific URs to mediate viral suppression indicates the central role of an antigen-specific response in the antiviral activity of CD8+ cells in infected individuals. Although controversy exists as to the mechanism of CD8+ cell suppression of viral replication, our data clearly demonstrate that CD8+ cells from uninfected individuals are capable of exerting antiviral activity when redirected by a HIV-1-specific UR. This strongly suggests that lysis plays a key role in the phenomenon but does not exclude involvement of other mechanisms mediated by soluble inhibitory factors. Others have shown that CTLs may also produce soluble factors active against hepatitis B virus (40, 41), and our recent data suggest that CTLs produce soluble factors active against HIV-1 (18).
UR-T cells circumvent several potential limitations inherent in active immunization strategies for inducing HIV-1-specific CTL responses (21). These cells act without HLA restriction and are not constrained by the repertoire of epitopes determined by the available HLA molecules. Furthermore, because binding of the HIV-1 envelope to CD4 is an essential step in the viral life cycle, CD4-ζ UR-T cells may be less prone to escape than natural CTLs, which target only single epitopes. Our data show that UR-T cells are able to recognize a wide variety of HIV-1 strains of differing phenotypes in other cell types. We have also observed that UR-T cells but not natural CTLs are capable of inhibiting viral replication in the cell line T2 (data not shown), an antigen-processing-deficient derivative of T1 cells (42).
Help from CD4+ T cells may be an important issue to be addressed in the use of these cells. HIV-1-infected individuals have been demonstrated to have poor HIV-1-specific helper responses, presumably due to loss of HIV-1-specific CD4+ cells during early infection (43). UR-transduced CD4+ cells have been shown to produce high levels of cytokines such as interleukin 2, suggesting that coinfusion of UR-modified CD4+ cells may enhance survival and activity in vivo of CD8+ UR-T cells (21).
The therapeutic potential of UR-T cells will depend on the elucidation of several aspects of HIV-1 immunopathogenesis. The clarification of why vigorous cellular immunity in many infected individuals ultimately fails will be a key issue. Some possible mechanisms for the eventual failure of native HIV-1-specific CTLs may be overcome by UR-directed CTLs, such as loss of virus-specific help (43), viral escape (29, 34–37), and antagonist (38) mutations and clonal exhaustion (44). Other factors, such as generalized immunosuppression (45) and immune-privileged sites (46), may not be remediable. Moreover, UR-T cells may traffic differently than naturally occurring CTLs and may have altered survival in vivo (47) as a result of ex vivo manipulations. Clarification of such issues await the results of human clinical trials that are now underway.
Acknowledgments
This work was supported by Public Health Service Grants F32 AI 09280-01, AI 28568, and AI 30914 from the National Institute of Allergy and Infectious Diseases.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CTL, cytotoxic lymphocyte; UR, universal receptor; PBMC, peripheral blood mononuclear cell; RT, reverse transcriptase; MOI, multiplicity of infection.
References
- 1.Langlade-Demoyen P, Ngo-Giang-Huong N, Ferchal F, Oksenhendler E. J Clin Invest. 1994;93:1293–1297. doi: 10.1172/JCI117085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto L A, Sullivan J, Berzofsky J A, Clerici M, Kessler H A, Landay A L, Shearer G M. J Clin Invest. 1995;96:867–876. doi: 10.1172/JCI118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowland-Jones S, Sutton J, Ariyoski K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, Takiguchi M, Schultz T, McMichael A, Whittle H. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker B D, Chakrabarti S, Moss B, Paradis T J, Flynn T, Durno A G, Blumberg R S, Kaplan J C, Hirsch M S, Schooley R T. Nature (London) 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 7.Carmichael A, Jin X, Sissons P, Borysiewicz L. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. J Exp Med. 1995;181:1356–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferbas J, Kaplan A H, Hausner M A, Hultin L E, Matud J L, Liu Z, Panicali D L, Nerng-Ho H, Detels R, Giorgi J V. J Infect Dis. 1995;172:329–339. doi: 10.1093/infdis/172.2.329. [DOI] [PubMed] [Google Scholar]
- 10.Harrer T, Harrer E, Kalams S, Barbosa P, Trocha A, Johnson R, Elbeik T, Feinberg M, Buchbinder S, Walker B. J Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- 11.Rinaldo C, Huang X-L, Fan Z, Ding M, Beltz L, Panicali D, Mazzara G, Liebmann J, Cottrill M, Gupta P. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinchmann J E, Gaudernack G, Vartdal F. J Immunol. 1990;144:2961–2966. [PubMed] [Google Scholar]
- 13.Chen C H, Weinhold K J, Bartlett J A, Bolognesi D P, Greenberg M L. AIDS Res Hum Retroviruses. 1993;9:1079–1086. doi: 10.1089/aid.1993.9.1079. [DOI] [PubMed] [Google Scholar]
- 14.Mackewicz C, Levy J A. AIDS Res Hum Retroviruses. 1992;8:1039–1050. doi: 10.1089/aid.1992.8.1039. [DOI] [PubMed] [Google Scholar]
- 15.Toso J F, Chen C-H, Mohr J R, Piglia L, Oei C, Ferrari G, Greenberg M L, Weinhold K J. J Infect Dis. 1995;172:964–973. doi: 10.1093/infdis/172.4.964. [DOI] [PubMed] [Google Scholar]
- 16.Walker C M, Moody D J, Stites D P, Levy J A. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 17.Wiviott L D, Walker C M, Levy J A. Cell Immunol. 1990;128:628–634. doi: 10.1016/0008-8749(90)90054-u. [DOI] [PubMed] [Google Scholar]
- 18.Yang O O, Kalams S A, Trocha A, Cao H, Luster A, Johnson R P, Walker B D. J Virol. 1997;71:3120–3128. doi: 10.1128/jvi.71.4.3120-3128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel M, Kirchoff F, Czajak S C, Sehgal P K, Desrosiers R C. Science. 1992;238:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 20.Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 21.Roberts M R, Qin L, Zhang D, Smith D H, Tran A-C, Dull T J, Groopman J E, Capon D J, Byrn R A, Finer M H. Blood. 1994;84:2878–2889. [PubMed] [Google Scholar]
- 22.Finer M H, Dull T J, Qin L, Farson D, Roberts M R. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 23.Sykulev Y, Cohen R J, Eisen H N. Proc Natl Acad Sci USA. 1995;92:11990–11992. doi: 10.1073/pnas.92.26.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sykulev Y, Joo M, Vturina I, Tsomides T J, Eisen H N. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 25.Brower R C, England R, Takeshita T, Kozlowski S, Margulies D H, Berzofsky J A, Delisi C. Mol Immunol. 1994;31:1285–1293. doi: 10.1016/0161-5890(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 26.Yang O O, Kalams S A, Rosenzweig M, Trocha A, Koziel M, Walker B D, Johnson R P. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salter R D, Howell D N, Cresswell P. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 28.Wilson C C, Wong J T, Girard D, Merril D P, Dynan M, An D, Kalams S A, Johnson R P, Hirsch M S, D’Aquila R T, Walker B D. J Infect Dis. 1995;172:88–96. doi: 10.1093/infdis/172.1.88. [DOI] [PubMed] [Google Scholar]
- 29.Johnson R P, Trocha A, Yang L, Mazzara G P, Panicali D L, Buchanan T M, Walker B D. J Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- 30.Walker B D, Flexner C, Birch L K, Fisher L, Paradis T J, Aldovini A, Young R, Moss B, Schooley R T. Proc Natl Acad Sci USA. 1989;86:9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson V A, Walker B D. In: Techniques in HIV Research. Aldovini A, Walker B D, editors; Aldovini A, Walker B D, editors. New York: Stockton; 1990. pp. 92–94. [Google Scholar]
- 32.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S Y. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 33.Gartner S, Popovic M. In: Techniques in HIV Research. Aldovini A, Walker B D, editors; Aldovini A, Walker B D, editors. New York: Stockton; 1990. pp. 53–66. [Google Scholar]
- 34.Dai L C, West K, Littaua R, Takahashi K, Ennis F A. J Virol. 1992;66:3151–3154. doi: 10.1128/jvi.66.5.3151-3154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi H, Merli S, Putney S D, Houghten R, Moss B, Germain R N, Berzofsky J A. Science. 1989;246:118–121. doi: 10.1126/science.2789433. [DOI] [PubMed] [Google Scholar]
- 36.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengede E, Kloetzel P-M, Neefjes J, Koszinowski U, Melief C. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 37.Phillips R E, Rowland J S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R, Rizza C R, McMichael A J. Nature (London) 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 38.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A, Giagrande P, Phillips R E, McMichael A J. Nature (London) 1994;369:403–406. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 39.Zinkernagel R M, Althage A. J Exp Med. 1977;145:644–651. doi: 10.1084/jem.145.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 41.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salter R D, Cresswell P. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miedema F, Meyaard L, Koot M, Klein M R, Roos M T, Groenink M, Fouchier R A, Van’t Wout A B, Tersmette M, Schellekens P T, Schuitemaker H. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 44.Moskophidis D, Lechner F, Pircher H, Zinkernagel R M. Nature (London) 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 45.Denner J, Norley S, Kurth R. AIDS. 1994;8:1063–1072. [PubMed] [Google Scholar]
- 46.Griffith T S, Brunner T, Fletcher S M, Green D R, Ferguson T A. Science. 1995;270:1189–1192. doi: 10.1126/science.270.5239.1189. [DOI] [PubMed] [Google Scholar]
- 47.Riddell S R, Elliott M, Lewinsohn D A, Gilbert M J, Wilson L, Manley S A, Lupton S D, Overell R W, Reynolds T C, Corey L, Greenberg P D. Nat Med. 1996;2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]