Sequential gene promoter methylation during HPV-induced cervical carcinogenesis (original) (raw)
Abstract
We aimed to link DNA methylation events occurring in cervical carcinomas to distinct stages of HPV-induced transformation. Methylation specific-multiplex ligation-dependent probe amplification (MS-MLPA) analysis of cervical carcinomas revealed promoter methylation of 12 out of 29 tumour suppressor genes analysed, with MGMT being most frequently methylated (92%). Subsequently, consecutive stages of HPV16/18-transfected keratinocytes (_n_=11), ranging from pre-immortal to anchorage-independent phenotypes, were analysed by MS-MLPA. Whereas no methylation was evident in pre-immortal cells, progression to anchorage independence was associated with an accumulation of frequent methylation events involving five genes, all of which were also methylated in cervical carcinomas. TP73 and ESR1 methylation became manifest in early immortal cells followed by RARβ and DAPK1 methylation in late immortal passages. Complementary methylation of MGMT was related to anchorage independence. Analysis of nine cervical cancer cell lines, representing the tumorigenic phenotype, revealed in addition to these five genes frequent methylation of CADM1, CDH13 and CHFR. In conclusion, eight recurrent methylation events in cervical carcinomas could be assigned to different stages of HPV-induced transformation. Hence, our in vitro model system provides a valuable tool to further functionally address the epigenetic alterations that are common in cervical carcinomas.
Keywords: MS-MLPA, Ms-SNuPE, MSP, DNA methylation, cervical cancer, transformation, biomarker
Worldwide carcinoma of the uterine cervix is a leading cause of cancer-related death for women (Pisani et al, 2002). The majority of cervical carcinomas are squamous cell carcinomas (SCCs), whereas adenocarcinomas (AdCAs) account for the remaining 15–20% of cases (Fu and Reagan, 1989; Pisani et al, 2002).
Development of cervical cancer is causally related to infection with high-risk human papillomaviruses (hrHPVs), predominantly types 16 and 18 (zur Hausen, 2002; Munoz et al, 2003). Although hrHPVs can be detected in virtually all cervical SCCs and AdCAs (Walboomers et al, 1999; Zielinski et al, 2003; Castellsague et al, 2006), progression of an hrHPV-positive premalignant lesion to invasive cancer is a rare event. Consistent with the multistep nature of human carcinogenesis, additive host cell alterations drive progression to invasive cancer (Snijders et al, 2006). Insight in these additional events may provide novel biomarkers for risk assessment of hrHPV-infected women. These events may involve chromosomal alterations affecting structure and expression of (candidate) oncogenes and tumour suppressor genes, as well as epigenetic alterations. The latter, including both histone modifications and DNA methylation, provide one mode of tumour suppressor gene silencing (Esteller and Herman, 2002). DNA methylation generally refers to the addition of a methyl group to the 5′ position of a cytosine base preceding a guanine. Methylation of CpG-rich sequences, so-called CpG islands, which are often present in gene promoter sequences, usually inhibits gene transcription.
Molecular markers based on DNA methylation, that is methylation markers, are of particular interest as recent studies indicate that DNA methylation can be easily detected in cervical scrapes using sensitive PCR-based methods like methylation-specific PCR (MSP). Moreover, positive MSP results in cervical scrapes represented methylation of respective genes in the underlying epithelium (Reesink-Peters et al, 2004; Feng et al, 2005).
To date, a number of studies have described aberrant methylation of established or candidate tumour suppressor genes in cervical carcinoma tissues, which include genes involved in apoptosis, WNT signalling, Ras-signalling and tumour invasion and metastasis (reviewed by Duenas-Gonzalez et al, 2005). In several of these studies, methylation of up to 16 gene promoters has been tested on cervical cancer biopsies, showing that up to 93% of them were positive for at least one of the methylation markers tested (Dong et al, 2001; Virmani et al, 2001; Narayan et al, 2003; Gustafson et al, 2004; Widschwendter et al, 2004; Feng et al, 2005; Zambrano et al, 2005; Wisman et al, 2006). However, for most of the genes studied, it is still unclear to what extent and at what stage promoter methylation reflects a functionally important step in the transformation process. This information will greatly contribute to the design of a clinically optimal marker panel for high-grade premalignant cervical lesions and cervical cancer in terms of both sensitivity and specificity.
In order to decipher functionally important steps in HPV-mediated transformation, a longitudinal in vitro model system of hrHPV-transfected keratinocytes (Steenbergen et al, 1996) can, next to cervical cancer cell lines, be of great value. We have previously transfected primary human keratinocytes with full-length HPV types 16 or 18, resulting in four immortal keratinocyte cell lines, that is FK16A and FK16B containing HPV16, and FK18A and FK18B containing HPV18 (Steenbergen et al, 1996). With increasing passage, these cell lines revealed increasing severity of dysplastic features in organotypic cultures that are reminiscent of the various stages of premalignant cervical lesions (Steenbergen et al, 1998b). Moreover, consecutive passages showed accumulation of chromosomal alterations overlapping with those found in cervical (pre)malignant lesions (Steenbergen et al, 1998a, 1998b, 2005; Wilting et al, 2006) and shared altered expression of certain genes with these (pre)cancer stages (Steenbergen et al, 1996, 2002; van Duin et al, 2003). It is currently, however, unknown to what extent this system also provides a valuable model for studying epigenetic alterations that are common in cervical carcinomas.
A novel method to assess the methylation status of multiple genes is methylation specific multiplex ligation-dependent probe amplification (MS-MLPA). MS-MLPA is a PCR-based technique allowing the semiquantitative detection of changes in DNA promoter methylation of multiple genes in a single reaction (Nygren et al, 2005). Discrimination between methylated and unmethylated targets is based on the annealing of probes containing a recognition site for the methylation-sensitive restriction enzyme _Hha_I.
In this study, we first assessed, by MS-MLPA, the genes out of the 29 (candidate) tumour suppressor genes that are methylated in cervical SCCs and AdCAs. Next, we determined at what stage during HPV-mediated transformation methylation of identified targets became manifest.
MATERIALS AND METHODS
Tissue specimens, cell lines and DNA isolation
Frozen specimens of 16 SCCs and eight AdCAs were collected during the course of routine clinical practice at the VU University Medical Center in Amsterdam. Serial cryosections were made, the outer of which were used for histomorphological assessment. All specimens included contained >70% tumour cells. The inner sections were used for DNA extraction and HPV typing as described previously (Wilting et al, 2006). This study followed the ethical guidelines of the Institutional Review Board of the VU University Medical Center.
Two primary keratinocyte cell lineages (EK05-1 and EK94-2) were cultured using conditions described before (Steenbergen et al, 1996). The cell lines FK16A, FK16B, FK18A and FK18B have been established previously by transfection of primary foreskin keratinocytes with either full-length HPV16 (FK16A and FK16B) or HPV18 (FK18A and FK18B) (Steenbergen et al, 1996). The cells were grown in serum-free keratinocyte growth medium (Life Technologies, Breda, The Netherlands), supplemented with bovine pituitary extract (50 _μ_g ml−1), epidermal growth factor (5 ng ml−1), penicillin (100 U ml−1), streptomycin (100 _μ_g ml−1) and L-glutamin (2 nM). Cells were harvested using 10 mM trypsin (Life Technologies).
Anchorage-independent cell clones of FK16A and FK18B were obtained by culturing late immortal cells in soft agarose, as described previously (Steenbergen et al, 2004), resulting in the outgrowth of a limited number of colonies. Of each cell line, a single colony was isolated, referred to as FK16ASA and FK18BSA and expanded further. Cervical cancer cell lines SiHa, HeLa and CaSki were obtained from the American Type Tissue Culture Collection (ATCC, Manassas, VA, USA). The six low passage cervical cancer cell lines 778, 808, 866, 879 and 915 were kindly provided by Professor Dr PL Stern and cultured as described previously (Brady et al, 1999).
From cultured cells, DNA was isolated using the QIAmp tissue kit (Qiagen, Hilden, Germany).
Methylation specific-multiplex ligation-dependent probe amplification
The MS-MLPA was performed as published previously (Nygren et al, 2005), using two probe mixtures (P041A and P041B). In total, the probe mixtures contained a panel of probes specific for 29 candidate tumour suppressor genes (Table 1). Some of the genes (i.e. APC, ATM, BRCA2, CHFR, CDH13, CDKN1B, CDKN2A, CDKN2B, ESR1, FHIT, GSTP1, HIC1, MLH1, PTEN, RARβ, RASSF1A, STK11, TP73, CADM1 (previously referred to as TSLC1), VHL) were represented by two or three probes that each recognised a different _Hha_1 restriction site in the promoter region of the respective genes. In addition, each probe mix contained control probes that lack the _Hha_1 restriction site and were used for quantification purposes.
Table 1. Genes represented by the probe mixtures (41A and 41B).
| 41A | 41B | ||
|---|---|---|---|
| TIMP3 | Tissue inhibitor of metalloproteinases-3 | BRCA1 | Breast cancer 1 |
| APC | Adenomatosis polyposis coli | BRCA2 | Breast cancer 2 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A; p16 | ATM | Ataxia telangiectasia mutated |
| MLH1 | mutL homologue 1 | TP53 | Tumour protein 53 |
| ATM | Ataxia telangiectasia mutated | PTEN | Phosphatase and tensin homologue deleted on chromosome 10 |
| RARB | Retinoic acid receptor, beta | SMARCA3 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a3 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B; p15 | CHFR | Checkpoint with forkhead and ring finger domains |
| HIC1 | Hypermethylated in cancer 1 | CDH13 | Cadherin 13 |
| CHFR | Checkpoint with forkhead and ring finger domains | TP73 | Tumour protein p73 |
| CASP8 | Caspase 8 | STK11 | Serine/threonine kinase 11 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B; p27 | VHL | von Hippel–Lindau |
| PTEN | Phosphatase and tensin homologue deleted on chromosome 10 | GSTP1 | Glutathione _S_-transferase pi |
| BRCA2 | Breast cancer 2 | HIC1 | Hypermethylated in cancer 1 |
| CD44 | CD44 molecule | ESR1 | Estrogen receptor 1 |
| RASSF1A | RAS association domain family 1A | RB1 | Retinoblastoma 1 |
| DAPK1 | Death-associated protein kinase 1 | FHIT | Fragile histidine triad gene |
| VHL | von Hippel–Lindau | STK11 | Serine/threonine kinase 11 |
| ESR1 | Estrogen receptor 1 | CADM1 | Cell adhesion molecule 1 |
| RASSF1A | RAS association domain family 1A | MGMT | _O_6-methylguanine-DNA methyltransferase |
| TP73 | Tumour protein p73 | CDKN1B | Cyclin-dependent kinase inhibitor 1B; p27 |
| FHIT | Fragile histidine triad gene | APC | Adenomatosis polyposis coli |
| CADM1 | Cell adhesion molecule 1 | CDKN2B | Cyclin-dependent kinase inhibitor 2B; p15 |
| CDH13 | Cadherin 13 | CDKN2A | Cyclin-dependent kinase inhibitor 2A; p16 |
| GSTP1 | Glutathione _S_-transferase pi | RASSF1A | RAS association domain family 1A |
| MLH1 | MutL homologue 1 | RARB | Retinoic acid receptor, beta |
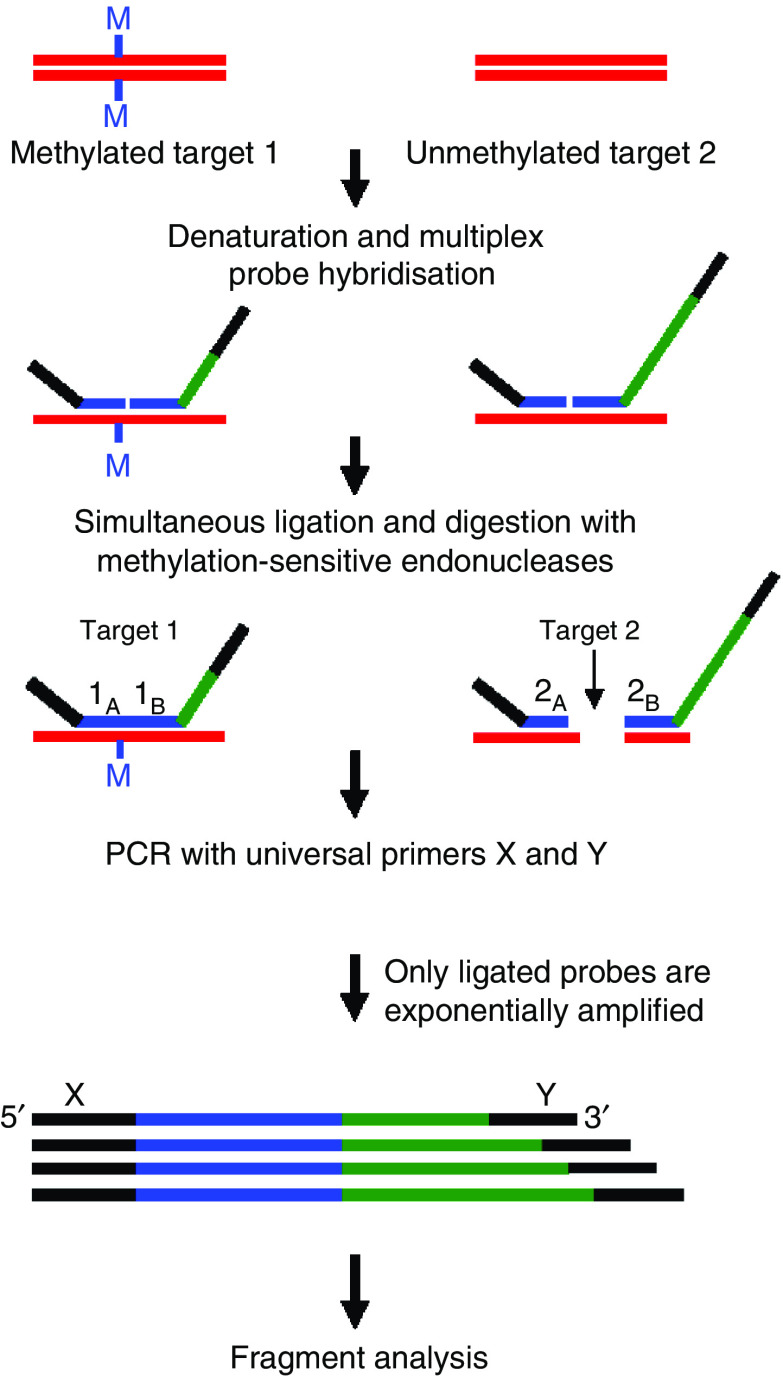
The MS-MLPA procedure is summarised in Figure 1. For each sample, 100 ng input DNA was used. After 10 min denaturation at 98°C, SALSA MLPA buffer and MS-MLPA probe mix P041A or P041B was added to the DNA, and the mixture was incubated for 1 min at 95°C, followed by hybridisation for 16 h at 60°C. After hybridisation, the samples were diluted at room temperature with H2O and 3 _μ_l ligase buffer A to a final volume of 20 _μ_l and divided over two series of tubes. Samples were heated to 49°C, after which a volume of 10 _μ_l of a mix containing 0.25 _μ_l ligase, 1.5 _μ_l ligase buffer B and 5 U _Hha_1 (Promega, Leiden, The Netherlands) was added. To the second series of tubes, an identical mix was added in which _Hha_1 was replaced by 50% glycerol. Ligation and digestion were performed simultaneously for 45 min at 49°C, which was followed by a 5 min incubation at 98°C to inactivate the enzymes.
Figure 1.
Overview of the MS-MLPA procedure (Nygren et al, 2005). The gene-specific probes spanning a recognition site for the restriction enzyme _Hha_1 are hybridised to the target DNA and subsequently ligated and digested with the methylation-sensitive enzyme _Hha_1. Undigested probes, that is probes of which the recognition sequence is methylated, will be amplified. If the CpG site is unmethylated, the DNA/probe complex will be digested and no amplification will take place. For each DNA sample, the MS-MLPA was performed with and without _Hha_1 digestion.
To amplify the ligation products, 5 _μ_l was added to a PCR mix containing 10 × SALSA PCR buffer, 1 U SALSA polymerase and SALSA FAM PCR primer-dNTP mix (all provided by MRC Holland, Amsterdam, The Netherlands) and the following PCR protocol was used: 1 min at 95°C; 35 cycles (30 s at 95°C, 30 s at 60°C, 1 min at 72°C); 20 min at 72°C.
For fragment analysis, 8.75 _μ_l Formamide (Sigma-Aldrich, Zwijndrecht, The Netherlands), 0.25 _μ_l Gene Scan-500 ROX Size Standard (PE Applied Biosystems, Foster City, CA, USA) and 1 _μ_l PCR product was run on an ABI PRISM 3100 Avant (ABI PRISM 3100-Avant Genetic Analyzer by PE Applied Biosystems). Analysis was performed using ABI 3100 Gene Scan 3.7 software. Each amplified fragment was normalised by dividing the area under the peak by the mean of the two flanking control fragments. The percentage of methylation was calculated by dividing normalised peaks in the _Hha_1-digested reaction by the normalised peaks in the undigested control reaction. Methylation below the threshold level of 10% was considered background (Nygren et al, 2005). All samples were analysed twice and scored positive for methylation when in both experiments the percentage of methylation for each individual probe was above the threshold. In case of multiple probes for a single gene promoter, the gene was scored positive when ⩾1 probe showed methylation.
Methylation-sensitive single-nucleotide primer extension (Ms-SNuPE)
Primers used to generate the specific MGMT PCR product for methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) analysis were 5′-GTATTAGGAGGGGAGAGATT-3′ and 5′-TCTATACCTTAATTTACCAAATAACCC-3′. The PCR was performed in a total volume of 25 _μ_l containing 25 ng bisulphite-modified DNA. PCR conditions were 8 min at 95°C, 35 cycles (30 s at 95°C, 30 s at 54°C, 45 s at 72°C); 4 min at 72°C. PCR products were separated on 1% agarose gels and isolated using the GeneClean III kit (Qbiogene, Irvine, CA, USA).
The Ms-SNuPE reactions were performed mainly as described before with some modifications (Gonzalgo and Jones, 1997). Conditions for primer extension reactions were: at 95°C for 1 min, 48°C for 1 min and 72°C for 1 min. The primers used for the Ms-SNuPE analysis were 5′-GGGATTTTTATTAAGCGGG-3′ and 5′-GGGATTTTTATTAAGTGGG-3′.
The reaction was performed in a total volume of 10 _μ_l, containing 4 _μ_l of purified PCR product and 6 _μ_l PCR mix consisting of: 10 × PCR buffer (MRC-Holland), 10 pmol of each Ms-SNuPE primer, 1 _μ_Ci of either [32P]dCTP or [32P]dTTP and 1 U of Taq polymerase (MRC-Holland).
Stop solution was added and samples were denatured for 4 min at 95°C. Of the sample, 1.5 _μ_l was loaded on a 15% polyacrylamide gel (7 M urea).
Radioactivity was quantitated using a phosphoimager. The percentage of methylation is equivalent to the value of C/(C+T). The specimens were rated positive when methylation level reached 5%.
Statistical analysis
Methylation percentages for individual genes in SCCs and AdCAs were compared using _χ_2 statistical testing. A two-sided _P-_value of ⩽0.05 was considered significant.
RESULTS
Promoter methylation profiles in cervical SCCs and AdCAs
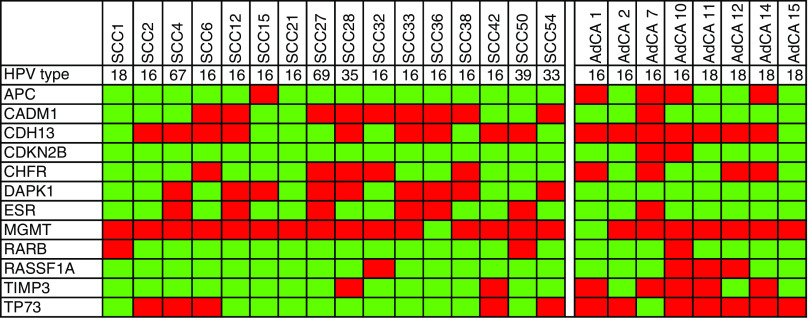
To firstly identify epigenetic alterations associated with cervical cancer, we analysed promoter methylation of 29 (candidate) tumour suppressor genes by MS-MLPA in 16 SCCs and eight AdCAs. All carcinomas contained DNA of high-risk HPV types (HPV16 in 10 SCCs and four AdCAs, HPV18 in one SCC and four AdCAs, HPV33, 35 and 39 each in one SCC) or not yet classified HPV types (HPV67 and 69, each in one SCC). An overview of all genes that were methylated in the individual SCCs and AdCAs is shown in Figure 2. In addition, a summary of the overall frequencies of methylation found in SCCs and AdCAs is detailed in Table 2. Twelve of the genes showed a positive MS-MLPA result in one or more of the carcinomas tested. Squamous cell carcinomas revealed frequent promoter methylation, that is in >40% of cases, of CDH13 (nine out of 16: 56.3%), DAPK1 (nine out of 16: 56.3%), MGMT (15 out of 16: 93.8%) and CADM1 (nine out of 16: 56.3%). In AdCAs, frequent promoter methylation of APC (four out of eight: 50%), CDH13 (seven out of eight: 87.5%), CHFR (four out of eight: 50%), MGMT (seven out of eight: 87.5%), TIMP3 (five out of eight: 62.5%) and TP73 (seven out of eight: 87.5%) was detected.
Figure 2.
Summary of MS-MLPA results on cervical SCCs (_n_=16) and AdCAs (_n_=8). Dark boxes indicate the presence of promoter methylation; light boxes represent unmethylated CpGs. For each SCC and AdCA, the HPV type present is also shown.
Table 2. Methylation frequencies in SCCs and AdCAs.
| Gene | SCC | AdCa | _P_-value |
|---|---|---|---|
| APC | 6.3% (1/16) | 50% (4/3) | 0.01 |
| CADM1 | 56.3% (9/16) | 6.25% (1/8) | 0.04 |
| CDH13 | 56.3% (9/16) | 87.5% (7/8) | 0.13 |
| CDKN2B | 0% (0/0) | 25% (2/0) | 0.04 |
| CHFR | 31.3% (5/16) | 50% (4/0) | 0.37 |
| DAPK1 | 56.3% (9/16) | 0% (0/0) | 0.01 |
| ESR1 | 37.5% (0/16) | 12.5% (1/8) | 0.20 |
| MGMT | 93.8%(15/16) | 87.5% (7/8) | 0.60 |
| RAR_β_ | 12.5% (2/16) | 12.5% (1/8) | 1.00 |
| RASSF1A | 6.3% (1/16) | 37.5% (3/0) | 0.05 |
| TIMP3 | 12.5% (2/16) | 62.5% (5/0) | 0.01 |
| TP73 | 31.3% (5/16) | 87.5% (7/0) | 0.01 |
MGMT was most frequently methylated in both tumour histotypes, that is in 92% of all carcinomas. On the other hand, methylation of DAPK1 (_P_=0.007) and CADM1 (_P_=0.04) was significantly higher in SCCs, whereas APC (_P_=0.01), CDKN2B (_P_=0.04), RASSF1A (_P_=0.05), TIMP3 (_P_=0.01) and TP73 (_P_=0.02) methylation was more common in AdCAs (Table 2).
No statistically significant different methylation profiles were found between HPV16-positive carcinomas and carcinomas containing other hrHPV types.
Accumulation of methylation events during HPV-mediated transformation in vitro
To determine at which stage during HPV-mediated transformation methylation of the various genes becomes manifest, we performed MS-MLPA analysis on an in vitro model system of HPV-transformed keratinocytes and cervical cancer cell lines. In previous studies, we and others have shown that at least four phenotypes can be distinguished during HPV-mediated transformation: (1) extended but still finite lifespan (pre-immortal), (2) immortalisation (3) anchorage-independent growth and (4) tumorigenicity (Chen et al, 1993; Steenbergen et al, 1996, 2005). We analysed two isolates of primary keratinocytes as normal controls and one HPV18-transfected cell culture (FK18B) representing the pre-immortal stage. Moreover, early passages (passage 39–61) and late passages (passage 75–124) of two HPV16 and two HPV18 immortalised cell lines (FK16A, FK16B, FK18A and FK18B) were included, as well as anchorage-independent clones of HPV16 (FK16ASA)- and HPV18 (FK18BSA)-containing cell lines. Finally, nine hrHPV-positive cervical cancer cell lines (SiHa, HeLa, CaSki, 778, 808, 866, 873, 879 and 915), of which the latter six were tested at low passage, were included as representatives of the tumorigenic stage. A schematic representation of the cell lines analysed is shown in Figure 3.
Figure 3.
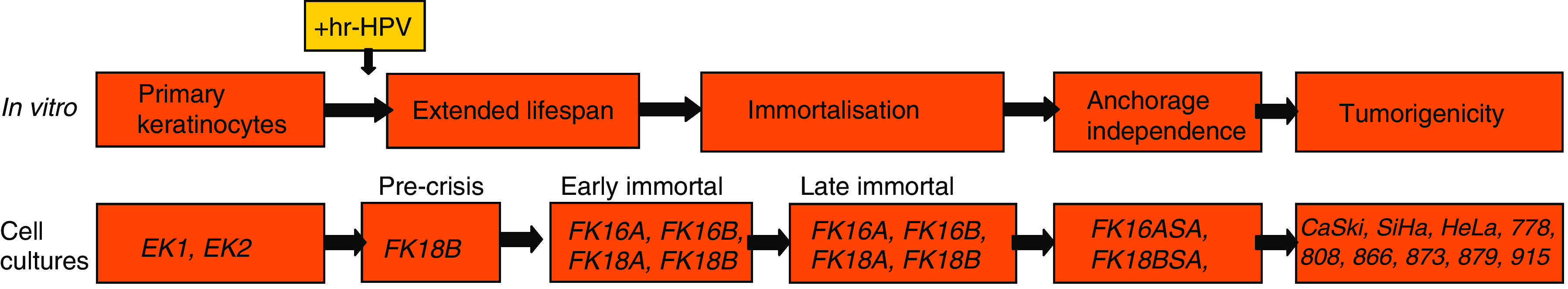
Schematic representation of the multistep process of HPV-mediated transformation in vitro, aligned with the hrHPV-transformed keratinocytes and cervical cancer cell lines used in this study.
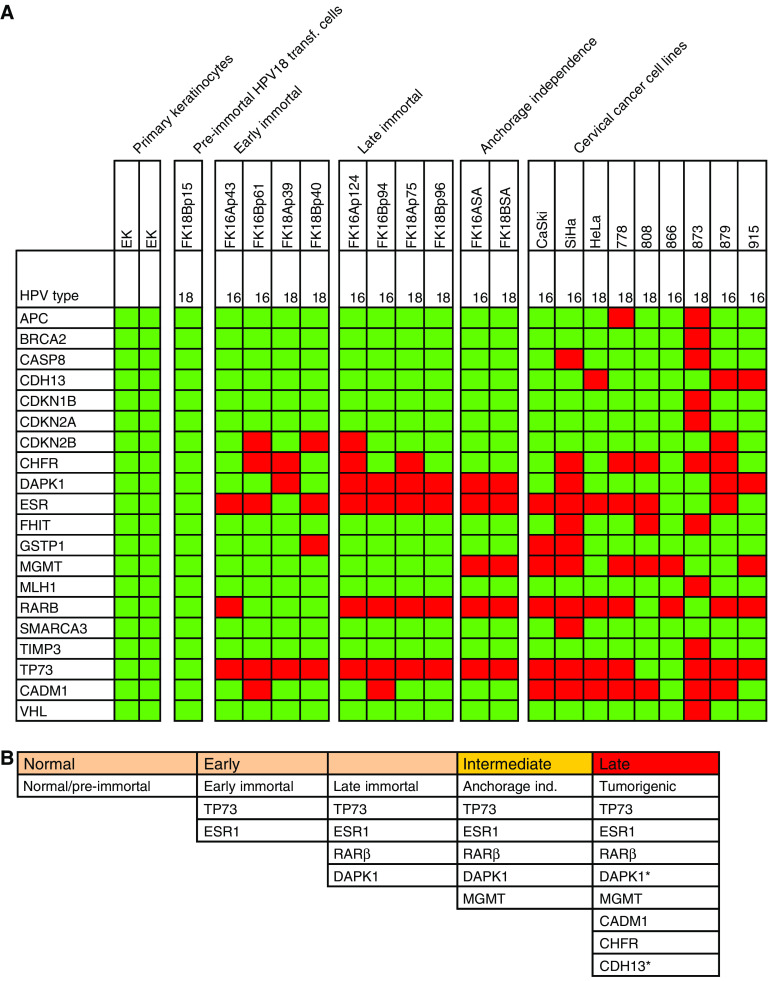
None of the 29 candidate tumour suppressor genes showed promoter methylation in two cultures of primary keratinocytes, nor in the pre-immortal passage of the cell line FK18B (Figure 4). At the early immortal stage, promoter methylation of TP73 was evident in all four cell lines and methylation of ESR1 in FK16A, FK16B and FK18B cell lines. Additive methylation of RARβ and DAPK1 became apparent at later immortal passages of all cell lines. Supplementary methylation of MGMT was associated with anchorage-independent growth of FK16A and FK18B. Cervical carcinoma cell lines, showed next to markers present in FK cell lines, also revealed frequent methylation of CADM1 and CHFR.
Figure 4.
(A) Summary of MS-MLPA results on primary keratinocytes (EK), HPV-transformed cell lines (FK16A, FK16B, FK18A, FK18B; p=passage number) and cervical carcinoma cell lines (CaSki, SiHa, HeLa, 778, 808, 866, 873, 879, 915). Only genes (20 out of 29) that were found to be hypermethylated in at least one of the cell lines are shown. HPV types are listed for each cell line. (B) Longitudinal scheme of epigenetic events associated with hrHPV-mediated transformation as detected in >50% of hrHPV-transformed cell lines and cervical carcinoma cell lines. *DAPK1 and CDH13 methylation was detected in 33% of carcinoma cell lines. Dark boxes indicate methylation positive, light boxes indicate methylation negative.
In Figure 4A an overview of the MS-MLPA results on all cell lines is shown. In Figure 4B, all genes that were found to be methylated in more than 50% of all hrHPV-transformed cell lines at the various stages of progression and in cervical carcinoma cell lines are summarised. Notably, all eight genes that were frequently methylated in the HPV-transfected cell lines and cervical cancer cell lines (i.e. TP73, ESR1, RARβ, DAPK1, MGMT, CADM1, CDH13 and CHFR) overlapped with those found to be methylated in the cervical carcinoma specimens.
Confirmation of MS-MPLA results on MGMT promoter by Ms-SNuPE
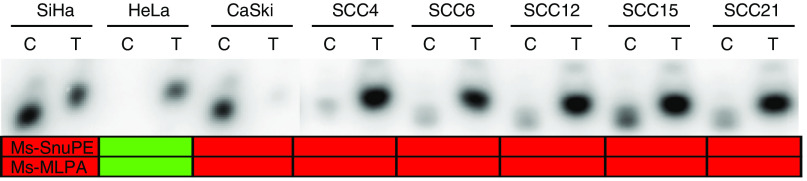
To confirm MS-MLPA results, we selected the MGMT gene for further analysis, since this gene revealed the highest frequency of positive MS-MLPA test results in cervical carcinoma specimens. For this purpose, we used Ms-SNuPE analysis, by which methylation differences at specific CpG sites can be assessed in a quantitative manner (Gonzalgo and Jones, 1997). In this method, DNA is treated with sodium bisulphite followed by amplification of the target sequence using primers specific for bisulphite-converted DNA. The subsequent primer extension reaction utilises an internal primer which anneals to the PCR product and terminates immediately 5′ of the cytosine to be assayed, 32P-labelled dCTP and dTTP and Taq polymerase. This is followed by denaturing polyacryalamide gel electrophoresis and phosphorimage analysis to quantitate the ratio of C versus T. To confirm our MS-MLPA results, primers were specifically designed to examine the same CpG dinucleotide as analysed by MS-MLPA (nt −459 relative to the transcription start site).
Methylation-sensitive single-nucleotide primer extension analysis of all cell lines confirmed MS-MLPA results for MGMT in all cases (Figure 5). Cell lines SiHa and CaSki were positive in both settings while the remaining cell lines were negative for MGMT promoter methylation. Also in cervical cancer specimens, Ms-SNuPE results were highly comparable to MS-MLPA results (Figure 5). In all except two of the cancer specimens, MGMT promoter methylation revealed identical results by MS-MLPA and Ms-SNuPE. The two exceptions were SCC 42 (MS-MLPA positive, Ms-SNuPE negative) and SCC 36 (MS-MLPA negative, Ms-SNuPE positive). It should, however, be noted that in both cases percentages of methylation measured with either or both techniques were near the cutoff levels of the assays.
Figure 5.
Representative Ms-SNuPE results for MGMT promoter methylation in a subset of cell lines and SCC specimens. Dark boxes indicate the sample was methylated and light boxes indicate the samples was unmethylated. Methylation specific-multiplex ligation-dependent probe amplification results for the same CpG dinucleotide in the MGMT promoter are depicted below for comparison.
DISCUSSION
Methylation specific-multiplex ligation-dependent probe amplification analysis of 29 tumour suppressor genes potentially targeted by methylation resulted in the identification of 12 methylated gene promoters in cervical carcinomas, eight of which could subsequently be associated with consecutive stages of HPV-mediated transformation in vitro.
The MGMT promoter was most frequently methylated (i.e. in 92% of carcinomas). Next to the common marker MGMT, also histotype-specific markers could be identified. DAPK1 and CADM1 were significantly more frequently methylated in SCCs while methylation of APC, CDKN2B, RASSF1A, TIMP3 and TP73 was significantly more frequent in AdCAs.
The detection of distinct methylation profiles between SCCs and AdCAs is in concordance with literature data, showing higher frequencies of DAPK1 promoter methylation in SCCs compared with AdCAs (Dong et al, 2001; Narayan et al, 2003; Yang et al, 2004; Kang et al, 2006) and increased rates of APC, RASSF1A and TIMP3 promoter methylation in AdCAs compared with SCCs (Dong et al, 2001; Narayan et al, 2003; Jeong et al, 2006; Kang et al, 2006; Wisman et al, 2006).
Based on the analysis of consecutive passages of HPV-transfected keratinocytes, we could assign methylation of the different gene promoters as detected in cervical carcinomas to distinct stages of transformation. Moreover, we showed that despite the recent finding that HPV18 E7 targets DNA methyl transferase 1 (DNMT1) involved in de novo methylation (Burgers et al, 2007), none of the 29 genes were found to be methylated in the pre-immortal cells expressing hrHPV E6 and E7. Only following immortalisation, an accumulation of methylated genes was detected, suggesting that inactivation of these genes is associated with a growth advantage of the hrHPV-containing keratinocytes. To what extent these genes (except CADM1; Steenbergen et al, 2004) are functionally involved in the different steps during the transformation process remains to be determined.
Methylation of both the TP73 and ESR1 promoter followed by RARβ and DAPK1 promoter methylation were identified as rather early events associated with immortalisation. The recent demonstration of an inverse relation between ESR1 protein expression and the severity of the cervical lesion (Bekkers et al, 2005) supports our observation of ESR1 promoter methylation being a rather early event during cervical carcinogenesis.
MGMT promoter methylation appeared as an intermediate event, which is in line with earlier studies showing MGMT promoter methylation in 26% of invasive cancers and 29% of high-grade CIN lesions, compared with only 3% of low-grade CIN lesions (Virmani et al, 2001).
Methylation of CHFR, CDH13 and CADM1 was only observed in cervical carcinomas and cervical carcinoma cell lines and can as such be designated as relative late events. To the best of our knowledge, this is the first study showing methylation of CHFR to be associated with cervical carcinogenesis. CHFR promoter methylation has, however, been described in other types of cancer, such as breast (Tokunaga et al, 2006), gastric (Kang et al, 2004) and colorectal cancer (Brandes et al, 2005).
The detection of CADM1 promoter methylation in cervical cancer cell lines is in concordance with our previous study showing reduced CADM1 mRNA expression associated with promoter methylation in nearly all cervical cancer cell lines, and not in HPV-immortalised cells (Steenbergen et al, 2004).
Validation of our MS-MLPA results by Ms-SNuPE analysis, an alternative method to specifically assess methylation of a single CpG dinucleotide, confirmed the presence of MGMT methylation (at position −459 relative to the transcription start site), in 92% of carcinomas. The frequency of MGMT methylation as detected by both techniques was different from most previous studies on cervical carcinomas, in which by MSP analysis MGMT methylation frequencies varying from 6.7 to 38% have been reported (Dong et al, 2001; Virmani et al, 2001; Yang et al, 2004; Lin et al, 2005). Interestingly, application of the same MSP assay as described in these previous studies, by which methylation at the region from +3 to +137 relative to the transcription start site was analysed, showed evidence for methylation in only 31% of our carcinomas (data not shown). This apparent discrepancy may, at least in part, be explained by heterogeneity of MGMT promoter methylation in cervical carcinomas.
On the other hand, MSP analysis of the cell lines for DAPK1, ESR1, RARβ and CADM1 promoter methylation showed a 78–89% concordance between the MS-MLPA and MSP results (data not shown). The absence of complete concordance may be explained by the fact that MS-MLPA is only based on a single CpG site compared to on average 4–6 CpG sites in MSP assays. Moreover, different CpG dinucleotides were targeted by MS-MLPA and MSP, indicating that, similar to MGMT, in the few discrepant cases heterogeneous methylation patterns may exist within the individual gene promoters.
Taken together, the present data illustrate the significance of our in vitro model. In our previous studies we already showed that both morphologically and genetically, the HPV-transfected cell lines closely resembled cervical (pre)malignant lesions (Steenbergen et al, 1996, 1998a, 2002; van Duin et al, 2003). The current study indicates that also with respect to DNA methylation, this model system nicely mimics cervical carcinogenesis in vivo. Consequently, it provides a valuable tool to analyse the functional involvement of the identified genes in the respective phenotypical alterations during HPV-induced transformation.
Next to future functional studies, the linkage of the different methylation events to distinct stages of HPV-induced malignant transformation provided first insight in the potential clinical applications of these markers. Early and intermediate methylation events may provide markers for better risk assessment of hrHPV-positive women with (ab)normal cytology. On the other hand, late methylation events or a combination of the various events may be used at the time of diagnosis for cancer staging and grading or selection and monitoring of therapy. Future clinical validation studies, for which specific guidelines have been proposed by Pepe et al (2001), on cervical (pre)malignant and cervical scrapes, will ultimately reveal the diagnostic value of the different methylation markers. It should be taken into account that the specificity and sensitivity of the different markers may need to be determined for individual target population as frequencies of promoter methylation can vary among different ethnic groups (Enokida et al, 2005; Das et al, 2006).
Gene alterations previously detected in our in vitro model system, such as GATA-3, hTERT, MGP and CADM1, were also found in clinical samples (Steenbergen et al, 2001, 2002, 2004; de Wilde et al, 2007). Therefore, we believe that the methylation events identified in the in vitro model system provide potential markers for cervical cancer detection as well, which is underlined by their large overlap with methylated gene promoters found in cervical carcinomas.
In conclusion, MS-MLPA has proven to be a powerful tool to identify genes targeted by DNA methylation in cervical carcinomas. In addition, we were able to link promoter methylation of eight of the 12 identified markers to distinct stages of HPV-induced malignant transformation. This resulted in more insight into the natural sequence of methylation events as well as novel candidates for future functional and clinical marker validation studies.
Acknowledgments
SMW was supported by the Centre of Medical Systems Biology (CMSB). RMO was supported by the Dutch Cancer Society (KWF). RDMS was supported by a fellowship of the Royal Netherlands Academy of Arts and Sciences. JGvR was supported by the Research Council for Earth and Life Sciences (ALW) with financial aid from the Netherlands Organization for Scientific Research (NWO).
References
- Bekkers RLM, van der Avoort IAM, Melchers WJG, Bulten J, de Wilde PCM, Massuger LFAG (2005) Down regulation of estrogen receptor expression is an early event in human papillomavirus infected cervical dysplasia. Eur J Gynaecolog Oncol 26: 376–382 [PubMed] [Google Scholar]
- Brady CS, Duggan-Keen MF, Davidson JA, Varley JM, Stern PL (1999) Human papillomavirus type 16 E6 variants in cervical carcinoma: relationship to host genetic factors and clinical parameters. J Gen Virol 80: 3233–3240 [DOI] [PubMed] [Google Scholar]
- Brandes JC, van EM, Wouters KA, Weijenberg MP, Herman JG (2005) CHFR promoter hypermethylation in colon cancer correlates with the microsatellite instability phenotype. Carcinogenesis 26: 1152–1156 [DOI] [PubMed] [Google Scholar]
- Burgers WA, Blanchon L, Pradhan S, de LY, Kouzarides T, Fuks F (2007) Viral oncoproteins target the DNA methyltransferases. Oncogene 26: 1650–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellsague X, Diaz M, de SS, Munoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS, Snijders PJ, Meijer CJ, Bosch FX (2006) Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. J Natl Cancer Inst 98: 303–315 [DOI] [PubMed] [Google Scholar]
- Chen TM, Pecoraro G, Defendi V (1993) Genetic analysis of in vitro progression of human papillomavirus-transfected human cervical cells. Cancer Res 53: 1167–1171 [PubMed] [Google Scholar]
- Das PM, Ramachandran K, Vanwert J, Ferdinand L, Gopisetty G, Reis IM, Singal R (2006) Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde J, Wilting SM, Meijer CJLM, van de Wiel MA, Ylstra B, Snijders PJF, Steenbergen RDM (2007) Gene expression profiling to identify markers associated with deregulated hTERT in HPV-transformed keratinocytes and cervical cancer. Int J Cancer (in press) [DOI] [PubMed]
- Dong SM, Kim HS, Rha SH, Sidransky D (2001) Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res 7: 1982–1986 [PubMed] [Google Scholar]
- Duenas-Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E (2005) Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Pookot D, Li LC, Tabatabai ZL, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R (2005) Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer 20;116: 174–181 [DOI] [PubMed] [Google Scholar]
- Esteller M, Herman JG (2002) Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 196: 1–7 [DOI] [PubMed] [Google Scholar]
- Feng QH, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi LF, Lu H, McIntosh MW, Young AM, Kiviat NB (2005) Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst 97: 273–282 [DOI] [PubMed] [Google Scholar]
- Fu YS, Reagan JW (1989) Pathology of the Uterine Cervix, Vagina and Vulva pp 288–335. W.B. Saunders and Co.: Philadelphia [Google Scholar]
- Gonzalgo ML, Jones PA (1997) Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res 25: 2529–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson KS, Furth EE, Heitjan DF, Fansler ZB, Clark DP (2004) DNA methylation profiling of cervical squamous intraepithelial lesions using liquid-based cytology specimens: an approach that utilizes receiver-operating characteristic analysis. Cancer 102: 259–268 [DOI] [PubMed] [Google Scholar]
- Jeong DH, Youm MY, Kim YN, Lee KB, Sung MS, Yoon HK, Kim KT (2006) Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer 16: 1234–1240 [DOI] [PubMed] [Google Scholar]
- Kang HC, Kim IJ, Park JH, Shin Y, Park HW, Ku JL, Yang HK, Lee KU, Choe KJ, Park JG (2004) Promoter hypermethylation and silencing of CHFR mitotic stress checkpoint gene in human gastric cancers. Oncol Rep 12: 129–133 [PubMed] [Google Scholar]
- Kang S, Kim JW, Kang GH, Lee S, Park NH, Song YS, Park SY, Kang SB, Lee HP (2006) Comparison of DNA hypermethylation patterns in different types of uterine cancer: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. Int J Cancer 118: 2168–2171 [DOI] [PubMed] [Google Scholar]
- Lin ZH, Gao MH, Zhang XL, Kim YS, Lee ES, Kim HK, Kim I (2005) The hypermethylation and protein expression of p16(INK4A) and DNA repair gene _O_-6-methylguanine-DNA methyltransferase in various uterine cervical lesions. J Cancer Res Clin Oncol 131: 364–370 [DOI] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de SS, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348: 518–527 [DOI] [PubMed] [Google Scholar]
- Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV (2003) Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A (2005) Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res 33(14): e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y (2001) Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 93: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Pisani P, Bray F, Parkin DM (2002) Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int J Cancer 97: 72–81 [DOI] [PubMed] [Google Scholar]
- Reesink-Peters N, Wisman GBA, Jeronimo C, Tokumaru CY, Cohen Y, Dong SM, Klip HG, Buikema HJ, Suurmeijer AJH, Hollema H, Boezen HM, Sidransky D, van der Zee AGJ (2004) Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: a feasibility study. Mol Cancer Res 2: 289–295 [PubMed] [Google Scholar]
- Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ (2006) HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol 208: 152–164 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, de Wilde J, Wilting SM, Brink AATP, Snijders PJF, Meijer CJLM (2005) HPV-mediated transformation of the anogenital tract. J Clin Virol 32: S25–S33 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, Hermsen MAJA, Walboomers JMM, Meijer GA, Baak JPA, Meijer CJLM, Snijders PJF (1998a) Non-random allelic losses at 3p, 11p and 13q during HPV-mediated immortalization and concomitant loss of terminal differentiation of human keratinocytes. Int J Cancer 76: 412–417 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, Kramer D, Braakhuis BJM, Stem PL, Verheijen RHM, Meijer CJLM, Snijders PJF (2004) TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst 96: 294–305 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, Kramer D, Meijer CJLM, Walboomers JMM, Trott DA, Cuthbert AP, Newbold RF, Overkamp WJI, Zdzienicka MZ, Snijders PJF (2001) Telomerase suppression by chromosome 6 in a human papillomavirus type 16-immortalized keratinocyte cell line and in a cervical cancer cell line. J Natl Cancer Inst 93: 865–872 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, OudeEngberink VE, Kramer D, Schrijnemakers HFJ, Verheijen RHM, Meijer CJLM, Snijders PJF (2002) Down-regulation of GATA-3 expression during human papillomavirus-mediated immortalization and cervical carcinogenesis. Am J Pathol 160: 1945–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen RDM, Parker JN, Isern S, Snijders PJF, Walboomers JMM, Meijer CJLM, Broker TR, Chow LT (1998b) Viral E6-E7 transcription in the basal layer of organotypic cultures without apparent p21cip1 protein precedes immortalization of human papillomavirus type 16- and 18-transfected human keratinocytes. J Virol 72: 749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen RDM, Walboomers JMM, Meijer CJLM, vanderRaaijHelmer EMH, Parker JN, Chow LT, Broker TR, Snijders PJF (1996) Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene 13: 1249–1257 [PubMed] [Google Scholar]
- Tokunaga E, Oki E, Nishida K, Koga T, Yoshida R, Ikeda K, Kojima A, Egashira A, Morita M, Kakeji Y, Maehara Y (2006) Aberrant hypermethylation of the promoter region of the CHFR gene is rare in primary breast cancer. Breast Cancer Res Treat 97: 199–203 [DOI] [PubMed] [Google Scholar]
- van Duin M, Steenbergen RD, de WJ, Helmerhorst TJ, Verheijen RH, Risse EK, Meijer CJ, Snijders PJ (2003) Telomerase activity in high-grade cervical lesions is associated with allelic imbalance at 6Q14-22. Int J Cancer 105: 577–582 [DOI] [PubMed] [Google Scholar]
- Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF (2001) Aberrant methylation during cervical carcinogenesis. Clin Cancer Res 7: 584–589 [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12–19 [DOI] [PubMed] [Google Scholar]
- Widschwendter A, Muller HM, Fiegl H, Ivarsson L, Wiedemair A, Muller-Holzner E, Goebel G, Marth C, Widschwendter M (2004) DNA methylation in serum and tumors of cervical cancer patients. Clin Cancer Res 10: 565–571 [DOI] [PubMed] [Google Scholar]
- Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, Albertson DG, Coffa J, Schouten JP, van de Wiel MA, Meijer CJ, Steenbergen RD (2006) Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol 209: 220–230 [DOI] [PubMed] [Google Scholar]
- Wisman GB, Nijhuis ER, Hoque MO, Reesink-Peters N, Koning AJ, Volders HH, Buikema HJ, Boezen HM, Hollema H, Schuuring E, Sidransky D, van der Zee AG (2006) Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer 119: 1908–1914 [DOI] [PubMed] [Google Scholar]
- Yang HJ, Liu VWS, Wang Y, Chan KYK, Tsang PCK, Khoo US, Cheung ANY, Ngan HYS (2004) Detection of hypermethylated genes in tumor and plasma of cervical cancer patients. Gynecol Oncol 93: 435–440 [DOI] [PubMed] [Google Scholar]
- Zambrano P, Segura-Pacheco B, Perez-Cardenas E, Cetina L, Revilla-Vazquez A, Taja-Chayeb L, Chavez-Blanco A, Angeles E, Cabrera G, Sandoval K, Trejo-Becerril C, Chanona-Vilchis J, Duenas-Gonzalez A (2005) A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski GD, Snijders PJ, Rozendaal L, Daalmeijer NF, Risse EK, Voorhorst FJ, Jiwa NM, van der Linden HC, de Schipper FA, Runsink AP, Meijer CJ (2003) The presence of high-risk HPV combined with specific p53 and p16INK4a expression patterns points to high-risk HPV as the main causative agent for adenocarcinoma in situ and adenocarcinoma of the cervix. J Pathol 201: 535–543 [DOI] [PubMed] [Google Scholar]
- zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2(5): 342–350 [DOI] [PubMed] [Google Scholar]